Abstract
Long noncoding RNAs (lncRNAs) have been shown to play crucial roles in human cancers. However, the underlying biological functions and mechanisms of lncRNAs in cholangiocarcinoma (CCA) remain largely unknown. We aimed to characterize the transcriptional landscape of lncRNAs in CCA and identify lncRNAs that were able to serve as prognosis markers and therapeutic targets for CCA. Here, we investigated the transcriptional landscape and dysregulation of lncRNAs in CCA. LINC01714 was found to be recurrently downregulated in CCA tumor samples. Our results revealed that decreased LINC01714 expression was associated with the poor survival of CCA patients. Our observations revealed that LINC01714 suppressed the proliferation, migration, and invasion abilities of CCA cells both in vitro and in vivo. Furthermore, we found that LINC01714 physically interacted with Forkhead Box O3 (FOXO3) and increased the FOXO3 protein level. In addition, LINC01714 could decrease the phosphorylation level of FOXO3. Interestingly, LINC01714 was able to enhance the sensitivity to gemcitabine in CCA tumor cells through modulating phosphorylated FOXO3-Ser318. Our study revealed LINC01714 as a promising prognostic indictor for patients with CCA, provided insights into the molecular pathogenesis of CCA, and also showed that LINC01714 is a potential therapeutic combination for gemcitabine in CCA treatment.
Keywords: cholangiocarcinoma, long noncoding RNA, tumor suppressor, FOXO3, gemcitabine
Introduction
Cholangiocarcinoma (CCA) is one of the primary hepatobiliary malignancies,1,2 which is anatomically classified into intrahepatic cholangiocarcinoma (ICC) and extrahepatic cholangiocarcinoma (ECC), derived from cholangiocytes and epithelial cells lining in the intra-hepatic and extra-hepatic biliary ducts, respectively. CCA is an aggressive malignancy regardless of its originating sites, with a median survival of about 24 months from the date of diagnosis.3 Surgical resections are only clinically applicable for early-stage CCA patients.4 For patients with advanced-stage CCA, combinatorial chemotherapy with gemcitabine and cisplatin is the standard clinical practice. Despite current therapeutic strategies, the survival rates of CCA patients still remain at a low level. Hence, it is urgent to explore the underlying molecular mechanisms and develop potential diagnostic and therapeutic targets for CCA.
With the improvement of methods investigating transcriptome in recent years, the major portion of human genome has been found to be non-coding regions.5,6 Long noncoding RNAs (lncRNAs) are among these indispensable noncoding RNAs that have been proven to play crucial regulatory roles in human cancers.7, 8, 9 The lncRNAs typically exert their roles through interaction with proteins,10, 11, 12 serving as scaffolds or guides to modulate protein-protein or protein-DNA interaction.13,14 Several lncRNAs have been reported to play roles in CCA cells. For example, Shi et al.15 found that the lncRNA AFAP1-AS1 exerts oncogenic effects in CCA, promoting growth and metastasis of CCA cells. In addition, H19 and HULC were reported to regulate CCA cell migration and invasion via a competing endogenous RNA (ceRNA) manner.16 However, the comprehensive transcriptions and molecular mechanisms of lncRNAs during the carcinogenesis and development of CCA have not been depicted. The FOXO subgroup of forkhead transcription factors has been shown to play major roles in the tumorigenesis of various cancers,17,18 including CCA.19 Guan et al.19 reported that FOXO3 deficiency promotes CCA tumorigenesis and induces tumor cell resistance to cisplatin through activating NRF2 signaling. However, the molecular mechanisms, covering lncRNAs and/or FOXO3, underlying CCA development remain largely unknown.
In our present study, we first characterized the transcriptional landscape of lncRNAs in CCA. We identified hundreds of lncRNAs that showed dysregulation in CCA tumor samples. We found and further validated that the long intergenic non-coding RNA, LINC01714, was recurrently downregulated in CCA and that its low expression predicted the poor outcome for CCA patients. Functional assays in CCA cells revealed that LINC01714 could substantially suppress the proliferation, migration, and invasion of CCA tumor cells both in vitro and in vivo. LINC01714 was found to be associated with several key genes involved in tumor-related pathways. Our further analysis revealed that LINC01714 physically interacted with FOXO3 protein by binding the forkhead (FH) domain. LINC01714 could inhibit the phosphorylation of FOXO3-Ser318 in CCA cells. Notably, our study suggested that LINC01714 could enhance gemcitabine sensitivity in CCA cells through suppressing FOXO3-Ser318 phosphorylation status. Our study sheds light on the molecular mechanisms underlying CCA and provides insights into therapeutic strategy for CCA patients.
Results
Transcriptional Landscape and Dysregulation of lncRNAs in CCA
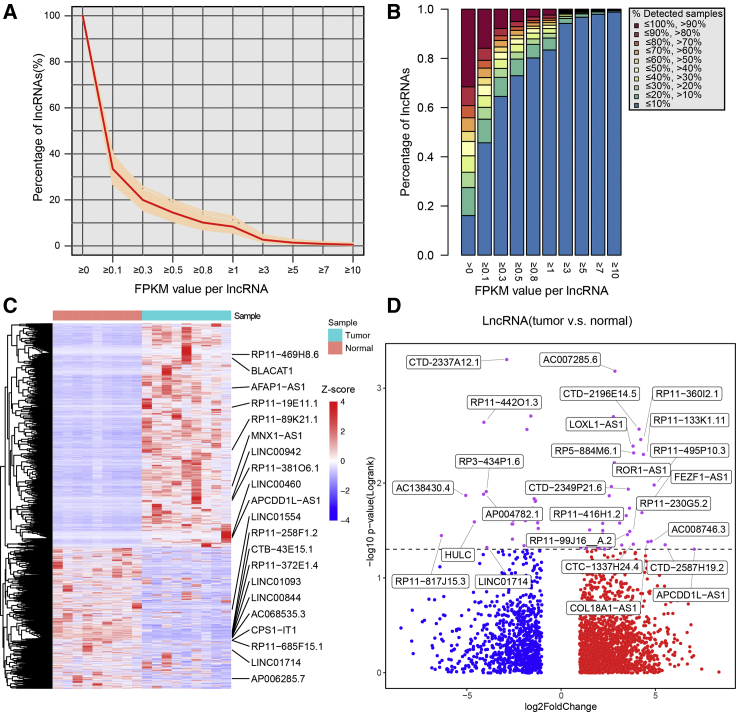
To investigate the comprehensive dysregulation and potential biological roles of lncRNAs in the carcinogenesis processes of CCA, an expression profile of samples from The Cancer Genome Atlas (TCGA) CCA cohort was obtained. In addition, lncRNAs annotated in GENCODE (release 22) were used to extract a lncRNA expression profile of CCA patients. Our analysis detected 14,058 lncRNAs that showed expression in at least one patient sample. In order to explore the expression distribution of lncRNAs in CCA, different cutoffs for lncRNA detection were used. On average, 66.53% lncRNAs showed expression levels that were lower than 0.1 fragments per kilobase of transcript per million mapped reads (FPKM), and 8.4% lncRNAs showed expression levels that were higher than 1 FPKM (Figure 1A). Across all expressed lncRNAs, an average of 16.1% lncRNAs were detected in less than 10% of all samples, and 31.6% lncRNAs were expressed in more than 90% samples (Figure 1B). These observations showed that most lncRNAs were extensively detected in CCA samples, despite their pervasively low expression. Furthermore, differential expression analysis revealed 751 upregulated and 569 downregulated lncRNAs in CCA tumor samples compared with adjacent normal samples (Figure 1C). Among these differentially expressed lncRNAs, the expression level of 79 lncRNAs showed significant association with CCA patient overall survival (Figure 1D). These lncRNAs may play important roles in CCA. The expression variation and survival association of these lncRNAs are listed in Table S1. Our results indicated that lncRNAs might be involved in the development and progression of CCA.
Figure 1.
The Expression Landscape of lncRNAs in CCA
(A) Overall distribution of lncRNA expression in CCA patients. (B) Detected sample distribution of lncRNAs with different expression levels in CCA samples. (C) Heatmap shows the expression variations of differentially expressed lncRNAs between CCA tumor and paired normal samples derived from the TCGA dataset. (D) Scatterplot shows the expression difference (tumor vs. normal fold change) and significance of survival difference (log-rank test p value) of differentially expressed lncRNAs. Red dots represent upregulated lncRNAs, blue dots represent downregulated lncRNAs, and purple dots represent lncRNAs that showed significant survival difference between high- and low-expression samples.
LINC01714 Is Recurrently Downregulated and Associated with Poor Survival Outcome in CCA Patients
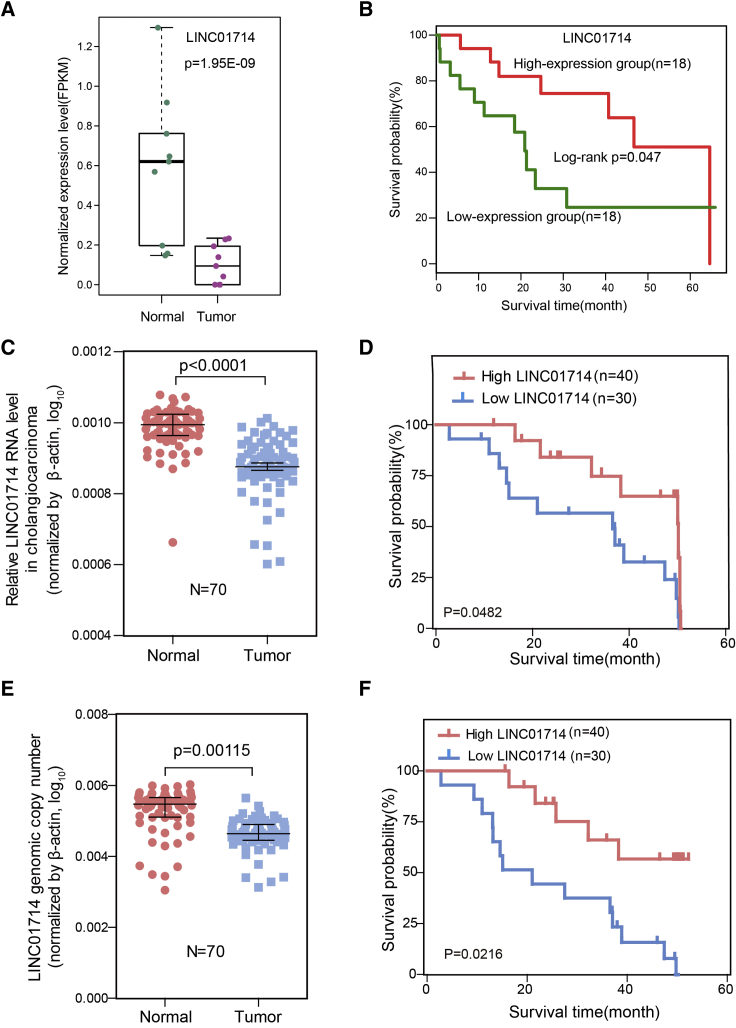
To identify lncRNAs that play crucial biological roles in CCA, we verified the top 10 long intergenic non-coding RNAs that showed both differential expression and significant survival association in clinical samples derived from an independent CCA cohort (named cohort 1 in this study; see Materials and Methods). LINC01714 showed significant downregulation in TCGA CCA tumor samples, compared with paired adjacent normal samples (Figure 2A; p = 1.95E−09, Student’s t test), and the low expression of LINC01714 was significantly associated with poor survival outcome among CCA patients in the TCGA cohort (Figure 2B; p = 0.047, log-rank test). The downregulation of LINC01714 was also observed in cohort 1 (Figure 2C). In addition, the expression variation of LINC01714 was explored in 33 TCGA cancer types. The significant downregulation of LINC01714 was observed only in CCA and liver cancer samples (Figure S1), which indicated the specific functions of LINC01714 in CCA. Furthermore, our analysis revealed that the low expression of LINC01714 was able to predict poor survival outcome of CCA patients in cohort 1 (Figure 2D). Moreover, CCA samples showed significant deletion of LINC01714 copy number (Figure 2E). The copy number deletion also indicated poor survival outcome of CCA patients in cohort 1. These observations showed that LINC01714 could be a potential predictor for CCA patient outcome and might participate in the progression of CCA carcinogenesis or development.
Figure 2.
LINC01714 Is Clinically Associated with CCA Patient Outcome
(A) The expression level (FPKM value) of LINC01714 between paired CCA tumor and adjacent normal samples quantified from TCGA RNA-seq data. (B) Kaplan-Meier analysis of the association between LINC01714 expression level and the overall survival of CAA patients in the TCGA dataset. (C) Boxplot shows the relative expression of LINC01714 (70 matched CCA tumor/non-tumor samples in cohort 1). (D) Kaplan-Meier plot indicates the survival difference between CCA patients that show high and low expression of LINC01714. High expression and low expression in groups of CCA patients were divided by the median level of LINC01714 expression. (E) The genomic copy numbers of LINC01714 in tumor and non-tumor samples. (F) Kaplan-Meier plot indicates the survival difference between CCA patients that show high and low copy numbers of LINC01714. High and low copy number groups of CCA patients were divided by the median level of LINC01714 genomic copy number. The RNA levels of LINC01714 in (C) and (D) were quantified by quantitative real-time PCR. Values are indicated as the median with interquartile range in (A), (C), and (E). Statistical analyses were performed using the Student’s t test, as in (A), (C), and (E), and the log-rank test, as in (B), (D), and (F).
Furthermore, the sequence of the LINC01714 gene was retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov). We found that the LINC01714 gene expressed three isoforms (Figure S2A). The 5′ and 3′ rapid amplification of complementary DNA end assays (5′ RACE and 3′ RACE) revealed that the full length of LINC01714 dominant isoform is about 497 bp (Figures S2B and S2C). LINC01714 showed expression variations across different CCA cell lines, wherein CCLP1 cells expressed the highest LINC01714 and HuCCT1 expressed the lowest (Figure S2D). LINC01714 also was expressed in both cytoplasm and nucleus (Figure S2E). The LINC01714 gene was predicted to possess no protein-coding potential by the LNCipedia database (Figure S3A).20 Moreover, in vitro transcription and translation assay revealed that neither the sense nor the antisense transcript of LINC01714 was able to encode protein, which demonstrated that LINC01714 was a bona fide noncoding RNA (Figure S3B).
LINC01714 Suppresses Growth, Migration, and Invasion Abilities of CCA Cells In Vitro and In Vivo
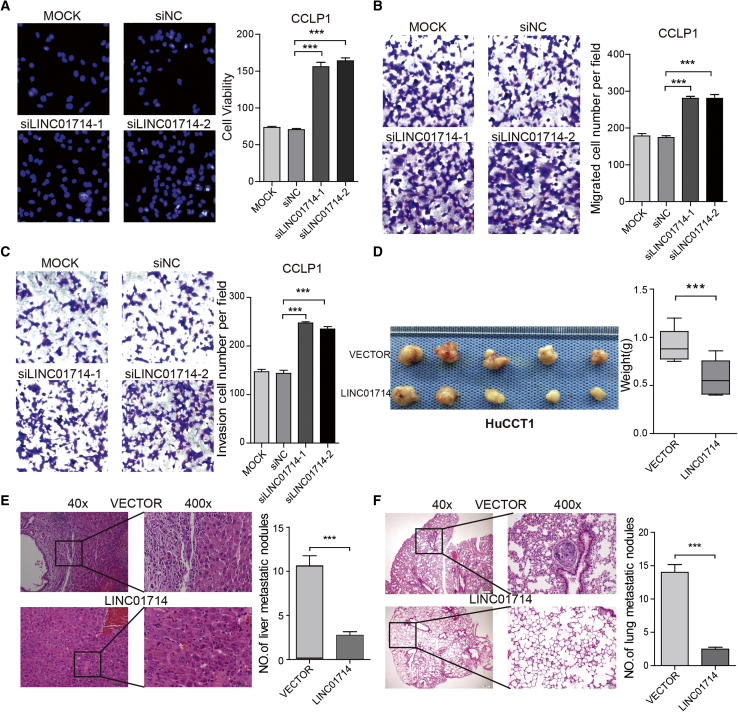
To further explore the roles of LINC01714, functional assays were performed to determine the biological effects of LINC01714 on CCA tumor cells. The accumulation of LINC01714 expression significantly impaired the viability of HuCCT1 cells (Figure S4A), while the knockdown of LINC01714 resulted in significant increased cell viability of CLLP1 cells (Figure 3A). Additionally, overexpression of LINC01714 significantly suppressed the migratory and invasive abilities of HuCCT1 cells (Figures S4B and S4C). Further knockdown of LINC01714 by independent small interfering RNAs (siRNAs) brought about the increase of cell migration and invasion in CLLP1 cells (Figures 3B and 3C). These results showed that LINC01714 suppressed the proliferative, migratory, and invasive abilities of CCA tumor cells in vitro.
Figure 3.
LINC01714 Suppresses Proliferation Migration and Invasion of CCA Tumor Cells In Vitro
(A) Cell viability assays for CCLP1 cells transfected with LIHC01714 siRNAs or mock controls. (B) Cell migration assays for CCLP1 cells transfected with LIHC01714 siRNAs or mock controls. (C) Cell invasion assays for CCLP1 cells transfected with LIHC01714 siRNAs or mock controls. (D) LINC01714 suppressed the CCA tumor growth in the nude mouse model bearing subcutaneous tumor xenografts from the LINC01714-transfected HuCCT1 cell line. (E) Representative images of H&E staining in the metastatic liver loci and statistical data comparing the LINC01714 group with the vector control group. (F) Representative images of H&E staining in the metastatic lung loci and statistical data comparing the LINC01714 group with the vector control group. Values are indicated as median with interquartile range in (D) and as mean ± SEM in (A)–(C), (E), and (F). **p < 0.01; ***p < 0.001.
To further examine the inhibition effects of LINC01714 on CCA tumor cells in vivo, HuCCT1 cells with transfection of expressed LINC01714 or vector control were transplanted into nude mice through subcutaneous injection. Both the volumes and weights of tumors were dramatically lower in the LINC01714 group than those in the control groups (Figure 3D). The effects of LINC01714 on cell migration and metastasis were also evaluated. The metastatic nodules in both liver and lung were significantly decreased in the LINC01714 group (Figures 3E and 3F; Figure S4D). In addition, the hematoxylin and eosin (H&E) staining results showed a dramatic decrease of metastatic foci derived from cells of the LINC01714 group in liver and lung tissue sites (Figures 3E and 3F). Taken together, these observations demonstrated that LINC01714 suppresses the tumorigenicity and tumor development of cholangiocyte both in vitro and in vivo.
The notable inhibitory effects of LINC01714 on CCA tumorigenicity and development drove us to investigate the possible underlying molecular mechanisms. RNA sequencing (RNA-seq) analysis was performed following LINC01714 knockdown to examine the variations of gene expression profiles in CCLP1 cells. Compared with control, our analysis identified hundreds of dysregulated genes in CCLP1 cells transfected with two independent LINC01714 siRNAs (Figure S5A). Functional annotation of these dysregulated genes revealed that they were involved in tumor-related biological processes, including metabolic process, cell cycle, and metastasis process (Figure S5B). Further gene set enrichment analysis (GSEA) showed that LINC01714 knockdown in CCA cells mainly affected the insulin signaling pathway, vascular endothelial growth factor (VEGF) signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway, which all play crucial roles in tumor (Figure S5C). Quantitative real-time PCR was used to verify the remarkable mRNA changes of selected genes involved in tumor growth and metastasis (Figure S6A). The protein levels of downregulation and upregulation were also verified by western blot in HuCCT1 and CCLP1 cells (Figure S6B). Our results suggested that LINC01714 might exert its roles in CCA through the regulation of key tumor-related genes in CCA cells.
LINC01714 Interacts with FOXO3 and Increases Its Transcription in CCA Cells
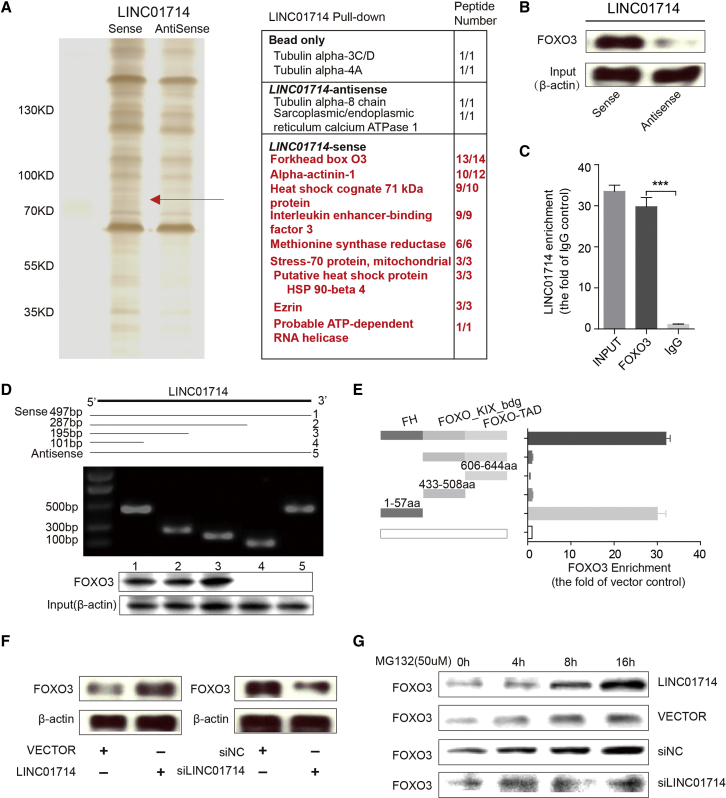
To obtain a better understanding of how LINC01714 performs its activities in cholangetic carcinogenesis and tumor progression, RNA pull-down assays were conducted to identify LINC01714-related proteins in CCA cells. The mass spectrometry analyses following LINC01714 pull-down experiments revealed specific protein bands around 70 kDa (Figure 4A). Based on the filtrations of high confidence scores (no less than 100 in mass spectrometric assays) and absence in corresponding antisense groups, nine proteins that might interact with LINC01714 were obtained (listed in the table in Figure 4A). Our quantitative real-time PCR results confirmed that FOXO3 protein was specifically associated with sense, but not antisense, LINC01714 (Figure 4B). The LINC01714-FOXO3 interaction was further verified by RNA immunoprecipitation (RIP) assays, wherein LINC01714 was significantly enriched in FOXO3 antibody but not immunoglobulin G (IgG) control (Figure 4C). In order to determine the specific LINC01714 fragment that binds the FOXO3 protein, a series of deletions were constructed to map the truncated LINC01714 fragments with FOXO3 protein. The results of deletion-mapping analyses showed that the 1- to 195-nt fragment of LINC01714 was required for its interaction with FOXO3 protein (Figure 4D). Moreover, our RIP assays revealed that the FH DNA-binding domain (1–57 aa) of FOXO3 was responsible for binding with LINC01714 (Figure 4E). Specifically, the interaction between FOXO3 and LINC01714 was significantly abolished under the deletion of the FOXO3 FH domain. The overexpression of LINC01714 increased the protein level of FOXO3, whereas knockdown of LINC01714 reduced the FOXO3 protein level (Figure 4F). The overexpression of LINC01714 enhanced the accumulation of FOXO3 protein in the cells treated with a proteasome inhibitor, MG132 (Figure 4G). The observation suggested that LINC01714 might inhibit the proteasome-dependent degradation of FOXO3 protein in CCA cells. In summary, LINC01714 could physically interact with FOXO3 and was positively associated with the FOXO3 protein level.
Figure 4.
LIHC01714 Physically Interacts with FOXO3 in CCA Cells
(A) LINC01714 pull-down assay analyzed by SDS-PAGE. (B) Western blot analysis of the FOXO3 protein retrieved from LINC01714 pull-down assay. (C) The quantitative real-time PCR results of RIP assays using an anti-FOXO3 antibody. (D) Immunoblotting detection of FOXO3 protein in the pull-down samples. The full-length sense and antisense biotinylated-LINC01714 (#1 and #5, respectively) and truncated biotinylated-LINC01714 sequences (#2 includes 1–287 bp, #3 includes 1–195 bp, and #4 includes 1–101 bp) were analyzed. β-actin serves as input control. (E) RIP assays for different domains of FOXO3 protein. Quantitative real-time PCR was used to determine the enrichment of LINC01714 binding with each FOXO3 domain. (F) Western blot analysis of FOXO3 protein level in LINC01714- or si-LIHC01714-transfected cells. (G) The FOXO3 protein levels were measured in LINC01714, si-LINC01714, or control cells. Cells were treated with MG132 (50 μmol/L) for 0, 4, 8, or 16 h before protein harvest. Values are indicated as mean ± SEM in (C) and (E). **p < 0.01; ***p < 0.001.
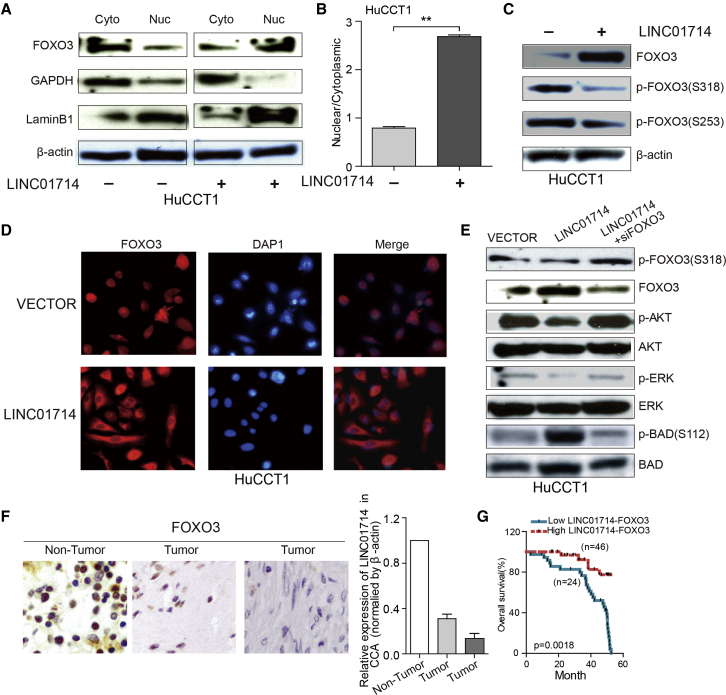
LINC01714 Reduces FOXO3 Phosphorylation in CCA Cells
Given that LINC01714 physically interacts with FOXO3, we next examined the molecular consequence of the interaction. The overexpression of LINC01714 significantly increased the FOXO3 protein level in nucleus compared with that in cytoplasm (Figures 5A and 5B). In immunofluorescence staining assays, the increasing FOXO3 protein level was also observed in LINC01714-overexpressed HuCCT1 cells (Figure 5D). Next, we examined whether LINC01714 exerted impacts on the phosphorylation of FOXO3 protein in CCA cells. Our analysis found that overexpression of LINC01714 significantly decreased the phosphorylation level of FOXO3-Ser318, while no obvious impact on FOXO3-Ser253 phosphorylation status was found (Figure 5C). To examine whether the influence of LINC01714 on phosphorylation status was specific to FOXO3, we further checked the phosphorylation level of AKT, ERK, and BAD proteins. The phosphorylation level of BAD protein, but not that of AKT or ERK protein, was increased in LINC01714-overexpressed HuCCT1 cells (Figure 5E). However, after the influence of FOXO3 siRNAs, LINC01714-overexpressed HuCCT1 did not show any increased level of phosphorylated BAD protein. This observation indicated that LINC01714 might influence the phosphorylation of BAD protein through interacting with FOXO3. To explore the pathological significance of the LINC01714-FOXO3 combination, immunohistochemical staining of FOXO3 in CCA tumor and corresponding non-tumor samples was analyzed. Consistent with the reduction of the FOXO3 protein level in CCA tumor samples, a decreased mRNA level of LINC01714 was also detected in the corresponding samples (Figure 5F). Notably, CCA patients with high LINC01714-FOXO3 levels showed better overall survival compared with those with low LINC01714-FOXO3 levels (Figure 5G). Our analysis revealed that LINC01714 specifically inhibited the phosphorylation status of FOXO3 protein.
Figure 5.
LINC01714 Regulates the Phosphorylation Level of FOXO3 in CCA Cells
(A) Immunoblotting analysis of the FOXO3 distribution between cytoplasm and nucleus in LINC01714 (+) and LINC01714 (−) HuCCT1 cells. GAPDH and lamin B1 serve as controls in cytoplasm and nucleus, respectively. (B) Barplots show the statistical data of FOXO3 distribution between cytoplasm and nucleus in HuCCT1 cells. (C) Immunoblotting results show the phosphorylation status of FOXO3-S318 and FOXO3-S253 in HuCCT1 cells with or without LINC01714 transfection. (D) Immunofluorescence staining of HuCCT1 cells after transfection with LINC01714 or vector control. (E) Immunoblotting analysis for the phosphorylation status of AKT, ERK, and BAD proteins in HuCCT1 with LINC01714, LINC01714+siFOXO3, or control vector transfection. (F) Representative immunohistochemistry (IHC) images of FOXO3 expression levels in tumor and non-tumor samples. Barplots show the mRNA levels of LINC01714 in corresponding tumor and non-tumor samples. (G) Kaplan-Meier analysis of the influence of the LINC01714-FOXO3 combination on CCA patient survival. Values are indicated as mean ± SEM in (B) and (F). **p < 0.01; ***p < 0.001.
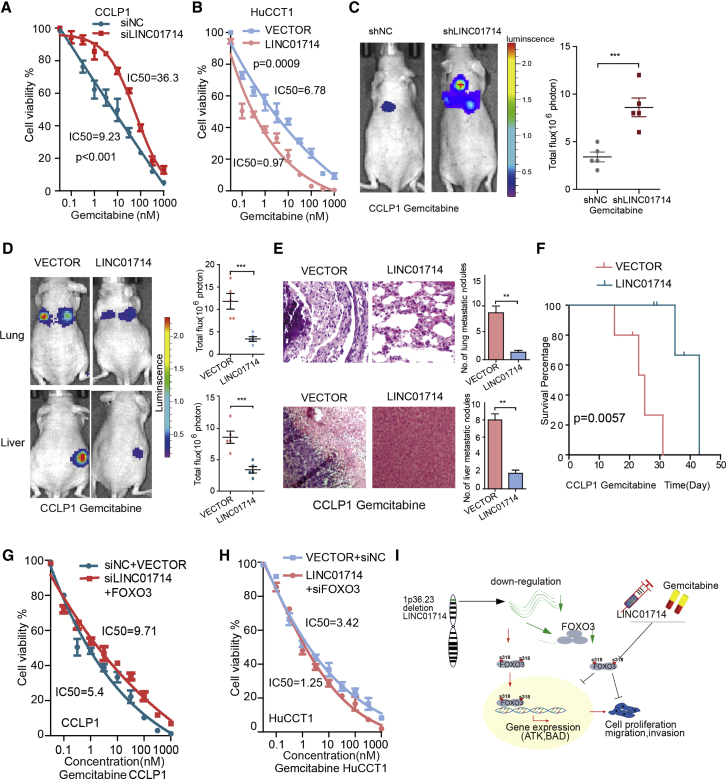
LINC01714 Enhances Drug Sensitivity of Gemcitabine in CCA Tumor Cells
To further explore the clinical significance of LINC01714 for CCA patients, we investigated its effects on treatment with gemcitabine, which is usually used in the chemotherapy for CCA patients. The knockdown of LINC01714 dramatically increased the half-maximal inhibitory concentrations (ICs50) in gemcitabine-treated CCLP1 cells (Figure 6A). Moreover, the overexpression of LINC01714 significantly reduced the IC50 in gemcitabine-treated HuCCT1 cells (Figure 6B). Remarkably, the tumor expanded dramatically in the nude mouse model bearing subcutaneous tumor xenografts from LINC01714-knockdown CCLP1 cell lines with gemcitabine treatment (Figure 6C). In mice in which LINC01714 was expressed, the tumor was repressed and decreased in both weight and volume (Figures S7A–S7C). Moreover, the number of both lung and liver metastatic nodules showed a significant decrease (Figures 6D, 6E, and S8A). We also observed that gemcitabine-treated mice with overexpressed LINC01714 have a longer survival time than those with control vector (Figure 6F). These observations indicated that LINC01714 was able to enhance gemcitabine sensitivity in CCA tumor cells. We further investigated whether FOXO3 impacts the enhancement of LINC01714 on gemcitabine. LINC01714 knockdown combined with FOXO3 overexpression could not induce a notable IC50 increase for gemcitabine-treated CCLP1 cells (Figure 6G). Compared with control samples, LINC01714-knockdown samples showed a reduction of the FOXO3 protein level and an increased phosphorylated FOXO3-Ser318 level (Figures S8D and S8E). Furthermore, the dramatic drop of IC50 was not observed in LINC01714+siFOXO3 HuCCT1 cells with gemcitabine treatment (Figure 6H). LINC01714-overexpression samples showed an increased FOXO3 protein level and a decreased FOXO3 phosphorylated FOXO3-Ser318 level (Figure S8B). The increase of the FOXO3 protein level and decrease of the FOXO3 phosphorylation level caused by the overexpression of LINC01714 was further validated by western blot assays in gemcitabine-treated CCLP1 cells (Figures S8C and S8D). These results suggested that LINC01714 exerted its enhancement on gemcitabine in CCA cells through phosphorylated FOXO3 protein.
Figure 6.
LIHC01714 Is a Candidate Therapeutic Target for CCA
(A) IC50 analysis for gemcitabine-treated CCLP1 cells with si-LINC01714 and corresponding control transfection. (B) IC50 analysis for gemcitabine-treated HuCCT1 cells with LINC01714 and corresponding control transfection. (C) LINC01714 suppressed the liver metastasis of CCA tumor in the nude mouse model bearing subcutaneous tumor xenografts from gemcitabine-treated CCLP1 cell lines. (D) Thermographic assessment of liver or lung tumor in LINC01714 overexpression and control mouse groups. (E) Representative data of H&E staining in the tumor foci of the liver or lung samples obtained from nude mice after injection with LINC01714-overexpression cells. (F) Survival analysis between mouse groups with gemcitabine-treated CCLP1 cell lines with LINC01714 or control transfection. (G) IC50 analysis for gemcitabine-treated CCLP1 cells with siLINC01714+FOXO3 and corresponding control transfection (Student’s t test p < 0.001). (H) IC50 analysis for gemcitabine-treated HuCCT1 cells with LINC0174+siFOXO3 and corresponding control mice groups (Student’s t test p = 0.0002). (I) Mechanism model depicting briefly how LINC01714 enhances gemcitabine sensitivity through reducing FOXO3 phosphorylation in CCA cells. Values are indicated as mean ± SEM in (C). **p < 0.01; ***p < 0.001.
Discussion
lncRNAs have been shown to play indispensable roles in human cancers. Certain RNAs have been found to participate in the process of tumor development in human cancers,21, 22, 23 including CCA.15,16 However, the general dysregulation of lncRNA transcription and their molecular roles in CCA have still been unclear. Our study first portrayed the transcriptional landscape of lncRNAs in CCA. Most lncRNAs were expressed at relatively low levels but were detected in a large portion of CCA samples. To date, the TCGA CCA cohort is the most abundant resource that includes genomic, transcriptomic, and clinical data, nut all TCGA RNA-seq data were generated from poly(A) RNA extraction. Total RNA extraction from CCA samples will surely result in more accurate and diverse lncRNA detection. We identified hundreds of lncRNAs that showed differential expression in CCA tumor samples. Among them, 79 lncRNAs also showed a significant association with patient survival. We only give evidence of 10 lncRNAs that showed the most expression variation in independent CCA cohorts. These 10 lncRNAs showed the potential to take part in the CCA progression, although other lncRNAs of less significance might also exert functions in CCA cells. LINC01714 was found to be recurrently downregulated in CCA tumor samples, and its low expression could predict poor survival outcome for CCA patients. These observations were found in both the TCGA and two other independent CCA cohorts, indicating our solid results. These observations suggested LINC01714 as a promising predictor for the survival outcome of CCA patients.
Our experiments in CCA cells and xenograft mice revealed that LINC01714 suppressed the growth, migration, and invasion abilities of CCA tumor cells. We found that LIINC01714 interacted with FOXO3 and inhibited the phosphorylation status of FOXO3-Ser318. Targeting the LINC01714 or FOXO3 suppressor in CCA tumor cells might be a prospective therapeutic strategy in clinical treatment for CCA patients.
Gemcitabine is used for patients with advanced-stage CCA in current clinical practice.24, 25, 26 Our IC50 analysis in CCLP1 and HuCCT1 cells revealed that LINC01714 could enhance the drug sensitivity of gemcitabine in CCA cells. Moreover, LINC01714 exerts its impact on gemcitabine through interaction with FOXO3 phosphorylation. This suggested that combinatorial use of gemcitabine and LINC01714 transcriptional promoters might produce better therapeutic effects for advanced-stage CCA patients. Our findings provide insights into clinical treatment for CCA patients.
In summary, our present study found that LINC01714 was recurrently downregulated in CCA tumor samples and predicted the survival outcome of patients with CCA. LINC01714 suppressed the growth, migration, and invasion of CCA cells both in vitro and in vivo. LINC01714 physically interacts with FOXO3 protein and inhibits its phosphorylation level in CCA cells. The sensitivity of gemcitabine could be enhanced by LINC1714 through modulating phosphorylated FOXO3-Ser318, suggesting LINC01714 as a candidate for combinatorial chemotherapy with gemcitabine for CCA patients.
Materials and Methods
Cell Lines and Human Clinical Samples
The CCA cell lines HuCCT1 and CCLP1 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cholangetic tissues of CCA cohorts (70 paired patients with CCA and corresponding adjacent normal samples were involved in this cohort) were obtained from the surgical specimen archives of Zhongshan Hospital, Fudan University, Shanghai, China.
Quantitative Real-Time PCR Assay
TRIzol Reagent (Invitrogen, Carsbad, CA, USA) was used to extract total RNA from the clinical tissue and cell lines in this study. Then, total RNA was reverse transcribed into cDNA in the LifePro Thermal Cycler (Hangzhou Bioer Technology, Hangzhou, China) according to the instructions of the reverse transcriptase kit (Takara Bio, Dalian, China). The quantitative real-time PCR was utilized to determine relative RNA levels, which were measured on the 7900 Real-Time PCR System with the SDS v.2.3 software sequence detection system (Applied Biosystems, Waltham, MA, USA) using the SYBR Green (Takara Bio) method. The relative RNA levels were normalized by using β-actin as the internal control in each sample and calculated by utilizing the 2−ΔΔCt relative quantification method.27
RNA-Seq
Total RNA was collected using TRIzol Reagent (Invitrogen); then, rRNAs were removed by using the RiboMinus Eukaryote Kit (QIAGEN, Valencia, CA, USA). We next prepared the RNA-seq libraries using the NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs, Beverly, MA, USA) according to the manufacturer’s instructions. The ribosome-depleted RNA libraries were subjected to the Illumina HiSeq 3000 (Illumina, San Diego, CA, USA) for sequencing. The raw sequencing reads were first processed to clip adaptor sequences and low-quality bases by Trimmomatic software.28 Afterward, all filtered reads were aligned to the human reference genome (hg38) using the splice-aware aligner HISAT2.29 The Cufflinks program30 was used to calculate the gene expression level in FPKM units.
TCGA Datasets
The RNA-seq V2 expression data and corresponding clinical information of TCGA CCA cohorts (36 tumor and 9 adjacent normal samples) were obtained from the genomic data commons (GDC) data portal (https://portal.gdc.cancer.gov/).31 The raw sequencing reads were aligned to hg38 by the GDC using a two-pass method with STAR aligner.32 Furthermore, the HTseq count tool33 was used to calculate the reads mapped to all genes annotated in the GENCODE database (v.22),34 including both protein-coding and long noncoding genes. Read counts were then normalized to FPKM values.
Differential Expression Analysis
Nine paired tumor/normal samples were included in the TCGA CCA cohorts. The gene read counts of tumor and paired adjacent normal samples were used for differential expression analysis by DESeq2.35 In differential analysis, lncRNA genes that showed a false discovery rate (FDR) ⩽ 0.05 (Benjamini-Hochberg corrected p value) and |log2 fold-change| ≥ 1 were identified as significantly differentially expressed.
Survival Analysis
Survival analysis was performed as described in a previous study.36 Briefly, the expression levels of each lncRNA across all tumor samples in each sample group were used to explore whether they were associated with the prognosis status of tumor patients. For each lncRNA, CCA tumor samples were divided into two groups according to the median value of expression. Then the log-rank test was used to compare the survival duration difference between two groups.
Northern Blot Assays
The Ambion Northern Max-Gly Kit (Austin, TX, USA) was used to detect LINC01714 RNA levels in tissue and cells. Briefly, the nylon membrane (nitrocellulose [NC] membrane) with positive charge was utilized to electrophorese and siphon the extracted total RNA samples. UV cross-linking was conducted to fix the RNA on the NC membrane. Then, the DIG Northern Starter Kit (Roche, Indianapolis, IN, USA) was used with a digoxin-labeled LINC01714-specific oligonucleotide probe to detect LINC01714.
Western Blot Assays
SDS-PAGE was used to separate proteins and transfer proteins to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA), which were blocked with 5% nonfat milk and incubated with corresponding primary antibodies followed by horseradish-peroxidase-conjugated secondary antibodies. Then, the chemiluminescence and enhanced chemiluminescence (ECL) reagents (Pierce Biotechnology, Rockford, IL, USA) were utilized for visualization of the immunoreactivity, and the densitometry was measured using the Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA).
RNAi and Lentivirus Construction
The siRNA oligonucleotides used for LINC01714 or FOXO3 in this study were purchased from RiboBio (Guangzhou, China). The human LINC01714 sequence was amplified from cDNAs. pWPXL-LINC01714 was generated by cloning LINC01714 sequence into the BamHI and EcoRI sites of pWPXL lentiviral vectors. In addition, the open reading frame sequence of FOXO3 was inserted into the pWPXL vector to construct FOXO3 expression vectors. pWPXL, pWPXL-LIHC01714, or pWPXL-FOXO3 was transfected along with the packaging plasmid psPAX2 and the envelope plasmid pMD2G into HuCCT1 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The virus particles were collected 48 h after transfection and then were infected into CCA cells with recombinant lentivirus-transducing units using 1 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA).
In Vitro Cell Migration and Invasion Assays
The invasion assays were performed in Millicell chambers that were coated with 30 μg Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Similar operations were conducted in the migration assays but without coated membrane. The cells (5 × 104 and 1 × 105, for migration and invasion assays, respectively) were added to the upper chambers. DMEM containing 10% fetal bovine serum (FBS) was placed into the lower chambers as a chemoattractant. The cells were then incubated at 37°C with 5% CO2 for 24 h. After the incubation, we fixed the cells that migrated or invaded through the filters with 20% methanol. Fixed cells were then stained with 0.1% crystal violet. We randomly selected five fields to count the cell numbers by using an inverted microscope (Olympus, Tokyo, Japan).
In Vivo Metastasis Assays
Nude mice (female BALB/c-nu/nu mice) were purchased from the Experimental Animal Center of the Shanghai Cancer Institute (Shanghai, China) for our in vivo metastasis assays. HuCCT1 cells (1 × 106 pWPXL-VECTOR or pWPXL-LINC01714 stable HuCCT1 cells) that were suspended in 0.2 mL serum-free DMEM were injected subcutaneously into each mouse (10 mice for each group) through the right axilla. The tumor growth was monitored. The mice were sacrificed after 60 days, and then livers and lungs were dissected. The liver and lung tissues derived from the mice were fixed with phosphate-buffered neutral formalin and prepared for the following histological examination. H&E staining was utilized to determine the number of metastatic foci in liver or lung tissues under a binocular microscope (Leica, Wetzlar Lottehaus, Germany). The tumor volume was measured as length × square width × 0.5. Experiments performed in this part were all under the regulations of the Shanghai Medical Experimental Animal Care Commission.
RNA Pull-Down Assays and Mass Spectrometry Analyses
First, LINC01714 and antisense LINC01714 RNAs were transcribed in vitro and labeled with the Biotin RNA Labeling Mix (Roche, Indianapolis, IN, USA). The RNA samples were treated with RNase-free DNase I (Takara Bio, Shiga, Japan) and then purified with the RNeasy Mini Kit (QIAGEN, Frederick, MD, USA). Second, to format an appropriate secondary structure, RNA structure buffer was used to pre-treat the biotinylated RNAs. Then, the pre-treated biotinylated RNAs were incubated with 1 mg protein extracts at 4°C for 1 h. After the incubation, 40 μL streptavidin-linked magnetic beads (Thermo Fisher Scientific, Rockford, IL, USA) were utilized to perform the pull-down at room temperature for 2 h. Next, the mixture of beads, RNA, and proteins was washed in 1× washing buffer (5 mM Tris-HCl, 1 M NaCl, 0.5 mM EDTA, and 0.005% Tween 20) five times. The precipitation and dilution were conducted in 60 μL protein lysis buffer; then the proteins were separated by using gel elecrophoresis, and the visualization was shown after silver staining according to the manufacturer’s instructions. Finally, the retrieved proteins were measured on SDS-PAGE gels for mass spectrometry analysis (Shanghai Applied Protein Technology, Shanghai, China) or western blot.
RIP Assays
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Danvers, MA, USA) was used to conduct the RIP assays in this study. In brief, lysis buffer (0.5 mL) was utilized to lyse cells in 10-cm dishes with protease inhibitors and RNase inhibitor (Thermo Fisher Scientific, Rockford, IL, USA) according to the manufacturer’s instructions. The lysed cells were then subjected to centrifuge at 12,000 rpm for 30 min. Then the supernatants were incubated with Protein G Dynabeads (Thermo Fisher Scientific, Carlsbad, CA, USA) and indicated antibodies. After incubation at 4°C for 12 h, the beads were washed thrice with wash buffer and then twice with PBS. Both the wash buffer and PBS contained RNase inhibitor. The Total RNA Isolation Kit (Thermo Fisher Scientific) was used to extract co-precipitated RNAs, which were then subjected to quantitative real-time PCR assays.
Immunoblotting Analysis
The lysis buffer (Beyotime Biotechnology, Shanghai, China) and protease inhibitors (Roche, Indianapolis, IN, USA) were used to lyse cells (5 × 106). The bicinchoninic acid (BCA) method was used to determine the protein concentrations (Pierce, Thermo Fisher Scientific, Rockford, IL, USA). SDS-PAGE was utilized to analyze the samples after centrifugation at 4°C for 15 min. The samples then were transferred to polyvinylidene (PVDF) membranes (Immobilon-P membrane; Millipore, Danvers, MA, USA), and horseradish-peroxidase (HRP)-conjugated secondary antibodies were used in the following immune blotting analysis. Specifically, TBS plus Tween 20 with skim milk (5%) was used to block the membranes at 4°C before the probing was conducted. Finally, the assay results were visualized by using the Enhanced Chemiluminescence Plus Western Blotting Detection Systems (GE Healthcare, Hartford, CT, USA) and the Amersham ImageQuant LAS-4000 EPUV Mini Luminescent Image Analyzer with ECL chromogenic substrates.
In Vitro Cellular IC50 Assays
Flat-bottomed plates (96 wells) were used to seed si-LINC01714 CCLP1 cells, pWPXL-LINC01714 HuCCT1 cells, and corresponding vector cells. Ten concentration gradients of gemcitabine were performed to determine the IC50 values in 100 μL suspended cells (5,000 cells) in each plate well. The Cell Counting Kit-8 (CCK-8) assays (Dojindo, Kyushu, Japan) were used to evaluate the cell viability after 48-h culture.
Statistical Analysis and Plots
Data are indicated as the mean ± SEM (standard error of the mean) or median with interquartile range. The difference between two groups was evaluated by Student’s t test. The association between LINC01714 RNA levels and overall survival of CCA patients was estimated using the log-rank test. Additionally, the chi-square test was used to assess the functional impact of LINC01714 on CCA cell metastasis in vivo. The heatmap was generated by using the pheatmap R package, wherein “complete” method with “euclidean” distance was used to cluster the lncRNAs. In addition, the “geom_point” function was applied in the ggplot2 R package to generate a volcano plot of differentially expressed lncRNAs. All statistical calculations and plots in this study were performed in the R environment.37 Unless specially stated, we considered a statistical test with a p value < 0.05 significant.
Author Contributions
Houbao Liu, Han Liu, and J.Z. designed and supervised the study. S.S., J.W., and B.Z. performed data analysis and functional experiments. S.S., Y.T., M.L., Y.W., X.N., and T.S. collected and managed patient samples. S.S., J.W., Houbao Liu, Han Liu, and J.Z. wrote and reviewed the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81602033 and 81872352), Shanghai Top Priority Clinical Medical Center and Key Discipline Construction Plan (2017ZZ02007), and the Foundation of Science and Technology Commission of Shanghai Municipality, China (16411952000).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.028.
Contributor Information
Houbao Liu, Email: liu.houbao@zs-hospital.sh.cn.
Han Liu, Email: liu.han@zs-hospital.sh.cn.
Jiwei Zhang, Email: joveus1024@gmail.com.
Supplemental Information
References
- 1.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blechacz B., Gores G.J. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatto M., Bragazzi M.C., Semeraro R., Napoli C., Gentile R., Torrice A., Gaudio E., Alvaro D. Cholangiocarcinoma: update and future perspectives. Dig. Liver Dis. 2010;42:253–260. doi: 10.1016/j.dld.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Rizvi S., Borad M.J., Patel T., Gores G.J. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin. Liver Dis. 2014;34:456–464. doi: 10.1055/s-0034-1394144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uszczynska-Ratajczak B., Lagarde J., Frankish A., Guigó R., Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018;19:535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Zhang J., Liu X., Li S., Wang Q., Chen D., Hu Z., Yu T., Ding J., Li J. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018;9:1572. doi: 10.1038/s41467-018-04006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Li Z., Liu L., Wang Q., Li S., Chen D., Hu Z., Yu T., Ding J., Li J. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171–187. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 13.Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.R. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S., Kopp F., Chang T.-C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y., Mendell J.T. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X., Zhang H., Wang M., Xu X., Zhao Y., He R., Zhang M., Zhou M., Li X., Peng F. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394–58404. doi: 10.18632/oncotarget.16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W.-T., Ye H., Wei P.-P., Han B.-W., He B., Chen Z.-H., Chen Y.Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myatt S.S., Lam E.W.F. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Ao X., Ding W., Ponnusamy M., Wu W., Hao X., Yu W., Wang Y., Li P., Wang J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan L., Zhang L., Gong Z., Hou X., Xu Y., Feng X., Wang H., You H. FoxO3 inactivation promotes human cholangiocarcinoma tumorigenesis and chemoresistance through Keap1-Nrf2 signaling. Hepatology. 2016;63:1914–1927. doi: 10.1002/hep.28496. [DOI] [PubMed] [Google Scholar]
- 20.Volders P.-J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., Vandesompele J., Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y., Yang Y., Wang F., Moyer M.-P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 22.Xu M.-D., Wang Y., Weng W., Wei P., Qi P., Zhang Q., Tan C., Ni S.J., Dong L., Yang Y. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin. Cancer Res. 2017;23:2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
- 23.Bian Z., Zhang J., Li M., Feng Y., Wang X., Zhang J., Yao S., Jin G., Du J., Han W. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin. Cancer Res. 2018;24:4808–4819. doi: 10.1158/1078-0432.CCR-17-2967. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Rahman O., Elsayed Z., Elhalawani H. Gemcitabine-based chemotherapy for advanced biliary tract carcinomas. Cochrane Database Syst. Rev. 2018;4:CD011746. doi: 10.1002/14651858.CD011746.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabriel E., Gandhi S., Attwood K., Kuvshinoff B., Hochwald S., Iyer R. Gemcitabine and capecitabine for advanced biliary cancer. J. Gastrointest. Oncol. 2017;8:728–736. doi: 10.21037/jgo.2017.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.O., Oh D.-Y., Hsu C., Chen J.-S., Chen L.-T., Orlando M., Kim J.S., Lim H.Y. Gemcitabine plus cisplatin for advanced biliary tract cancer: a systematic review. Cancer Res. Treat. 2015;47:343–361. doi: 10.4143/crt.2014.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A., Staudt L.M. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., Hu Z., Zhao Y., Huang S., He X. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology. 2019;69:359–375. doi: 10.1002/hep.30158. [DOI] [PubMed] [Google Scholar]
- 37.R Development Core Team . R Foundation for Statistical Computing; 2016. R: a language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.