Abstract
Corneal transparency is maintained by a monolayer of corneal endothelial cells. Defects in corneal endothelial cells (CEnCs) can be rectified surgically through transplantation. Fuchs’ endothelial corneal dystrophy (FECD) is the foremost cause of endothelial dysfunction and the leading indication for transplantation. Increased sensitivity of CEnCs to oxidative stress is thought to contribute to the pathogenesis of FECD through increased apoptosis. In part, this is thought to be due to loss of NRF2 expression: a global regulator of oxidative stress. We demonstrate that expression of the redox sensor, peroxiredoxin 1 (PRDX1) is selectively lost from CEnCs in FECD patient samples. We reveal that expression of PRDX1 is necessary to control the response of CEnCs to agents that cause lipid peroxidation. Iron-dependent lipid peroxidation drives non-apoptotic cell death termed ferroptosis. We establish that the inhibitor of ferroptosis, ferrostatin-1 rescues lipid peroxidation and cell death in CEnCs. Furthermore, we provide evidence that the transcription factor NRF2 similarly regulates lipid peroxidation in CEnCs.
Keywords: PRDX1, Corneal endothelial cells, Fuchs' endothelial corneal dystrophy, Lipid peroxidation, Ferroptosis
Abbreviations: AnV, Annexin V; ARE, antioxidant response element; CEnC, Corneal Endothelial cell; CH, cumene hydroperoxide; DM, Descemet's membrane; FECD, Fuchs' endothelial corneal dystrophy; Fer-1, ferrostatin-1; GPX4, Glutathione peroxidase 4; GSH, Glutathione; NRF2, nuclear factor-like 2; PI, Propidium iodide; PRDX, peroxiredoxin; ROS, reactive oxygen species
Graphical abstract

Highlights
-
•
PRDX1 is a key regulator of lipid peroxidation.
-
•
Expression of PRDX1 is lost in Fuchs' endothelial corneal dystrophy.
-
•
Oxidative stress reduces PRDX1 expression in cultures of CEnCs.
1. Introduction
The cornea is a transparent, refractive structure of the eye composed of an epithelial layer, Bowman's layer, an inner stromal layer, Descemet's membrane (DM) and corneal endothelium. The corneal endothelium is a monolayer of hexagonal corneal endothelial cells (CEnCs) attached to DM [1,2]. The corneal endothelium is essential for maintaining transparency of the cornea, via the regulation of corneal hydration. This is achieved through a pump-leak mechanism: nutrients from the aqueous humor are allowed to passively diffuse through the ‘leaky’ endothelium into the stroma and at the same time an ionic pump drives fluid from the stroma into the aqueous humor [1]. Together this mechanism ensures optimal corneal transparency and hydration.
Corneal endothelial cells have limited capacity to regenerate in vivo. If the functional integrity of the corneal endothelium is compromised, low visual acuity and ultimately loss of sight will occur. Fuchs’ endothelial corneal dystrophy (FECD) is a common cause of endothelial dysfunction. Currently, surgical intervention is required to treat advanced stage disease. FECD is characterised by a visible decrease of CEnC density, abnormal CEnC morphology with polymegethism and pleomorphism of the cells, together with excrescence of extracellular matrix named guttae present at the posterior part of DM [3]. Loss of CEnCs in FECD is presumed to derive from an increase in apoptosis. Furthermore, it has been demonstrated that there is an increase in apoptotic CEnCs surrounding the largest guttae [4]. Although several genetic loci have been implicated in FECD, oxidative stress is thought to contribute to the pathogenesis of the disease [5]. Interestingly, the peroxiredoxin (PRDX) family of redox sensors is known to be down regulated in FECD tissue [6]. However, their exact role has not been determined. The transcription factor; nuclear factor-like 2 (NRF2), a master regulator of antioxidant response that promotes PRDX expression via binding to an antioxidant response element (ARE) [7], has been reported to be down regulated in FECD [8]. This is consistent with the hypothesis that loss of redox control contributes to the pathogenesis of FECD [9].
Ubiquitously expressed, the PRDX family of peroxidases are capable of reducing hydrogen peroxide. Six isoforms of PRDX proteins exist in mammals, with differing subcellular localisation patterns. Interestingly, PRDX proteins have been reported to regulate multiple signalling pathways that use peroxide as a second messenger [10]. Exactly how PRDX controls oxidative stress in corneal endothelial cells has not been characterised. Western blot analysis of PRDX expression has revealed that PRDX2, PRDX3, and PRDX5 protein levels are significantly down regulated in FECD [6]. However, exactly how this impacts the pathogenesis of FECD is not fully elucidated.
In addition, it is not known if increased reactive oxygen species (ROS) is an early indicator of cell death in CEnC or, is a consequence of cells undergoing apoptosis.
Oxidative stress is known to induce a plethora of effects in cells. This includes DNA damage and lipid peroxidation resulting in apoptosis. However, in addition to apoptosis, the discovery of caspase-independent, non-apoptotic cell death has resulted in a shift in how cell death is regulated during disease [11]. Initially described for certain cancer lines, iron-dependent, ferroptotic cell death has emerged as an important regulator in degenerative disease [12,13]. Ferroptosis is distinct from apoptosis and is defined by excessive toxic lipid peroxidation. Glutathione peroxidase 4 (GPX4) has been demonstrated to be an essential regulator of ferroptotic cell death [14]. As GPX4 is regulated by glutathione (GSH); ferroptosis can be induced pharmacologically by small molecules such as erastin. Erastin binds and inhibits SLC7A11: the subunit of cystine/glutamate transporter. The inhibition of the importation of cysteine, leads to the depletion of GSH and thus inactivation of GPX4. The loss of GPX4 activity leads to overwhelming lipid peroxidation and ultimately cell death [14]. Interestingly, NRF2 has been demonstrated to regulate genes involved in iron synthesis, regulators of ferroptosis, including SLC7A11 and, promote resistance to ferroptosis [[15], [16], [17]]. However, to date there has not been any link to the loss of NRF2, lipid peroxidation and ferroptosis in human CEnCs.
In this study we sought to characterise the expression pattern and role(s) of PRDX in normal and in corneal endothelial cells from patients with FECD. We demonstrated that the expression of PRDX1 in CEnC is sensitive to oxidative stress. Furthermore, we have shown a significant loss of PRDX1 expression in patient derived FECD tissue. We have also established that PRDX1 controls the sensitivity of CEnCs to stimuli that induce cell death through enhanced lipid peroxidation. We further demonstrate that expression of both PRDX1 and NRF2 is required to regulate lipid peroxidation. However, GPX4 appears largely redundant. In addition, we have demonstrated that inhibitors of ferroptosis block enhanced lipid peroxidation and restore cell viability in the absence of PRDX1. We propose the use of inhibitors of ferroptosis as a novel treatment of FECD.
2. Materials and methods
2.1. Chemicals
Ferrostatin-1 and erastin were purchased from Sigma-Aldrich (Merck, Darmstadt, Germany). We selected cumene hydroperoxide (CH) as the oxidative stress-inducing agent as a more stable alternative to H2O2. Furthermore CH is lipophilic and acts predominantly on cell membranes [18]. Moreover, we have previously demonstrated that CH induces lipid peroxidation in CEnCs [19]. CH was provided along with BODIPY® 581/591 C11, Image-iT™ lipid peroxidation kit (C10445, Thermo Fisher Scientific).
2.2. Human corneal tissue and cell culture
The Singhealth centralized institutional review board approved this study. The study was conducted according to the tenets of the declaration of Helsinki. Written and informed consent was obtained from patients undergoing surgical treatment for FECD.
Human CEnCs attached to DM from FECD patients was obtained from endothelial keratoplasty surgery. Tissue was stored in organ culture media prior to processing.
Primary human corneal endothelial cells (hCEnCs) were obtained from cadaveric corneal tissue unsuitable for transplantation procured through Lions Eye Institute for Transplant and Research (Tampa, FL, USA). Isolated hCEnCs were expanded using a dual media approach until passage 2 or passage 3 in 6 cm plates as previously described [20]. Prior to experiments hCEnCs were isolated using TrypLE™ Express and seeded on 12 or 24 well plates pre-coated with FNC coating mix® (AthenaES, Baltimore, USA) at high density (1.5–2 x 104 cells/cm2) for 24–48 h in M5 media consisting of Human endothelial-SFM (Life Technologies) supplemented with 5% serum (EquaFetal® Atlas Biologicals, Fort Collins, CO, USA).
The human corneal endothelial cell line, B4G12-CEnC [21], was maintained at 37 °C, 5% CO2 in DMEM (high glucose) supplemented with 10% bovine serum (Gibco; ThermoFisher Scientific). The oncogenic RAS mutant fibrosarcoma cell line, HT1080 (Kind gift from D Virshup, DUKE-NUS, Singapore), was selected as a positive control for ferroptotic cell death induced by the small molecule erastin [13]. HT1080 cells were similarly maintained at 37 °C, 5% CO2 in DMEM (high glucose) supplemented with 10% bovine serum (Gibco; Thermo Fisher Scientific).
2.3. Western blot analysis
Total cell lysates were prepared by directly lysing cells in RIPA buffer containing a cocktail of protease inhibitors (Abcam). Proteins were quantitated by BCA assay (Pierce, Thermo Fisher Scientific) and equivalent amounts loaded on 4–20% mini-PROTEAN® TGX™ Gels (BioRad). Gels were transferred to PVDF membranes and blocked in 5% non-fat milk. The following antibodies were used for immunoblotting: PRDX1(ab41906), PRDX2(ab109367), PRDX3(ab128953), PRDX4(ab184167), PRDX5(ab180123), PRDX6(ab16947) and GPX4 (ab125066, all from Abcam). NRF2 (D1Z9C, Cell Signalling Technology), and as controls GAPDH (BioLegend) or Lamin A/C (4C11, Sigma brand, Merck). Blots were washed in PBST (PBS+0.1% tween-20) and probed with HRP conjugated secondary antibodies (Cell signalling Technology) and visualised by chemiluminescence. Bands were quantified using ChemiDoc™ MP imaging system and image lab software (Bio-Rad). For Western blot analysis of FECD samples experiments were performed using pooled DM-CEnCs (experiment 1 (N = 5), age range 69–83; experiment 2 (N = 5), age range 62–86, experiment 3 (N = 5), age range 75–85). Age matched control DM-CEnCs were isolated from corneas procured form Lions Eye institute.
Nuclear and cytoplasmic extracts were extracted using NE-PER nuclear and cytoplasmic extraction kit (Pierce, Thermo Fisher Scientific) according to manufactures instructions.
2.4. siRNA transfections
Confluent cultures of HT1080 or B4G12 cells were harvested and seeded in 12 plates at 30-35 k/cm2. Cells were transfected with 10 pmol siRNA (Silencer® select validated siRNA, Ambion® by Life Technologies) in combination with Lipofectamine™ RNAiMAX transfection reagent (Thermo Fisher Scientific) within 2 h post seeding according to manufactures instructions. After 24 h, culture media was changed and cells re-transfected. Cells were analysed the following day.
The following siRNA reagents were used: Silencer® select PRDX1 (ID# s10007), GPX4 (ID# s6112), NFE2L2 (NRF2, ID# s9493) and as control Silencer® select negative control #1.
2.5. Flow cytometry
2.5.1. Apoptosis assay
Experiments were performed on siRNA transfected cells plate in duplicate or triplicate wells. On the day of experiment cells were treated with CH for 3 h at 37 °C in a tissue culture incubator. For Annexin V (AnV)/propidium iodide (PI) labelling, cells were washed in PBS and harvested by trypsinisation. Aliquots (100 μl) of cells were incubated with 5 μl of FITC-Annexin V and 10 μl of PI (BioLegend) for 15 min at room temperature in Annexin V binding buffer (BioLegend). Cells were re-suspended in 400 μl AnV binding buffer and immediately analysed on a BD FACSVerse™ flow cytometer.
Alternatively, for detection of activated caspase-3 and caspase-7 aliquots of harvested cells were labelled with the CellEvent™ Caspase 3/7 detection reagent (Thermo Fisher Scientific) according to manufacturer's instructions. Briefly, cells were treated with 0.5 μm staurosporine or 100 μm CH for 90 min at 37 °C in a tissue culture incubator. Harvested cells were resuspended in 500 μl PBS containing 1 μm caspase 3/7 detection reagent for 30 min at 37 °C. During the last 5 min of incubation SYTOX AADvanced dead cell stain was added at a final concentration of 2 μm. Cells were immediately analysed on a BD FACSVerse™ flow cytometer.
2.5.2. Lipid peroxidation assay
Cells were treated with CH for 2 h at 37 °C in a tissue culture incubator. Where indicated erastin (10 μm/ml) or ferrostatin-1 (2 μm/ml) was added at the same time as CH. BODIPY® 581/591 C11 (Thermo Fisher Scientific, 10 μm final concentration) was added during the last 30 min of culture. Cells were harvested by trypsinisation and immediately analysed by flow cytometry. C11 BODIPY was analysed by measuring fluorescence from both the green (FL-1) and red (FL-3) channels on a BD FACSVerse™ flow cytometer. To calculate the amount of lipid peroxidation, both the percentage gated (FL-1) was determined relative to controls, or, the mean fluorescence intensity (MFI) for both FL-1 and FL-3 channels were determined. The amount of lipid peroxidation was quantified by dividing the MFI(FL-1)/MFI(FL-3) from triplicate samples. In control cells the MFI(FL-3) is high resulting in a low FL-1:FL-3 ratio. However, following treatment with CH the MFI (FL-3) decreases whilst MFI (FL-1) increases resulting in a high FL-1:FL-3 ratio.
2.6. Cell viability assays
Cell viability was assessed using the xCelligence real-time cell analyzer (ACEA Bioscience). RNAi transfected HT1080 or B4G12 cells were seeded at 5 × 104 in each well of an E-Plate 96 (ACEA Biosciences) and left to attach for 24 h. Cells were treated with the indicated compounds. The electrical impedance readings of overall cell viability were recorded using the xCELLigence real time cell analyzer throughout the experiment. The percentage of viable cells was calculated for each time point relative to time zero which was set as the last impedance reading prior to addition of compounds. Data is representative of triplicate or quadruplicate wells from three independent experiments.
For primary human CEnCs, cells at passage 2 were dissociated into single cells and seeded at 2.5 × 104/cm2 for each well of a E-Plate 96 pre-coated with FNC for 24 h. Viability was calculated as above.
2.7. Real time PCR analysis
B4G12-CEnC were transfected with siRNA reagents as described above. Total RNA was extracted with PureLink RNA kit (Thermo Fisher Scientific) with on-column DNaseI digestion. Total RNA (500 ng) was reversed transcribed with high capacity cDNA reverse transcription kit (Applied Biosystems) according to manufactures protocol. Real time PCR was performed using the following TaqMan gene expression assays (Thermo Fisher Scientific). GAPDH, (Hs02758991_g1); PRDX1, (Hs00602020_mH); NFE2L2, (Hs00975961_g1) and SLC7A11 (Hs00921938_m1). PCR was performed using TaqMan Fast master mix in the Light Cycler 480 II (Roche). Relative expression was calculated using 2−ΔΔCT normalised to GAPDH.
2.8. Statistical analysis
All statistical analysis was performed using Prism 8 (Graphpad software).
3. Results
3.1. Absence of PRDX1 in corneal endothelial cells derived from FECD
We determined if PRDX expression is selectively lost in CEnCs from FECD corneal tissue. Since there is substantial CEnC loss in FECD patient DM compared to control DM-CEnC, we pooled DM-CEnC from 5 patient samples.
Consistent with previous reports [6], we observed that expression of PRDX2, PRDX3, and PRDX6 from FECD DM-CENC is down regulated compared to controls. Interestingly, we also observed a significant decrease in PRDX1 expression in FECD tissue samples (Fig. 1A). The loss of PRDX1 was independently verified with additional patient DM-CEnCs (Fig. 1B). However, for this pool of FECD derived DM-CEnCs expression of PRDX6 was normal. Similarly, PRDX5 expression was normal compared to controls. Expression of the CEnC marker Na+K+-ATPase was reduced in both experiments. To verify equivalent amount of protein was loaded on our gels, we used GAPDH as a loading control. Surprisingly, and in contradiction to our protein assays (see materials and methods) expression of GAPDH was reduced in FECD CEnCs (Fig. 1A and B). To confirm this, we repeated the analysis with additional patient derived FECD tissue pooled from 5 donors. (Fig. 1C). To account for reduced protein expression apparent in FECD we loaded a greater amount (~8X) of total protein lysate. Whilst this greatly restored expression of Na+K+-ATPase, GAPDH levels remained lower than controls. PRDX1 and PRDX6 expression remained undetectable, whereas expression of PRDX2 and PRDX5 were strongly reduced. Hence our data demonstrated there was heterogeneity in PRDX expression in pooled donors derived from FECD patient samples. However, PRDX1 expression was consistently deficient in FECD samples, suggesting PRDX1 may be the most relevant PRDX family member in relation to the pathogenesis of FECD.
Fig. 1.
PRDX expression in Fuchs’ endothelial corneal dystrophy (FECD), and normal CEnCs.Western blot analysis of PRDX expression from FECD patient CEnCs; isolated during endothelial keratoplasty surgery, compared to age matched controls. Three independent experiments are presented for comparison (A,B & C). PRDX protein levels are shown relative to GAPDH, (A&B) or, expression levels of the indicated proteins are shown relative to control CEnC (C). (D) Western blot of PRDX1-6 expression in donor derived cultures of human CEnCs (N=3) or B4G12-CEnCs treated with 100mm CH for 4 hours.
To address whether oxidative stress contributes to loss of PRDX expression, we treated CEnCs with CH. PRDX levels were determined by Western blot and as controls we used the CEnC cell line B4G12. As expected, CEnCs expressed all 6 members of the PRDX family (Fig. 1A). Interestingly, we could not detect PRDX2 expression in the B4G12-CEnC cell line despite detecting all other PRDX family members. Our data showed that PRDX1, PRDX2 and PRDX6 were sensitive to CH with down-regulation of their expression following a 4-h CH exposure. Neither PRDX3 nor PRDX4 was altered by CH treatment. PRDX5 expression was inconsistent. PRDX5 expression was down regulated in two donor-derived CEnC but up-regulated in a 3rd donor. Interestingly, PRDX expression in B4G12-CEnC was not affected by CH.
Hence, our data suggested that PRDX1 expression was lost in CEnCs derived from FECD. Furthermore, PRDX1 levels appeared to be sensitive to oxidative stress in cultures of donor derived CEnCs. For this reason, we concluded that DM-CEnCs from FECD patients may have reduced PRDX1 protein expression due to enhanced oxidative stress.
3.2. siRNA silencing of PRDX1 in B4G12-CEnC reveals a role for PRDX1 in lipid ROS
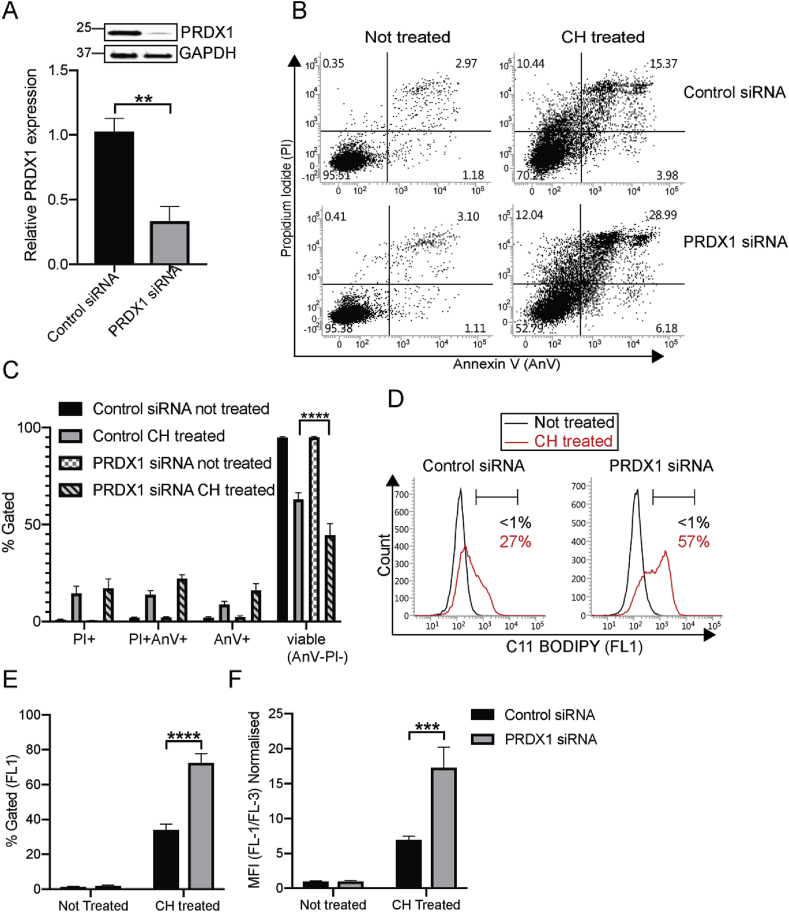
We reasoned that employing siRNA to selectively deplete PRDX1 would render B4G12-CEnCs more susceptible to oxidative stress and apoptosis. Western blot data demonstrated that PRDX1 protein was reduced by approximately 70% compared to control siRNA-transfected cells (Fig. 2A). To monitor cell death, we challenged B4G12-CEnCs with CH and monitored apoptosis with AnV/PI labelling. B4G12-CEnCs lacking PRDX1 appeared more susceptible to apoptosis than control siRNA transfected cells as judged by an overall increase in cells binding AnV and/or PI (Fig. 2B). However, over multiple experiments we could not detect a statistically significant change in the percentage of AnV+ or PI+ subpopulations (Fig. 2C). Interestingly, the percentage of cells judged viable (AnV−/PI−) were significantly reduced in the absence of PRDX1, suggesting cells deficient in PRDX1 have reduced viability, but without an apparent increase in a particular subpopulation of AnV and/or PI (Fig. 2B and C). Subsequently we tested the efficacy of CH to induce lipid peroxidation. Surprisingly we discovered a striking phenotype: PRDX1 knockdown B4G12-CEnCs displayed elevated lipid peroxidation compared to controls (Fig. 2D). This was revealed by a two-fold increase in the percentage of cells positive for C11-Bodipy (Fig. 2E). Similar results were obtained when we compared the ratio of the dual fluorescent C11-BODIPY between oxidised and non-oxidised forms (Fig. 2F).
Fig. 2.
Phenotypic analysis of B4G12-CEnCs after depletion of PRDX1 with siRNA. (A) Knockdown efficiency of PRDX1 using siRNA. Bar charts represent mean ± SEM, of PRDX1 protein levels normalised to GAPDH and relative to negative control siRNA (N = 4. **p = < 0.01, two-tailed t-test). (B) B4G12-CEnC cells were transfected with negative control or PRDX1 siRNA. B4G12-CEnC were left untreated or treated with 100 μm CH for 3 h at 37 °C. Harvested cells were stained with Annexin V and propidium iodide (PI). Representative FACS plots of viable (AnV-/PI-), early (AnV+/PI-) or late apoptotic (AnV+/PI+) and necrotic (AnV-/PI+) is shown. (C) Bar graphs represent percentage of AnV/PI subpopulations. Two-way ANOVA with Bonferroni's post-test was performed to find statistical significance (Data presented as mean ± SEM from technical and biological replicates from three independent experiments. ****p = 0.0001). (D) Lipid ROS was determined after 2-h treatment with 100 μm CH using C11 BODIPY. (E) Percentage of cells gated positive for C11 BODIPY relative to untreated controls. (F) Mean fluorescence intensity (MFI) of C11 BODIPY (green: FL-1 channel) normalised to C11 BODIPY (red: FL-3 channel), relative to not treated controls. Data (N = 5) expressed as mean ± SEM. ****p=<0.0001, ***p = 0.009 (Two-way ANOVA with Bonferroni's post-tests). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Therefore, our data suggests that B4G12-CEnCs lacking PRDX1 have increased sensitivity to CH as reflected by a reduced ability to control lipid peroxidation. Furthermore, our data suggest that loss of viability in response to CH is AnV independent.
3.3. GPX4 independent, ferroptosis-like response in corneal endothelial cells
Ferroptosis is a form cell death associated with lipid peroxidation [13]. Compared to staurosporine: a potent inducer of apoptosis, CH only weakly induced caspase activity in B4G12-CEnCs (Supplementary Fig. S1). This suggests that CH might induce cell death by a caspase independent mechanism such as ferroptosis. We determined if loss of GPX4 resembled PRDX1 deficient cells, with respect to induction of lipid peroxidation and ferroptotic cell death. Using siRNA we could achieve a 90% knockdown of GPX4 protein compared to siRNA-targeting PRDX1, or negative control siRNA (Fig. 3A). Silencing of GPX4 moderately affected induction of lipid peroxidation compared to B4G12-CEnCs transfected with negative control siRNA. However this was not significant (Fig. 3B). As a control for these experiments we used siRNA-targeting PRDX1. Consistent with our previous observation, loss of PRDX1 was sufficient to induce a significant increase in lipid peroxidation (Fig. 3B). Therefore, our data suggests that in response to CH, lipid peroxidation is predominantly regulated by PRDX1 and not GPX4 in B4G12-CEnCs. To confirm these findings, we targeted GPX4 in B4G12-CEnCs as well as HT1080 cells. We confirmed that siRNA mediated knockdown of GPX4 was comparable in HT1080 cells to B4G12-CEnCs (Supplementary Fig. S2A). Interestingly, GPX4 deficient HT1080 demonstrated an increased basal level of lipid peroxidation, consistent with previous reports [14] (Supplementary Fig. S2). However, basal lipid ROS levels in B4G12-CEnC cells were lower than HT1080 and not affected by GPX4 deficiency (Supplementary Fig. 2B). We confirmed that HT1080 were sensitive to ferroptosis induced by erastin. Treatment of HT1080 cells with erastin resulted in a time-dependent decrease in cell viability (Fig. 3C). Moreover, the effects of erastin were augmented by the selective depletion of GPX4 (Fig. 3C) resulting in approximately 2 times more cell death (Fig. 3D). However, in parallel experiments erastin did not induce ferroptotic cell death in B4G12-CEnCs (Fig. 3C). Interestingly, depleting GPX4 expression in B4G12-CEnC resulted in growth retardation of B4G12-CEnCs with loss of cell viability (Fig. 3C). The addition of erastin to GPX4 deficient B4G12 resulted in a rapid loss of viability; however, the amount of cell death was not statistically significant, compared to GPX4 deficient B4G12-CEnCs cultured for 24 h in the absence of erastin (Fig. 3C and D).
Fig. 3.
B4G12-CEnC are resistant to erastin mediated cell death. (A) Knockdown efficiency of GPX4 and PRDX1 using siRNA. Bar charts mean ± SEM, of PRDX1 and GPX4 protein levels normalised to GAPDH, relative to negative control siRNA (N = 3). (B) Lipid peroxidation was calculated as mean fluorescence intensity (MFI) of C11 BODIPY (FL-1) normalised to C11 BODIPY (FL-3 channel), relative to controls. Data representative of N = 3, *p=<0.05, two-way ANOVA with Bonferroni's post-tests. (C) HT1080 and B4G12-CEnC cells were transfected with control or GPX4 siRNA for 48 h. Cells were harvested and seeded on xCELLigence plates for 24 h s before exposure to 10 μm erastin. Cellular viability was assessed for an additional 24 h. Data represents mean ± SEM from triplicate samples (D). Viability of HT1080 or B4G12 was calculated from impedance readings of at hourly time points. Percent viability was calculated relative to time 0 (24hr time point prior to addition of compounds). Data represents mean ± SEM from three independent experiments (Two-way ANOVA with Bonferroni's post-tests). **p < 0.005, ****p=<0.0001.
Taken together our data suggest that GPX4 is required for the normal growth and viability of B4G12-CEnC.
3.4. PRDX1 regulates cell viability in B4G12-CEnCs
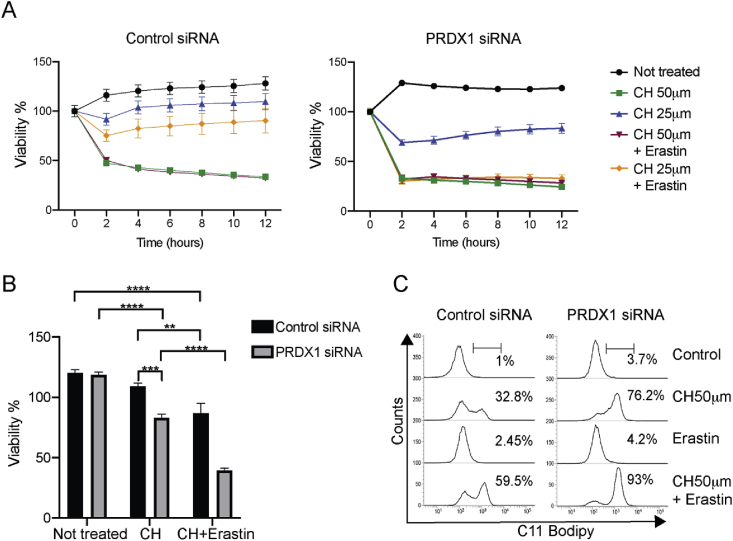
We next tested whether loss of PRDX1 would affect the viability of B4G12-CEnCs to respond to CH. The addition of 50 μm CH rapidly resulted in cell death for both control and PRDX1 deficient B4G12-CEnCs (Fig. 4A). Reducing the concentration of CH to 25 μm resulted in control siRNA transfected B4G12-CEnCs remaining predominantly viable. However, PRDX1 deficient B4G12-CEnCs remained susceptible to CH (Fig. 4A and B). Interestingly, erastin potentiated CH induced cell death. Once more, this effect was particularly evident when the concentration of CH was reduced. Whilst a concentration of 25 μm CH did not significantly reduce the viability of B4G12-CEnCs transfected with control siRNA, after treatment with CH in combination with erastin, viability was reduced by approximately 30% compared to untreated controls (Fig. 4A and B). In stark contrast, PRDX1 deficient cells were more susceptible to the lower dose of CH in combination with erastin as the amount of cell death was approximately 2-fold higher compared to control cells treated with CH + erastin (Fig. 4A and B). This data demonstrated that PRDX1 deficient cells were more sensitive to cell death in response to lipid peroxidation. To confirm this, we measured lipid peroxidation in B4G12-CEnCs with a combination of CH and erastin. Erastin alone did not induces any lipid peroxidation. However, in combination with CH the amount of lipid peroxidation was greater than CH alone (Fig. 4C).
Fig. 4.
Erastin potentiates CH mediated lipid peroxidation. (A) B4G12-CEnCs were transfected with respective siRNA for 48 h. Cells were harvested and seeded on xCELLigence plates for 24 h before exposure to indicated concentrations of CH ± 10 μm erastin. Cellular viability was calculated from impedance readings of B4G12-CEnCs at hourly time points. Percent viability was calculated relative to time 0 (24-h time point prior to addition of compounds) from triplicate samples. (B) Data from three independent experiments at 6-h post treatment with 25 μm CH ± 10 μm erastin was used to generate the bar chart figure. Data represents mean ± SEM (Two-way ANOVA with Bonferroni's post-tests). **p = 0.003, ***p = 0.005, ****p=<0.0001. (C) Representative FACS plot (N = 3) of C11 Bodipy fluorescence from B4G12-CEnCs treated with CH, erastin or CH + erastin.
3.5. Ferrostatin-1 rescues CH induced cell death in B4G12-CEnC and primary human CEnCs
We next treated B4G12-CEnCs with the ferroptosis inhibitor ferrostatin-1 (Fer-1). For both negative control siRNA and PRDX1 siRNA transfected B4G12-CEnC cells, Fer-1 effectively rescued CH dependent lipid peroxidation (Fig. 5A). Furthermore, Fer-1 rescued cell viability (Fig. 5B). Importantly, Fer-1 did not rescue H2O2 induced cell death: H2O2 does not provoke ferroptosis, as previously reported [13]. Similar results were obtained for primary human CEnCs (hCEnCs). Addition of Fer-1 to CH treated hCEnCs rapidly restored cell viability in CH treated hCEnCs (Fig. 5D). However, and in contrast to B4G12-CEnCs addition of erastin did not statistically potentiate CH induced cell death (Fig. 5D). As addition of CH to cultures of primary hCEnCs reduced expression of PRDX1, we examined whether PRDX1 downmodulation could be reversed by the addition of Fer-1. Interestingly, PRDX1 levels are restored to normal following the addition of Fer-1 to CH treated hCEnCs (Fig. 5E).
Fig. 5.
Ferrostatin-1 restores viability of corneal endothelial cells. (A) Ferrostatin-1 (Fer-1, 2 μm) was added to cultures at the same time as CH and lipid peroxidation analysed by the addition of C11-BODIPY. Representative (N = 3) FACS plots are shown. (B) Cell viability of siRNA transfected B4G12-CEnCs or untouched (C) B4G12-CEnCs were treated with CH at 50 μm or 25 μm or H2O2 (100 μm) in the presence or absence of Fer-1 and analysed as in Fig. 3. Data represents cell viability calculated 6-h post treatment from 4 independent experiments (mean ± SEM, two-way ANOVA with Bonferroni's post-tests; ****p < 0.0001). (D) Primary human CEnCs isolated from 6 independent donors were treated with 50 μm CH ± 10 μm erastin or 2 μm Fer-1 for 4 h. Data represents mean ± SEM, Two-way ANOVA with Bonferroni's post-tests; ****p < 0.0001). (E) Primary human CEnCs were treated with indicated reagents for 4 h. RIPA lysates were analysed for protein expression with anti-PRDX1 antibodies by Western blot. Relative PRDX1 expression was calculated by densitometry, relative to GAPDH expression. Data represents three independent experiments using three different, donor derived human CEnCs. Data represents mean ± SEM (ordinary one-way ANOVA with Bonferroni's post-tests, *p < 0.05).
3.6. NRF2 regulates lipid peroxidation in B4G12-CEnCs
Finally, we determined whether NRF2 influences lipid peroxidation in B4G12-CEnCs. Targeted disruption of NRF2 with siRNA resulted in a strong down regulation of NRF2 mRNA levels (Fig. 6A). To confirm this we probed protein lysates with anti-NRF2 antibodies. Whilst we failed to detect expression of NRF2 in total cell lysates from control siRNA transfected B4G12-CEnCs (negative data not shown), we could detect NRF2 expression in the nuclear fraction by Western blot (Fig. 6D and E). This data confirmed the effective siRNA mediated targeting of NRF2, as NRF2 specific siRNA strongly abrogated nuclear NRF2 expression (Fig. 6D and E). To confirm whether loss of NRF2 would impact ferroptosis we examined SLC7A11 expression. Consistent with previous reports, targeting NRF2 abrogated SLC7A11 expression (Fig. 6C). Interestingly, PRDX1 mRNA levels were only partially reduced by loss of NRF2 (Fig. 6B). However, similar to PRDX1 deficiency, loss of NRF2 caused a substantial increase in lipid peroxidation compared to controls (Fig. 6F and G) suggesting that in B4G12-CEnCs lipid peroxidation is predominantly controlled by the concerted actions of both NRF2 and PRDX1.
Fig. 6.
Knockdown of NRF2 increases lipid ROS in B4G12-CEnC cells.B4G12-CEnCs were depleted of PRDX1 or NRF2 by siRNA and left untreated or treated with 50mm CH for 6 hours. Quantitative PCR analysis of (A) PRDX1, (B) NRF2 and (C) SLC7A11 was performed to determine their mRNA levels. Data represents relative mRNA levels (fold change) normalised to GAPDH, relative to untreated control. (A) Targeted depletion of NRF2 resulted in a significant decrease in PRDX1 mRNA in untreated samples. Data (N=3) represents mean ± SEM (Two-way ANOVA with Bonferroni's post-tests, **p=0.002), ns= not significant. (B) Knockdown of NRF2 does not impact PRDX1 mRNA levels. (C) Significant reduction of SLC7A11 mRNA following NRF2 knockdown. (D) Western blot analysis of nuclear extracts from B4G12-CEnCs transfected with negative control or siRNA targeting NRF2. (E) Nuclear NRF2 expression was determined relative to lamin A/C. Data represents mean ± SEM, (N=3), **p=0.0067, two-tailed t-test. Lipid ROS was determined as in Fig. 2. (F) Percentage of cells gated positive for C11 BODIPY relative to untreated controls. (G) Mean fluorescence intensity (MFI) of C11 BODIPY (green: FL-1 channel) normalised to C11.
4. Discussion
Death of CEnCs in FECD is permanent since CEnCs have limited ability to regenerate in vivo [22]. FECD is predominantly a late onset progressive disease and the leading indication for keratoplasty surgery. A CTG tri-nucleotide expansion of an intronic sequence in the TCF4 gene correlates with disease severity [23,24]. However, increased susceptibility to oxidative stress, mitochondrial dysfunction and apoptosis is thought to play a prominent role in FECD [9,22]. We propose that increased oxidative stress drives the loss of PRDX1 expression and renders CEnCs susceptible to lipid peroxidation. We have demonstrated that with reduced expression of PRDX1 the B4G12-CEnC line has increased sensitivity to agents which cause lipid peroxidation. We have shown that CH induced cell death is reminiscent of that described for ferroptotic cell death [25]. Ferroptosis, defined as lethal, iron-dependent lipid peroxidation, that can be suppressed by Fer-1 as well as iron chelators. Our data suggests that CH strongly induces lipid peroxidation. Moreover, this can be suppressed by Fer-1 as well as iron chelators such as DFO (not shown). Agents such as erastin have been demonstrated to trigger ferroptosis via GPX4 inhibition. In stark contrast to cancer cell lines, erastin did not have any effects on B4G12-CEnCs. However, B4G12-CEnCs were sensitised to erastin when the level of GPX4 was reduced. Furthermore, erastin acted synergistically with CH to increase lipid ROS compared to CH alone. This suggested that erastin may only partially inhibit GPX4 in B4G12-CEnCs. Furthermore, this suggests that CH might induce lipid peroxidation by a distinct GPX4 independent pathway in CEnCs.
The degree of endothelial cell loss in FECD is related to several factors. This includes patient age, density and size of guttae as well as other clinical manifestations [22]. Previous reports have noted the down-regulation or complete loss of PRDX expression in FECD [6]. In particular loss of PRDX2 expression as well as significant downregulation of PRDX3,5 and PRDX6. PRDX1 was not analysed in that study [6]. The tissue specimens we analysed were isolated from patients with advanced FECD with significant endothelial cell loss. Therefore, to maximise protein yield we analysed PRDX expression from FECD tissue pooled from 5 donors. Endothelial cell loss in FECD affected the total cellular protein concentration we could extract in our lysates. However, as CEnCs are attached to DM we cannot rule out that our protein assays are skewed by protein coming from both CEnCs as well as DM. Indeed, there was a degree of heterogeneity with protein expression including the expression of the housekeeping protein, GAPDH. However, loss of PRDX1 was highly consistent. We believe that loss of PRDX1 and its role in regulating lipid ROS may well be novel with respect to CEnCs. It will be interesting to determine whether PRDX1 plays a similar role in other cell types.
In the absence of NRF2 it is reported that macrophages do not express PRDX1 in response to oxidative stress [17]. In the absence of NRF2, PRDX1 mRNA appeared reduced compared to controls (Fig. 6A). However, the addition of CH largely restored mRNA levels (Fig. 6A). Furthermore, we could not detect a significant reduction in PRDX1 protein levels following NRF2 depletion (ML unpublished observation/data not shown). This suggested that PRDX1 was not regulated by NRF2. Moreover it suggested that PRDX1 and NRF2 control lipid ROS via different pathways. As controls for these experiments we monitored a bona fide target of NRF2, SLC7A11. Expression of SLC7A11 mRNA was severely down regulated in the absence of NRF2. However, loss of SLC7A11 expression could not explain the sensitivity of NRF2 deficient B4G12-CEnCs to CH, as erastin mediated inhibition of SLC7A11 has no effects on B4G12-CEnCs. NRF2 controls multiple genes involved in the regulation of ferroptosis. Currently it is not known which genes are responsible for the sensitivity to CH with respect to heightened lipid ROS. Regardless, loss of NRF2 has been reported in FECD [26] with the suggestion this results in an increased sensitivity to apoptosis. However, in light of our data we would argue that both apoptosis and ferroptosis are driving significant cell death of CEnCs in FECD.
Loss of PRDX1 is likely to be independent to loss of NRF2 expression. Exactly how loss of PRDX1 triggers ferroptosis is not known. Multiple cellular functions have been ascribed to PRDX1 [10]. Originally thought to be a peroxide scavenging enzyme it is now clear that PRDX1 is more than an antioxidant with chaperone, tumor suppressor [27] and signal transductions functions being described [28]. Moreover, the localisation of PRDX1 is not restricted to the cytosol. A proportion of total cellular PRDX1 has been demonstrated to be associated in lipid domains at the plasma membrane. Interestingly, in this context PRDX1 has been demonstrated to regulate growth factor receptor signalling [29]. In addition, PRDX1 has been detected in the nucleus; where it has been demonstrated to protect telomeres from oxidative damage [30]. The role of PRDX1 in regulating lipid ROS is likely to attributed to one or more of its interacting partners. To date 210 possible interactions have been identified (https://thebiogrid.org/111089/table/homo-sapiens/PRDX1.html [31]). One putative interaction is PEBP1 which encodes the scaffold protein phosphatidylethanolamine-binding protein 1. Interestingly, PEBP1 has been shown to regulate ferroptotic cell death [32].
In conclusion, we infer that CEnCs in FECD patients are under continual oxidative stress. In part, this is due to the inability to activate an antioxidative response due to loss of NRF2 expression. In addition, we present evidence that the loss of PRDX1 could further contribute through the upregulation of lipid ROS.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We thank Professor David Virshup, DUKE-NUS, Singapore for kindly providing the HT-1080 cell line.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101417.
Contributor Information
Matthew Lovatt, Email: lovatt.matthew.jason@seri.com.sg.
Jodhbir S. Mehta, Email: Jodmehta@gmail.com.
Funding support
This work was supported by the Singapore National Medical Research Council (NMRC), Clinician Scientist Award (NMRC/CSA-INV/0004/2015). The funding body had no role in study design, nor the analysis and interpretation of data or the decision to publish.
Financial disclosure
All authors have no proprietary or commercial interests in any materials involved in this article.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bourne W.M. Biology of the corneal endothelium in health and disease. Eye. 2003;17:912–918. doi: 10.1038/sj.eye.6700559. [DOI] [PubMed] [Google Scholar]
- 2.Peh G.S.L., Beuerman R.W., Colman A., Tan D.T., Mehta J.S. Human corneal endothelial cell expansion for corneal endothelium transplantation: an overview. Transplantation. 2011;91:811–819. doi: 10.1097/TP.0b013e3182111f01. [DOI] [PubMed] [Google Scholar]
- 3.Vedana G., Villarreal G., Jun A.S. Fuchs endothelial corneal dystrophy: current perspectives. Clin. Ophthalmol. 2016;10:321–330. doi: 10.2147/OPTH.S83467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocaba V., Katikireddy K.R., Gipson I., Price M.O., Price F.W., Jurkunas U.V. Association of the gutta-induced microenvironment with corneal endothelial cell behavior and demise in fuchs endothelial corneal dystrophy. JAMA Ophthalmol. 2018 doi: 10.1001/jamaophthalmol.2018.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurkunas U.V., Bitar M.S., Funaki T., Azizi B. Evidence of oxidative stress in the pathogenesis of fuchs endothelial corneal dystrophy. Am. J. Pathol. 2010;177:2278–2289. doi: 10.2353/ajpath.2010.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurkunas U.V., Rawe I., Bitar M.S., Zhu C., Harris D.L., Colby K. Decreased expression of peroxiredoxins in Fuchs' endothelial dystrophy. Investig. Ophthalmol. Vis. Sci. 2008;49:2956–2963. doi: 10.1167/iovs.07-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes J.D., Dinkova-Kostova A.T. The NRF2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Decline in DJ-1 and Decreased Nuclear Translocation of NRF2 in Fuchs Endothelial. corneal dystrophy. 2012;vol 53:5806–5813. doi: 10.1167/iovs.12-10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurkunas U.V. Fuchs endothelial corneal dystrophy through the prism of oxidative stress. Cornea. 2018;37(Suppl 1):S50–S54. doi: 10.1097/ICO.0000000000001775. [DOI] [PubMed] [Google Scholar]
- 10.Rhee S.G., Kil I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 11.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerins M.J., Ooi A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxidants Redox Signal. 2018;29:1756–1773. doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan Z., Wirth A.-K., Chen D., Wruck C.J., Rauh M., Buchfelder M. NRF2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6 doi: 10.1038/oncsis.2017.65. e371–e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y. Transcription factor NRF2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 18.Vroegop S.M., Decker D.E., Buxser S.E. Localization of damage induced by reactive oxygen species in cultured cells. Free Radic. Biol. Med. 1995;18:141–151. doi: 10.1016/0891-5849(94)00107-u. [DOI] [PubMed] [Google Scholar]
- 19.Lovatt M., Adnan K., Peh G.S.L., Mehta J.S. Regulation of Oxidative Stress in Corneal Endothelial Cells by PRDX6. Antioxidants. 2018;vol 7:180. doi: 10.3390/antiox7120180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peh G.S.L., Chng Z., Ang H.-P., Cheng T.Y.D., Adnan K., Seah X.-Y. Propagation of human corneal endothelial cells: a novel dual media approach. Cell Transplant. 2015;24:287–304. doi: 10.3727/096368913X675719. [DOI] [PubMed] [Google Scholar]
- 21.Valtink M., Gruschwitz R., Funk R.H.W., Engelmann K. Two clonal cell lines of immortalized human corneal endothelial cells show either differentiated or precursor cell characteristics. Cells Tissues Organs (Print) 2008;187:286–294. doi: 10.1159/000113406. [DOI] [PubMed] [Google Scholar]
- 22.Matthaei M., Hribek A., Clahsen T., Bachmann B., Cursiefen C., Jun A.S. Fuchs endothelial corneal dystrophy: clinical, genetic, pathophysiologic, and therapeutic aspects. Annu Rev Vis Sci. 2019;5:151–175. doi: 10.1146/annurev-vision-091718-014852. [DOI] [PubMed] [Google Scholar]
- 23.Xing C., Gong X., Hussain I., Khor C.-C., Tan D.T.H., Aung T. Transethnic replication of association of CTG18.1 repeat expansion of TCF4 gene with Fuchs' corneal dystrophy in Chinese implies common causal variant. Investig. Ophthalmol. Vis. Sci. 2014;55:7073–7078. doi: 10.1167/iovs.14-15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soh Y.Q., Peh Swee Lim G., Htoon H.M., Gong X., Mootha V.V., Vithana E.N. Trinucleotide repeat expansion length as a predictor of the clinical progression of Fuchs' Endothelial Corneal Dystrophy. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng H., Stockwell B.R. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C., Chen Y., Kochevar I.E., Jurkunas U.V. Decreased DJ-1 leads to impaired NRF2-regulated antioxidant defense and increased UV-A-induced apoptosis in corneal endothelial cells. Investig. Ophthalmol. Vis. Sci. 2014;55:5551–5560. doi: 10.1167/iovs.14-14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann C.A., Krause D.S., Carman C.V., Das S., Dubey D.P., Abraham J.L. Essential role for the peroxiredoxin PRDX1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 28.Rhee S.G., Woo H.A., Kang D. The role of peroxiredoxins in the transduction of H2O2 signals. Antioxidants Redox Signal. 2018;28:537–557. doi: 10.1089/ars.2017.7167. [DOI] [PubMed] [Google Scholar]
- 29.Woo H.A., Yim S.H., Shin D.H., Kang D., Yu D.-Y., Rhee S.G. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Aeby E., Ahmed W., Redon S., Simanis V., Lingner J. Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep. 2016;17:3107–3114. doi: 10.1016/j.celrep.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 31.Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenzel S.E., Tyurina Y.Y., Zhao J., St Croix C.M., Dar H.H., Mao G. PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell. 2017;171:628–641. doi: 10.1016/j.cell.2017.09.044. .e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.