Abstract
Adult mammalian cardiomyocytes (CMs) retain a limited proliferative ability, which is insufficient for the repair of CM loss in ischemic cardiac injury. Regulation of the Hippo signaling pathway to promote endogenous CM proliferation has emerged as a promising strategy for heart regeneration. Previous studies have shown that the microRNA cluster miR302–367 negatively regulates the Hippo pathway, promoting CM proliferation. In this study, we identified another microRNA, miR-10b, that regulates the Hippo pathway and promotes cell proliferation in human embryonic stem cell-derived CMs (hESC-CMs). We observed that miR-10b expression was enriched in the early stage of CMs, but its expression was reduced over time. Overexpression of miR-10b promoted CM proliferation, while knockdown of miR-10b suppressed CM proliferation. Moreover, miR-10b protected CMs against apoptosis. miR-10b functions, in part, by directly targeting LATS1, which is a major component of the Hippo pathway. Our study suggests that miR-10b has promising potential for heart regeneration.
Keywords: miR-10b, human embryonic stem cells, cardiomyocytes, proliferation, Hippo pathway
Introduction
Heart failure due to cardiomyocyte (CM) loss after ischemic heart disease is the leading cause of death in developed countries.1,2 Adult mammalian CMs retain a limited endogenous renewal capacity,3,4 which is insufficient for the replacement of acute or chronic CM loss in ischemic cardiac injury.4,5 One attractive strategy to improve the outcome in heart failure is to regenerate damaged myocardium by producing more functional CMs. Several signaling pathways have been demonstrated to regulate CM proliferation, such as IGF1,6 perisotin,7 neuregulin,8 fibroblast growth factor, the Wnt/β-catenin signaling pathway,9 and Hippo signaling pathways.10,11 Among these signaling pathways, the Hippo signaling pathway plays a crucial role in the progression of heart failure by regulating the growth and death of CMs.12,13
The Hippo pathway is an evolutionarily conserved signaling pathway that controls organ size by regulating cell proliferation and apoptosis.14 In mammals, the main members of the Hippo pathway include MST1/2, Sav1, Lats1/2, and Mob1, which form a kinase cascade that phosphorylates and inhibits the downstream effector YAP, thereby preventing its nuclear entry.14 Several studies have shown that inactivation of Sav or Lats kinases or activation of Yap in mouse models induces CM renewal.10,11,15,16 Leach et al.15 treated mice with adeno-associated virus 9 (AAV9), which encodes a short hairpin RNA (shRNA) against Salv; the treatment recovered heart function and led to CM cell cycle re-entry, highlighting the promising potential of the Hippo pathway as a therapeutic target for the treatment of heart failure.
Interestingly, microRNAs (miRNAs) play a crucial role in the regulation of the Hippo pathway in the heart. miRNAs are a class of small noncoding single-stranded RNAs (∼22 nt) that regulate gene expression at the posttranscriptional level mainly through binding to the 3′ untranslated regions (UTRs) of their targets.17,18 It has been reported that the miRNA cluster miR302–367 negatively regulates the Hippo pathway, promoting CM proliferation and dedifferentiation.19 Here, we identified another miRNA, miR-10b, that regulates the Hippo pathway and promotes cell proliferation in human embryonic stem cell-derived CMs (hESC-CMs). We found that miR-10b expression was reduced as the CM proliferation decreased. Overexpression of miR-10b promoted CM proliferation. Furthermore, we showed that miR-10b functioned, at least in part, by targeting LATS1. Our study suggests that miR-10b is a promising molecule for heart regeneration.
Results
Proliferation of hESC-CMs Decreased Over Time
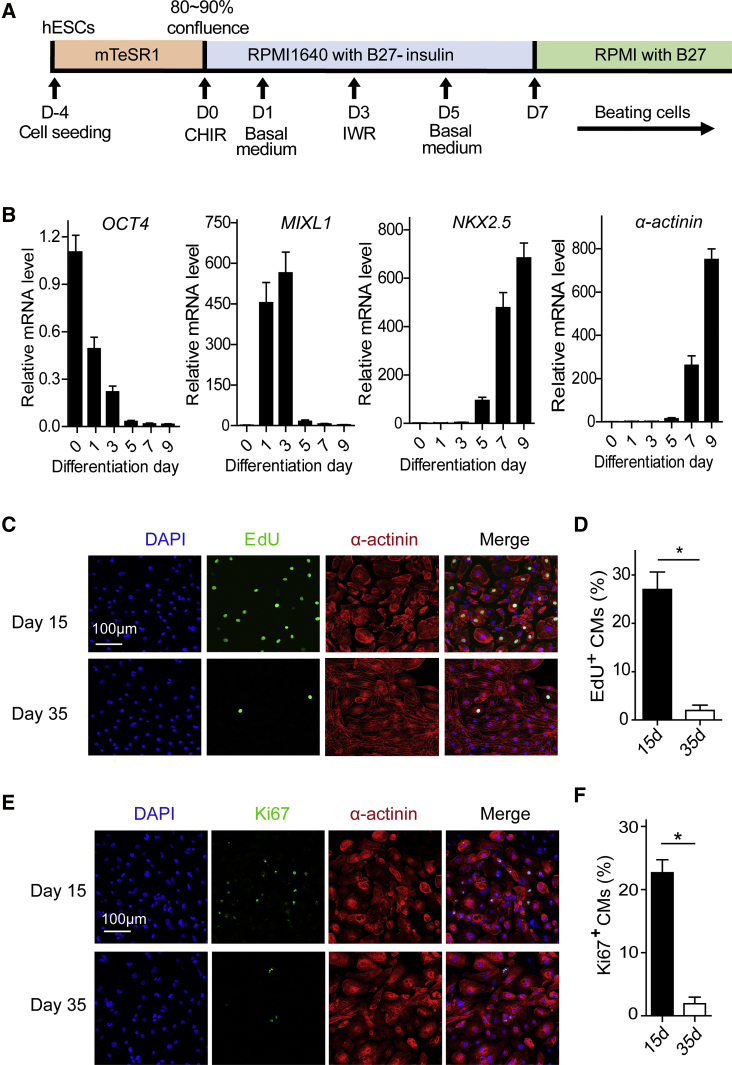
In this study, hESC-CMs were used to investigate the molecular mechanism of proliferation. We differentiated hESCs into CMs using a small molecule-based monolayer stepwise system (Figure 1A).20 Beating CMs were observed 8–10 days after induction. hECS-CMs were further purified with glucose-depleted culture medium containing abundant lactate.21 There are four major stages of CM differentiation in vitro: stem cells, mesodermal progenitor cells, cardiac progenitor cells, and CMs.22 Consistent with these four major stages, quantitative real-time PCR analysis revealed that the stem cell marker OCT4 significantly decreased over time, the mesodermal marker MIXL1 was transiently upregulated at days 1–3, and the cardiac markers NKX2.5 and α-actinin remarkably increased at day 7 (Figure 1B). We investigated the proliferation ability of CMs at days 15 and 35. EdU+ and Ki67+ staining revealed that hESC-CM proliferation dramatically decreased at day 35 (Figures 1C–1F), consistent with previous reports.23,24 These hESC-CMs provide us with a good opportunity to study how miRNAs regulate CM proliferation.
Figure 1.
Proliferation of hESC-CMs Decreases during the Maturation Process
(A) Schematic of the CM differentiation protocol using small molecules. (B) Relative expression levels of the marker genes, including OCT4, MIXL1, NKX2.5, and α-actinin, during CM differentiation as revealed by quantitative real-time PCR. GAPDH was used as an internal reference to calculate the relative mRNA level. (C) Evaluation of hESC-CM proliferation by EdU (green), α-actinin (red), and DAPI (blue) staining at day 15 and day 35. (D) Percentage of EdU+ hESC-CMs at day 15 and day 35 (n = 3). (E) Evaluation of hESC-CM proliferation by Ki67 (green), α-actinin (red), and DAPI (blue) staining at day 15 and day 35. (F) Percentage of Ki67+ hESC-CMs at day 15 and day 35 (n = 3). Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01.
Overexpression of miR-10b Promotes hESC-CM Proliferation
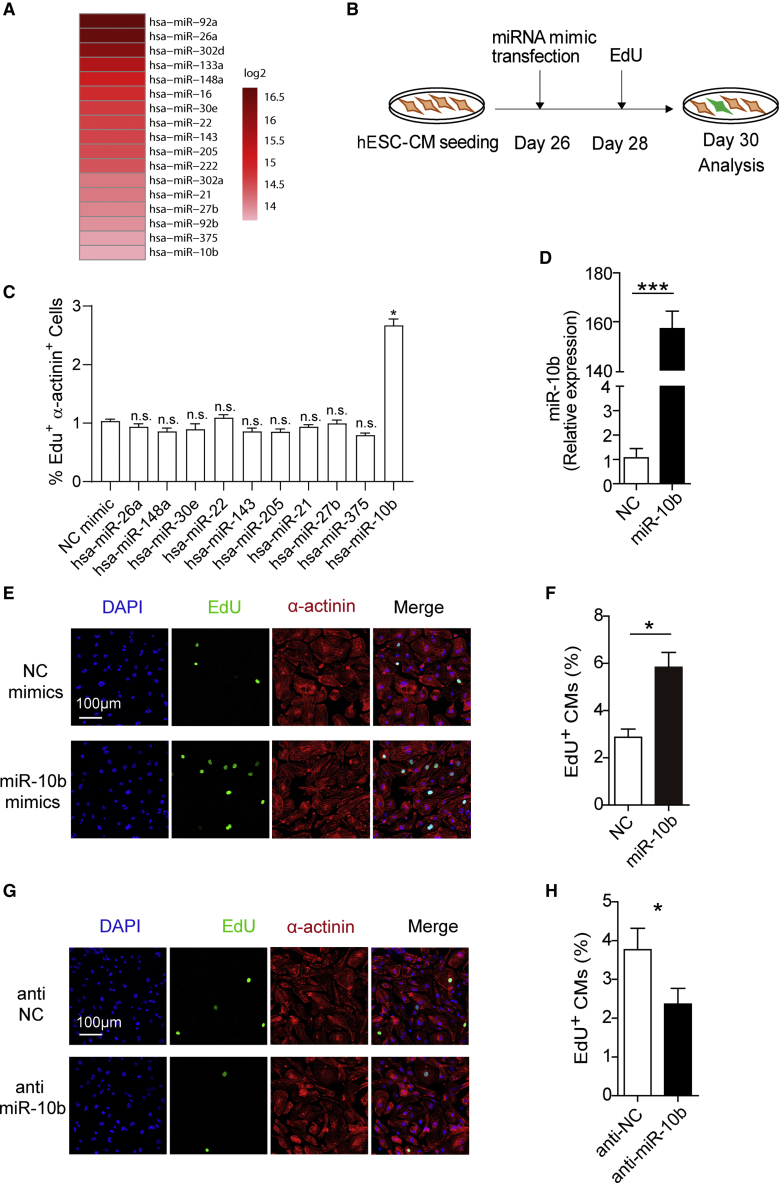
We investigated the abundance of each miRNA at the early stage of hESC-CMs (7 days after CM differentiation) by profiling miRNA expression at the genome-wide level, and the results revealed that there are 17 relatively abundant miRNAs at this stage (Figure 2A). Among these miRNAs, miR-92a/b, miR-302a/d, miR-222, miR-16, and miR-133a have been reported to play important roles in regulating CM proliferation.19,25, 26, 27, 28 Thus, we investigated the proliferative effect of the remaining miRNAs with primary immunofluorescence screening. We transfected 26-day-old hESC-CMs with either negative control (NC) mimics or miRNA mimics (Figure 2B). The results revealed that only miR-10b promoted CM proliferation (Figures 2C–2F; Figures S1A and S1B). Consistent with CM proliferation, qRT-PCR revealed that expression of a variety of cell cycle-related genes, including CDKN2A, CCNB, CCND, CDK1, NUSAP1, and PCNA, was changed after miR-10b transfection (Figure S1C). miR-10b overexpression affected the nucleation of hESC-CMs, as the percentage of mononucleated CMs increased and the percentage of binucleated CMs decreased after transfection (Figure S1D).
Figure 2.
miR-10b Overexpression Promotes hESC-CM Proliferation
(A) miRNA sequencing (miRNA-seq) analysis of cardiac progenitor cells identified a subset of highly expressed miRNAs. (B) Schematic diagram depicting the experimental design. (C) miRNA mimic screening for hESC-CM proliferation. Percentage of EdU+, α-actinin+ cells were calculated for each miRNA mimic transfection compared to NC mimic (n = 3). (D) miR-10b expression in hESC-CMs transfected with NC mimics or miR-10b mimics was analyzed by qRT-PCR (n = 3). (E) Evaluation of hESC-CM proliferation after transfection with NC mimics or miR-10b mimics by EdU (green), α-actinin (red), and DAPI (blue) staining. (F) Percentage of EdU+ CMs after transfection with NC mimics or miR-10b mimics (n = 3). (G) Evaluation of hESC-CM proliferation after transfection with anti-NC or anti-miR-10b by EdU (green), α-actinin (red), and DAPI (blue) staining. (H) Percentage of EdU+ CMs after transfection with anti-NC or anti-miR-10b (n = 3). Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01.
We further investigated the effects of miR-10b knockdown on hESC-CM proliferation. Transfection of miR-10b inhibitors (anti-miR-10b) in CMs decreased miR-10b expression, as revealed by qRT-PCR (Figure S2A), and decreased the proliferation capacity of hESC-CMs, as revealed by both EdU and Ki67 staining (Figures 2G and 2H; Figures S2B and S2C). After transfection with miR-10b mimic or anti-miR-10b, the size of hESC-CMs was not influenced (Figures S2D and S2E). Next, we investigated miR-10b expression during CM maturation. We performed qRT-PCR at four time points, day 7, day 15, day 25, and day 35, and the results revealed that miR-10b was downregulated over time (Figure S3A). Since miR-10a is homologous to miR-10b, we also investigated whether miR-10a can promote CM proliferation. The miR-10a expression did not change during CM differentiation (Figure S3B). Immunostaining results showed that miR-10a could not promote CM proliferation (Figures S3C–S3F).
Several studies have shown that mature adult CMs can reenter the cell cycle and form new CMs through dedifferentiation, proliferation, and redifferentiation.29,30 We investigated whether this was the case in hESC-CMs. We transfected hESC-CMs with either NC mimics or miR-10b mimics. Immunostaining of sarcomeric α-actinin revealed that miR-10b transfection resulted in a higher percentage of CMs with disorganized sarcomeric structures compared to control CMs (Figures S4A and S4B). qRT-PCR revealed that miR-10b transfection increased expression of the cardiac-progenitor-associated marker NKX2.5 (Figure S4C), which is the earliest detectable precardiac marker during heart regeneration.31 Collectively, these data suggest that miR-10b may induce hESC-CM dedifferentiation and proliferation.
miR-10b Protects hESC-CMs against Apoptosis
In addition to proliferation, we also investigated the effects of miR-10b on CM apoptosis. CMs were transfected with miR-10b or NC mimics followed by H2O2 treatment and annexin V-allophycocyanin (APC)/propidium iodide (PI) staining. Flow cytometry analysis showed that miR-10b transfection significantly reduced cell apoptosis compared to NC (Figures 3A and 3B). We further tested the expression of several apoptosis-related genes by qRT-PCR. The results showed that overexpression of miR-10b led to downregulation of apoptosis-inducing genes, including BAX, DAPK, and STK17B (Figure 3C). Taken together, these results indicate that overexpression of miR-10b protects hESC-CMs against apoptosis.
Figure 3.
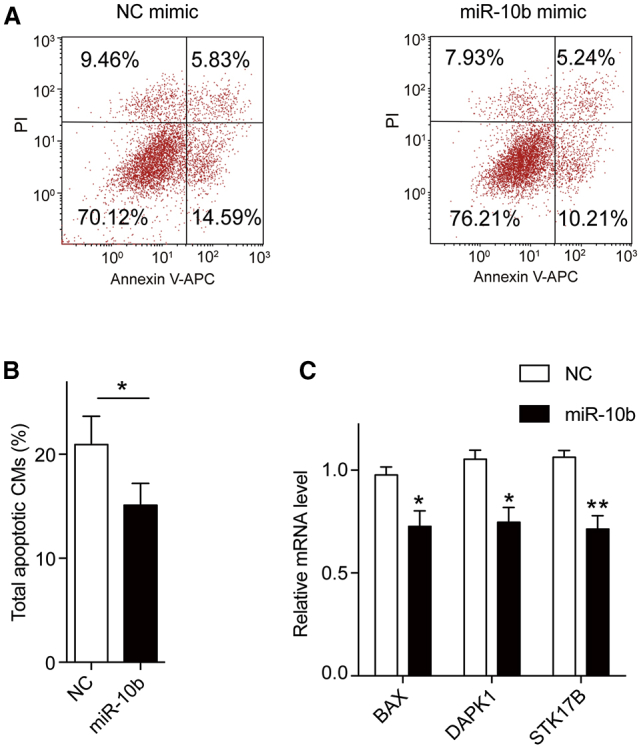
miR-10b Overexpression Decreases hESC-CM Apoptosis
(A) Apoptosis of hESC-CMs transfected with NC mimics or miR-10b mimics followed by H2O2 treatment was analyzed by flow cytometry. The annexin V-APC (x axis)-positive area represents the early stage of apoptotic cells, and the PI (y axis)-positive area represents the late stage of apoptotic cells. (B) Percentage of total apoptotic hESC-CMs transfected with NC mimics or miR-10b mimics (n = 3). (C) Relative expression levels of genes related to apoptosis (BAX, DAPK1, STK17B) in hESC-CMs transfected with NC mimics or miR-10b mimics. GAPDH was used as an internal reference to calculate the relative mRNA levels (n = 3). Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01.
miR-10b Enhances hESC-CM Proliferation by Targeting LATS1
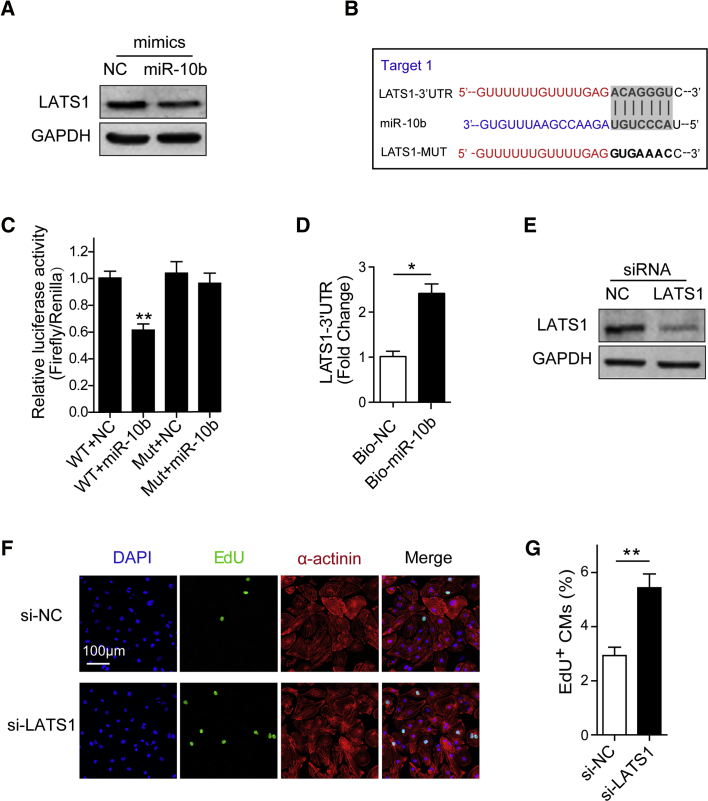
To investigate the molecular mechanism by which miR-10b regulates hESC-CM proliferation, we searched for miR-10b targets that were predicted by the miRNA target prediction tool miRanda or had been reported to promote cell proliferation and got a list of 13 potential targets. qRT-PCR analysis showed that overexpression of miR-10b decreased the mRNA level of CSMD1, TIP30, BABAM1, BCAR1, PTEN, MIB1, ZC3HC1, and LATS1 in hESC-CMs (Figure S5A).32, 33, 34, 35, 36 The targets were further narrowed down by using the dual-luciferase assay. We constructed luciferase reporter vectors containing wild-type (WT) 3′ UTR of potential targets. Overexpression of miR-10b led to decreased luciferase activity of the reporter with WT LATS1-3′ UTR, indicating that LATS1 could be the direct target of miR-10b (Figure S5B). Notably, two potential binding sites for miR-10b were predicted on LATS1, but only one site could decrease luciferase activity (Figure S5B). Western blot analysis further showed that overexpression of miR-10b decreased LATS1 expression at the protein level (Figure 4A). Thus, we hypothesized that miR-10b partly promotes human CM proliferation by inhibiting LATS1. To further test whether miR-10b directly regulates LATS1 expression, we constructed a luciferase reporter vector containing LATS1-3′ UTR with a mutated miR-10b binding site. The mutated binding site abolished the repression effects of miR-10b (Figures 4B and 4C). In addition, a biotin-avidin pull-down assay confirmed that miR-10b could directly bind to LATS1-3′ UTR (Figure 4D). Taken together, we identified LATS1 as a novel target for miR-10b.
Figure 4.
LATS1 is the Target of miR-10b
(A) Western blot analysis of LATS1 expression in hESC-CMs transfected with NC ormiR-10b mimics. (B) The predicted miR-10b binding site in the 3′ UTR of LATS1 mRNA (target 1). (C) Binding of miR-10b to the LATS1 3′ UTR was determined by a luciferase reporter assay (n = 3). (D) Biotin-labeled NC or miR-10b mimics were transfected into hESC-CMs. The 3′ UTR of LATS1 was pulled down by NC or miR-10b and quantified by qRT-PCR (n = 3). (E) Western blot analysis of LATS1 expression in hESC-CMs transfected with si-NC or si-LATS1. (F) Evaluation of hESC-CM proliferation after transfection with si-NC or si-LATS1 by EdU (green), α-actinin (red), and DAPI (blue) staining. (G) Percentage of EdU+ CMs transfected with si-NC or si-LATS1 (n = 3). Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01.
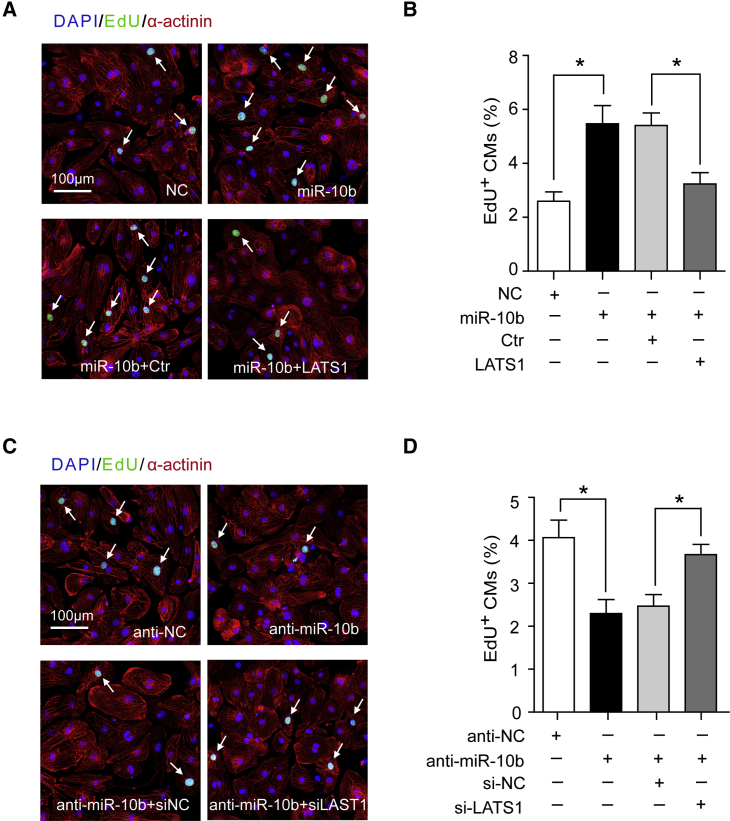
To determine whether reduced LATS1 expression promotes hESC-CM proliferation, a loss-of-function study was performed using small interfering RNA (siRNA) against LATS1. LATS1 knockdown was confirmed by qRT-PCR (Figure S6A) and western blot analyses (Figure 4E). Direct inhibition of LATS1 by siRNA promoted CM proliferation, as indicated by the significant increase in Ki67+ and EdU+ CMs (Figures 4F and 4G; Figures S6B and S6C). Furthermore, when miR-10b was overexpressed together with LATS1 in hESC-CMs, it blocked the effect of miR-10b overexpression, as indicated by the decrease in the number EdU+ CMs (Figures 5A and 5B). In contrast, knockdown of LATS1 with si-LATS1 reversed the reduction of hESC-CM regeneration induced by inhibiting miR-10b (Figures 5C and 5D). Taken together, these results suggest that LATS1 is a direct functional target of miR10b and that miR-10b promotes hESC-CM proliferation, at least in part, by downregulating LATS1.
Figure 5.
miR-10b Promotes hESC-CM Proliferation by Targeting LATS1
(A) Evaluation of hESC-CM proliferation after transfection with NC mimics, miR-10b mimics, miR-10b mimics + control (Ctr) (empty control plasmid), and miR-10b mimics + LATS1 (LATS1 expressing plasmid) by EdU staining. The white arrows indicate proliferative CMs. (B) Percentage of EdU+ CMs transfected with NC mimics, miR-10b mimics, miR-10b mimics + Ctr and miR-10b mimics + LATS1 (n=3). (C) Evaluation of hESC-CM proliferation after transfection with anti-NC, anti-miR-10b, anti-miR-10b + si-NC, and anti-miR-10b + si-LATS1 by EdU staining. The white arrows indicate proliferative CMs. (D) Percentage of EdU+ CMs transfected with anti-NC, anti-miR-10b, anti-miR-10b + si-NC, and anti-miR-10b + si-LATS1 (n=3). Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01.
miR-10b Promotes hESC-CM Proliferation by Inhibiting the Hippo Pathway
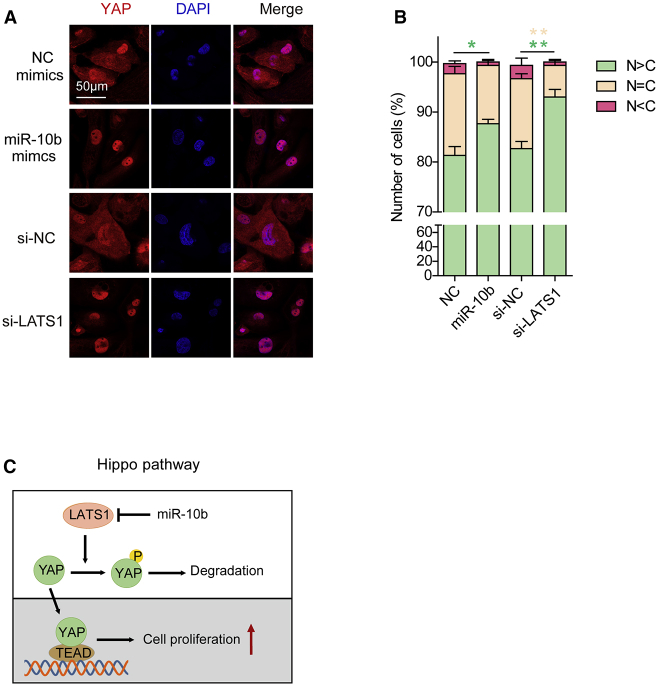
LATS1 is an essential component in the Hippo pathway that regulates cell proliferation.11 To determine whether miR-10b promotes hESC-CM proliferation through the Hippo pathway, we tested the subcellular localization of YAP in hESC-CMs after transfection of miR-10b mimics or si-LATS1. The immunostaining results showed that transfection of miR-10b mimics or si-LATS1 increased the ratio of nuclear- to cytoplasmic-localized YAP (Figures 6A and 6B). Therefore, we propose a model in which the overexpression of miR-10b promotes hESC-CM proliferation in part through regulation of the Hippo pathway via targeting LATS1 (Figure 6C).
Figure 6.
miR-10b Promotes hESC-CM Proliferation by Inhibiting the Hippo Pathway
(A) Immunofluorescence images of YAP (red) subcellular localization in hESC-CMs transfected with si-NC/si-LATS1 and NC/miR-10b mimics. Nuclei were stained with DAPI (blue). (B) Quantification of YAP subcellular localization. C, cytoplasm; N, nucleus. More than 100 cells were randomly selected in each calculation. Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM. *p < 0.05, **p < 0.01. (C) Proposed model of miR-10b promotion of hESC-CM proliferation through inhibiting the Hippo pathway.
Discussion
The adult mammalian heart exhibits a limited regenerative capacity after myocardial injury. Uncovering the genetic mechanisms underlying CM proliferation is a critical step in developing new strategies to repair the heart. In this study, we demonstrate that miR-10b can promote hESC-CM proliferation. miR-10b was enriched in the early stage of hESC-CMs, but its expression decreased over time. Consistent with the expression pattern of miR-10b, the proliferation capacity of hESC-CMs also decreased over time. Importantly, overexpression of miR-10b increased the proliferation capacity of hESC-CMs, indicating that miR-10b plays a crucial role in regulating hESC-CM proliferation.
miR-10b has been studied in angiogenesis, cancer, cell differentiation, and embryonic development. Hassel et al.32 and Wang et al.37 described the role of miR-10b in modulating the angiogenic behavior during development through targeting FLT1 and MIB1. miR-10b was implicated in mediating cancer metastasis and invasion by targeting, for example, CSMD1, TIP30, PTEN, and HOXB3.33, 34, 35, 36 miR-10b could also directly regulate Hox genes during Nile tilapia embryonic development.38 However, its role in CM regeneration still remains unknown. Our study first established the link between miR-10b and CM proliferation.
Several studies have shown that inhibition of the Hippo pathway is sufficient to promote the proliferation of endogenous CMs,11,15,16 indicating that manipulation of the Hippo pathway in the heart may be a promising treatment for heart failure. The miRNA cluster miR302–367 has been reported to regulate the Hippo pathway by targeting Mst1, Lats2, and Mob1b, the core components of the Hippo pathway.19 Overexpression of miR302–367 inhibits the Hippo pathway, resulting in reactivation of the CM cell cycle and increased regeneration of CMs. However, persistent reexpression of miR302–367 in the postnatal heart causes prolonged induction of an immature dedifferentiated state of CMs and heart failure.19 In contrast, transient treatment of mice with miR302–367 mimics promotes mouse cardiac regeneration without adverse effects on physiological function.19 Although a list of miRNAs has the capacity to promote CM proliferation,19,25,39 only the miR302–367 cluster has been shown to function by targeting the Hippo pathway in the heart.19 We demonstrated that miR-10b also targeted the Hippo pathway.
We used hESC-CMs to study the function of miR-10b. The results obtained from human CMs can be easily translated to clinical applications. However, the function of miR-10b at the organ level should also be investigated in future studies. In addition to the miR302–367 cluster and miR-10b, a recent large-scale screen using human CMs identified a list of miRNAs that potentially target the Hippo pathway to promote CM proliferation.40 Further studies of these miRNAs will provide a better understanding of the mechanisms underlying CM proliferation and allow this knowledge to finally be translated into clinical applications.
Materials and Methods
Cells Lines
hESCs were obtained from the human ESC line H9 (WA09 line, WiCell institute, NIH registry 0046). H9 cells were seeded on Matrigel (354277, Corning, NY, USA)-coated plates and were cultured with mTeSR1 medium (catalog no. 05850, STEMCELL Technologies, Canada) at 37°C, 5% CO2. The medium was changed every 24 h. hESC-CMs were cultured in RPMI 1640/B27 (with insulin), and the medium was changed every 48 h. HEK293T cells were maintained with DMEM containing 10% fetal bovine serum (FBS) and 1× penicillin/streptomycin.
Differentiation from hESCs to CMs
When the confluence of hESCs was approximately 80%–90%, we started differentiation into CMs and named this day D0. Cells were treated with 10 μM CHIR99021 (Sl263, Selleck, USA) in RPMI 1640/B27 (without insulin) for 24 h. At D1, the medium was replaced with RPMI 1640/B27 (without insulin), and 5 μM IWP-2 (686770-61-6, Sigma, Germany) in RPMI 1640/B27 (without insulin) was added to the plates at D3. After 2 days of culture, we replaced the medium with fresh medium for another 2 days. Cells were maintained with RPMI 1640/B27 (with insulin) from D7, and for 8–10 days after induction, beating cells could be observed.
Transfection
All of the miR-10b mimics, anti-miR-10b, si-LATS1, and their respective NCs were purchased from Shanghai GenePharma (China) and transfected into CMs using Lipofectamine RNAiMAX (13778-150, Life Technologies, USA) following the manufacturer’s instructions at a final concentration of 50 μM. All of the oligonucleotide sequences are listed in Table S1. Lipofectamine 3000 was used to transfect an empty control plasmid and human LATS1-expressing plasmid, which was a gift from Prof. Wufan Tao.
RNA Extraction and Quantitative Reverse Transcriptase PCR (qRT-PCR)
Total RNA was extracted from CMs at 48 h after transfection using the RNAsimple Total RNA kit (DP419, Tiangen, China) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA with a FastQuant RT kit including gDNase (KR106-02, Tiangen, China). Quantitative Reverse Transcriptase PCR (qRT-PCR) was performed to quantitatively detect mRNA levels using SuperReal Premix Plus-SYBR Green (FP205-02, Tiangen, China) on a LightCycler96 qPCR system, and the level of GAPDH was used as an internal control to normalize the gene-specific expression levels. The primers used for qRT-PCR are listed in Table S2. For quantitative detection of miRNA expression, the miScript reverse transcription kit (218061, QIAGEN, Germany) and miScript SYBR Green PCR kit (218073, QIAGEN, Germany) were used according to the manufacturer’s instructions. The U6 primers were used as an NC for normalization. The primer sequences are listed in Table S2.
Immunofluorescence
CMs were washed with PBS three times 72 h after transfection and then fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. After washing with PBS again, CMs were permeabilized with 0.3% Triton X-100 for 20 min. Next, CMs were blocked with 3% BSA in PBS for 1 h and then incubated with the following primary antibodies overnight at 4°C: α-actinin (mouse monoclonal, A7732, Sigma, USA; 1:500), Ki67 (rabbit monoclonal, 11882S, CST, USA; 1:300), and Yap (mouse monoclonal, sc-101199, Santa Cruz, CA, USA; 1:500). The next day, after washing with PBS, CMs were incubated with Alexa fluorogenic secondary antibodies (Alexa Fluor 568, ab175473, Abcam, UK; 1:1,000) for 1 h in the dark, and the cells were further stained with DAPI (D9542, Sigma, Germany; 1:1,000) to visualize nuclei. For EdU staining, 5 mM 5-ethynyl-2′-deoxyuridine (C10338, Life Technologies, USA) was added to the medium for 48 h before immunofluorescence. After incubation with secondary antibodies, the cells were further processed using the Click-IT EdU 555 imaging kit (C10338, Life Technologies, USA) according to the manufacturer’s instructions.
Luciferase Assay
We constructed a psi-check plasmid by inserting a fragment containing the binding site, miR-10b with the 3′ UTR of the target gene, into the psi-check vector. The fragment was amplified from the human genome using the designed primers (F/R) presented in Table S3. The fragment was cut with XhoI and NotI and then inserted into the psi-check vector. The mutant vector was constructed using the specific primers (F/R) presented in Table S3.
HEK293T cells were seeded into 12-well plates 24 h before transfection with the vectors and mimics or their controls. Transfection was performed using Lipofectamine 2000 (Life Technologies, USA) in the following groups: the psi-check vector (approximately 100 ng per well) and mimics (approximately 1.45 μL per well), psi-check vector and NC, mutant vector and mimics, and mutant vector and NC. Each group included four wells. At 48 h after transfection, the luciferase activities were measured using the Dual-Luciferase assay system from Promega according to the provided protocol.
Western Blot Analysis
Forty-eight hours after transfection, CMs were washed with cold PBS and lysed in cold lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-Cl [pH 7.4], 1 mM EDTA) with a protease inhibitor (04693132001, Roche, Switzerland). Protein samples were then loaded onto 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA) for approximately 2 h. Next, the membranes were blocked with 5% BSA in Tris-buffered saline with Tween 20 (TBST) at room temperature for 1 h, followed by incubation with the following primary antibodies at 4°C overnight: GAPDH (5174S, CST, USA) and LATS1 (C66B5, CST, USA). The next day, the membranes were incubated with anti-rabbit secondary antibodies (Abcam, ab6721) for 1 h after washing with TBST three times for 10 min each time. Images were acquired using a Tanon 5200 chemiluminescent imaging system after another three washes with TBST.
RNA Pull-Down Assay
Biotinylated miR-10b and its NC miRNA (both of which were biotin-labeled at the 3′ end) were purchased from GenePharma (Shanghai, China). hESC-CMs were transfected with NC miRNA and miR-10b using Lipofectamine RNAiMAX. Twenty-four hours after transfection, the cells were lysed with lysis buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl, 0.3% IGEPAL CA-630), and the lysate (approximately 500 μL) was mixed with prewashed streptavidin beads overnight at 4°C. The remaining lysate was stored at −80°C as an input. The next day, the mixture was washed with lysis buffer, and RNA was extracted with TRIzol reagent (Invitrogen). Then, quantitative real-time PCR was performed according to the standard protocol.
Cellular Apoptosis Assay
200 μM 30% H2O2 was added to the fresh medium to induce CM apoptosis, and the samples were collected 2 h later. Flow cytometry was used to detect the apoptosis rate using an annexin V-APC staining kit (eBioscience, China). CMs were digested using 0.25% trypsin (Gibco, USA) at 37°C for approximately 10 min after washing with PBS and collected; they were then centrifuged at 900 rpm for 3 min. Then, the samples were washed with cold PBS and 1× binding buffer (eBioscience, China), followed by resuspending in 200 μL of 1× binding buffer, staining with 10 μL of annexin V-APC, and incubation in the dark at room temperature for 10–15 min. After washing with the binding buffer again, the sample was stained with PI and kept in the dark at 4°C
Quantification and Statistical Analysis
Statistics and graphs were prepared using GraphPad Prism 7. Arithmetic means for each experiment are plotted, and error bars represent the standard error of the mean (SEM). The numbers of replicates (n) for each experiment are listed in the figure legend and refer to biological replicates. Statistical significance was calculated using Student’s t test for paired samples. Data are shown as the means ± SEM; ∗p < 0.05, ∗∗p < 0.01.
Author Contributions
Y.X., Q.W., N.G., and F.W. designed and performed the experiments; Y.W., F.L., F.Z., L.J., Z.H., J.G., and H.W. supervised the project. All the authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81870199 and 31521003), the National Basic Research Program of China (2015CB943300), and the Opening Program 2018 of the State Key Laboratory of Genetic Engineering (SKLGE1809), Beijing Natural Science Foundation no. Z190013.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.026.
Contributor Information
Junbo Ge, Email: junboge@126.com.
Hongyan Wang, Email: wanghy@fudan.edu.cn.
Yongming Wang, Email: ymw@fudan.edu.cn.
Supplemental Information
References
- 1.Roger V.L. Epidemiology of heart failure. Circ. Res. 2013;113:646–659. doi: 10.1161/CIRCRESAHA.113.300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakata Y., Shimokawa H. Epidemiology of heart failure in Asia. Circ. J. 2013;77:2209–2217. doi: 10.1253/circj.cj-13-0971. [DOI] [PubMed] [Google Scholar]
- 3.Senyo S.E., Lee R.T., Kühn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. (Amst.) 2014;13(3 Pt B):532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerr R.L., Huang S., Miraliakbar H.R., Clark R., Chien K.R., Ross J., Jr. Insulin-like growth factor-1 enhances ventricular hypertrophy and function during the onset of experimental cardiac failure. J. Clin. Invest. 1995;95:619–627. doi: 10.1172/JCI117706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kühn B., del Monte F., Hajjar R.J., Chang Y.S., Lebeche D., Arab S., Keating M.T. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 8.Baliga R.R., Pimental D.R., Zhao Y.Y., Simmons W.W., Marchionni M.A., Sawyer D.B., Kelly R.A. NRG-1-induced cardiomyocyte hypertrophy. Role of PI-3-kinase, p70S6K, and MEK-MAPK-RSK. Am. J. Physiol. 1999;277:H2026–H2037. doi: 10.1152/ajpheart.1999.277.5.H2026. [DOI] [PubMed] [Google Scholar]
- 9.Fan Y., Ho B.X., Pang J.K.S., Pek N.M.Q., Hor J.H., Ng S.Y., Soh B.S. Wnt/β-catenin-mediated signaling re-activates proliferation of matured cardiomyocytes. Stem Cell Res. Ther. 2018;9:338. doi: 10.1186/s13287-018-1086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin M., Kim Y., Sutherland L.B., Murakami M., Qi X., McAnally J., Porrello E.R., Mahmoud A.I., Tan W., Shelton J.M. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heallen T., Morikawa Y., Leach J., Tao G., Willerson J.T., Johnson R.L., Martin J.F. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Re D.P., Yang Y., Nakano N., Cho J., Zhai P., Yamamoto T., Zhang N., Yabuta N., Nojima H., Pan D., Sadoshima J. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J. Biol. Chem. 2013;288:3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto S., Yang G., Zablocki D., Liu J., Hong C., Kim S.-J., Soler S., Odashima M., Thaisz J., Yehia G. Activation of Mst1 causes dilated cardiomyopathy by stimulating apoptosis without compensatory ventricular myocyte hypertrophy. J. Clin. Invest. 2003;111:1463–1474. doi: 10.1172/JCI17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F.-X., Guan K.-L. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leach J.P., Heallen T., Zhang M., Rahmani M., Morikawa Y., Hill M.C., Segura A., Willerson J.T., Martin J.F. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature. 2017;550:260–264. doi: 10.1038/nature24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z., von Gise A., Zhou P., Gu F., Ma Q., Jiang J., Yau A.L., Buck J.N., Gouin K.A., van Gorp P.R. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquinelli A.E.N.R.G. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y., Liu Y., Wang T., Zhou N., Kong J., Chen L., Snitow M., Morley M., Li D., Petrenko N. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.3010841. 279ra238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Burridge P.W., Keller G., Gold J.D., Wu J.C. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snir M., Kehat I., Gepstein A., Coleman R., Itskovitz-Eldor J., Livne E., Gepstein L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H2355–H2363. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 24.Cui L., Johkura K., Takei S., Ogiwara N., Sasaki K. Structural differentiation, proliferation, and association of human embryonic stem cell-derived cardiomyocytes in vitro and in their extracardiac tissues. J. Struct. Biol. 2007;158:307–317. doi: 10.1016/j.jsb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., Xiao J., Zhu H., Wei X., Platt C., Damilano F., Xiao C., Bezzerides V., Boström P., Che L. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.J., Matkovich S.J., Dorn G.W., 2nd, van Rooij E., Olson E.N. miR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepilina A., Coon A.N., Kikuchi K., Holdway J.E., Roberts R.W., Burns C.G., Poss K.D.C. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 32.Hassel D., Cheng P., White M.P., Ivey K.N., Kroll J., Augustin H.G., Katus H.A., Stainier D.Y., Srivastava D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ. Res. 2012;111:1421–1433. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Q., Gong L., Wang J., Tu Q., Yao L., Zhang J.R., Han X.J., Zhu S.J., Wang S.M., Li Y.H., Zhang W. miR-10b exerts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1) BMC Cancer. 2016;16:806. doi: 10.1186/s12885-016-2801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang H., Gore J., Deitz S., Korc M. MicroRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene. 2014;33:4664–4674. doi: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S., Sun J., Lan Q. TGF-β-induced miR10a/b expression promotes human glioma cell migration by targeting PTEN. Mol. Med. Rep. 2013;8:1741–1746. doi: 10.3892/mmr.2013.1709. [DOI] [PubMed] [Google Scholar]
- 36.Chen H., Fan Y., Xu W., Chen J., Xu C., Wei X., Fang D., Feng Y. miR-10b inhibits apoptosis and promotes proliferation and invasion of endometrial cancer cells via targeting HOXB3. Cancer Biother. Radiopharm. 2016;31:225–231. doi: 10.1089/cbr.2016.1998. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Ling C.C., Li L., Qin Y., Qi J., Liu X., You B., Shi Y., Zhang J., Jiang Q. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016;110:140–150. doi: 10.1093/cvr/cvw023. [DOI] [PubMed] [Google Scholar]
- 38.Giusti J., Pinhal D., Moxon S., Campos C.L., Münsterberg A., Martins C. MicroRNA-10 modulates Hox genes expression during Nile tilapia embryonic development. Mech. Dev. 2016;140:12–18. doi: 10.1016/j.mod.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 40.Diez-Cuñado M., Wei K., Bushway P.J., Maurya M.R., Perera R., Subramaniam S., Ruiz-Lozano P., Mercola M. miRNAs that induce human cardiomyocyte proliferation converge on the hippo pathway. Cell Rep. 2018;23:2168–2174. doi: 10.1016/j.celrep.2018.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.