Abstract
Insulin-like growth factor 1 (IGF1) is a multifunctional cellular regulatory factor that can regulate cell growth and development by mediating growth hormone stimulation. However, the mechanism of IGF1 dysfunction in cardiomyocyte development is seldom reported. To study this, we employed the models of IGF1 knockdown in chicken embryo in vivo and in cardiomyocytes in vitro. We detected the antioxidant capacity, PI3K/Akt pathway, energy metabolism-related genes, and myocardial development-related genes. Our results revealed that the low expression of IGF1 can significantly suppress the antioxidant capacity and increase the ROS (P < 0.05) levels, activating the AMPK and PI3K pathway by inhibiting the expression of IRS1. We also found that myocardial energy metabolism is blocked through IGF1, GLUT, and IGFBP inhibition, further inducing myocardial developmental disorder by inhibiting Mesp1, GATA, Nkx2.5, and MyoD expression. Altogether, we conclude that low IGF1 expression can hinder myocardial development through the dysfunction of energy metabolism caused by ROS-dependent FOXO activation.
1. Introduction
Insulin-like growth factors (IGFs) are a group of polypeptides with growth-promoting function. The secretory cells are widely distributed in tissues such as the liver, kidney, lung, heart, brain, and intestine [1]. IGFs play an important role in cell proliferation, differentiation, individual growth, and development [2]. The IGF family has two subtypes: insulin-like growth factor 1 (IGF1) and insulin-like growth factor 2 (IGF2). The production of IGF1 is dependent on the growth hormone (GH), which is an important growth factor in life processes. Myocardial development is a complex process that is regulated by complex molecular networks composed of many development-related factors. Many studies have shown that various signal pathways are involved in the development of vertebrate hearts, including the bone morphogenetic protein (BMP), Wnt, Notch, and fibroblast growth factor 4 (FGF 4) signal transduction pathways. The BMP and Wnt signaling pathways play an important role in the development of early mesoderm cells into cardiomyocytes; they act on the cardiac-specific transcription factor GATA4 and Nkx2.5 through a signal cascade process, promoting the differentiation of cardiac precursor cells into cardiomyocytes [3, 4]. Musarò et al. demonstrated that localized synthesis of IGF1 is closely related to skeletal muscle hypertrophy, the molecular pathways of which are similar to those responsible for cardiac hypertrophy [5].
Insulin is a hormone secreted by islet β cells, and it is the only hormone that reduces blood sugar and promotes the synthesis of glycogen, fat, and protein in animals [6]. Insulin has been proven to regulate metabolism and growth in the body [7]. The insulin receptor (IR) is a tetramer formed by two alpha subunits and two beta subunits linked by disulfide bonds. The two alpha subunits are located on the outer side of the plasma membrane and have a binding site for insulin; the two beta subunits are transmembrane proteins that play a role in signal transduction. The IR family contains IR, insulin-like growth factor receptor (IGFR), and insulin receptor-related receptor (IRR). Intracellular signaling is initiated by activating intracellular tyrosine kinases through a series of structural conformational changes after IR binding to ligands, which exerts important physiological functions in the body [8]. The cardiac cell membrane is rich in IR, making cardiomyocytes a very important target organ for insulin action. Insulin plays a key role in the regulation of various aspects of cardiovascular metabolism through glucose metabolism, protein synthesis, and vascular tone. The IGF family can regulate cardiac lineage induction by expanding the mesodermal cell population [9]. Bisping et al. demonstrated that although IGF1 is unnecessary for cardiac structure and function, GATA4 must be activated by the IGF1 pathway to exert its function [10].
Conformational changes occur in the beta receptor subunit when insulin binds to IR to form a complex, and this leads to autophosphorylation and activation of tyrosine kinase (TK). The complex phosphorylates insulin receptor substrate (IRS) and activates the phosphatidylinositol 3-kinase (PI3K) pathway and mitogen-activated protein kinase (MAPK) pathway. Insulin augments cardiomyocyte contraction, increases ribosomal biogenesis and protein synthesis, stimulates vascular endothelial growth factor (VEGF), and thereby suppresses apoptosis, promoting cell survival and increasing blood perfusion of the myocardium principally through the PKB/Akt signaling pathway [11]. IGF1 can regulate the process of membrane assembly at the axonal growth cone by activating the PI3K pathway [12]. Zhu et al. found that IGF1 can upregulate VEGF-C in breast cancer by mediating the PI3K/Akt and MAPK/ERK1/2 signaling pathways [13]. Treating the smooth muscle cells of the saphenous vein with IGF1 can induce phosphorylation of PI3K-Akt/PKB and promote proliferation of saphenous vein smooth muscle cells [14].
Organisms can produce free radicals during normal metabolism, and excessive oxygen free radicals can cause damage to human tissues and cellular structures [15, 16]. The free radical balances can be maintained depending on the antioxidant system. The body can mediate the accumulation of excess reactive oxygen species (ROS) through some cell signal transduction, which enhances the expression of many protective proteins in the cell. IGF can sense the changes in ROS levels and thus affect the insulin pathway [17]. Papaiahgari et al. demonstrated that ROS can mediate the activation of nuclear factor erythroid 2-related factor 2 (Nrf2) through the PI3K/Akt pathway [18].
In our previous study, we demonstrated that selenium deficiency disrupted insulin responsiveness through inhibition of the PI3K/Akt pathway by producing excessive oxygen free radicals [19, 20]; meanwhile, selenium deficiency can downregulate the expression of IGF1. However, the role of IGF1 in myocardial development is still less reported; in our present study, we developed models for IGF1 knockdown in cardiomyocyte cultures (siRNA) in vitro and IGF1 knockdown in a chicken embryo model in vivo to detect the effect of IGF1 suppression on energy metabolism, insulin pathways, and myocardial development.
2. Materials and Methods
All procedures used in this study were approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (SRM-11).
2.1. Primary Cardiomyocyte Culture
Twelve-day-old chicken embryos were used to obtain primary cardiomyocytes for culture. Subsequent to surface disinfection (using 75% alcohol), the chest was dissected to collect the apical portion of the pericardium (approximately 1/3 of the heart), which was immediately transferred to phosphate-buffered solution (PBS) (4°C) and washed to remove fat, connective tissue, and blood clots. Subsequently, the myocardial tissue was cut into small pieces and washed 3 times with PBS. After enzymatic digestion with collagenase-II (0.1%) for 15 minutes on a constant temperature magnetic stirrer (37°C, 100 r/min) for acclimatization and centrifugation, an equal volume of Dulbecco's Modified Eagle's Medium (DMEM)/F12w containing 10% fetal bovine serum and 1x mycillin was added to terminate digestion. The pellet was digested until the small tissue fragments were completely digested. All supernatants were collected with 300 mesh and 500 mesh filters. The cell suspension was centrifuged at 600 rpm for 5 minutes and resuspended in DMEM/F12w twice in disposable Petri dishes for differential adhesion (the first was 1 h; the second was 1.5 h). Nonadherent cells (cardiomyocytes) were collected, centrifuged at 600 rpm, counted, plated in 6-well plates at 2 × 105, and incubated at 37°C, 5% CO2, in an adherent culture incubator for 48 h [21].
2.2. Establishment of the IGF1 Knockdown Model In Vitro
Cardiomyocytes attain 80% confluence after approximately 48 h of incubation. In chicken cardiomyocyte primary cultures, which are the same as described in our previous experimental method, IGF1 was knocked down using siRNA (sense 5′-GTTCGTATGTGGAGACAGA-3′, anti-sense 5′-TCTGTCTCCACATACGAAC-3′) subsequent to two washes with Opti-MEM (prewarmed). All cells were randomly divided into two groups: C (control group) and KD (knockdown group). For each group, 3 replicates were prepared with 6 × 105 cardiomyocytes per replicate (n = 3). Cells in the knockdown group were transfected with 3 μL of 20 μM siRNA and 3 μL of Lipofectamine RNAi MAX Reagent (Invitrogen) in 2 mL of Opti-MEM. Cells in the control group were treated with 2 mL of Opti-MEM (Invitrogen), which only contain the same volume of Lipofectamine RNAi MAX Reagent. Approximately 48 h posttransfection, the cells were harvested for analysis.
2.3. Establishment of the IGF1 Silence Model In Vivo
First, 50 μg of nucleic acid was diluted with pure water without endotoxin to a concentration of 1 μg/μL; the final concentration of glucose was 5%, and the final volume was 100 μL. Then, 25 μL of Entranster™ in vivo reagent was diluted with 50 μL of 10% glucose solution and supplemented with pure water. The final concentration of running glucose was 5%, and the final volume was 100 μL of liquid.
The diluted transfection reagent was added to the diluted nucleic acid solution to form a transfection complex, which was then left at room temperature for 15 min. Ninety hatching eggs were randomly divided into two groups (45 per group), viz., the normal group (N) and siRNA group (Si). The subgerminal cavity of each egg was injected with 1 μg siRNA and sealed with a sealing film. All of the eggs were incubated in a constant temperature incubator. The hearts were taken at 6, 8, and 10 days for subsequent experiments.
2.4. Detection of Intracellular ROS Accumulation
Posttransfection, ROS activities were measured using an ROS assay kit (Nanjing Jiancheng Bioengineering Institute, China). First, 10 μM DCFH-DA (2,7-dichlorofurescin diacetate) was added to the culture medium, containing the cell samples to be tested, which was then incubated at a constant temperature (37°C) for 45 min. Then, the medium was discarded, and PBS (37°C preheat) was used to wash the cells three times. Finally, the cells were collected for detecting the activities of ROS at the excitation wavelength of 500 ± 15 nm and emission wavelength of 530 ± 20 nm. The cardiomyocytes were visualized using fluorescence microscopy.
2.5. Determination of Oxidative Stress Markers
Cells were grown on 6-well plates at a density of 3 × 105 mL−1, collected with jets of saline, and centrifuged at 700 × g; then, the supernatant was collected. The hearts of chicken embryos were taken for homogenization in saline solution and centrifuged at 700 g for 20 min, and the supernatants were collected. The hydrogen peroxide (H2O2), glutathione (GSH), glutathione peroxidase (GSH-Px), catalase (CAT), malondialdehyde (MDA), induced nitric oxide synthase (iNOS), superoxide dismutase (SOD), and total antioxidant capability (T-AOC) contents were measured by detection kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer's protocols. The SOD activity was measured at 25°C using autooxidation of pyrogallol in 50 Mm Tris/HCl, pH 8, with 100 mM pyrogallol (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.6. Determination of the mRNA Expression of the Genes Related to IGF1, the PI3K/Akt Pathway, Insulin, and Cardiac Differentiation
Total RNA was isolated from heart tissues of three points in time and cardiomyocytes by using the TRIzol reagent according to the manufacturer's instructions (Roche, Basel, Switzerland). The dried RNA pellets were resuspended in 50 μL of diethyl pyrocarbonate-treated water. The concentration and purity of the total RNA were determined using a spectrophotometer. cDNA was synthesized from 5 μg of the total RNA using oligo-dT primers and Superscript II reverse transcriptase according to the manufacturer's instructions (Promega, Beijing, China). cDNA was diluted at a ratio of 1 : 5 with sterile water and stored at −80°C.
Primer Premier Software (PREMIER Biosoft International, USA) was used to design specific primers for IGF1 and AMP-activated protein kinase (AMPK), phosphatidylinositol 3-kinase (PI3K), c-Jun N-terminal kinase (JNK), threonine-protein kinase (Akt), forkhead box protein (FOXO), insulin-like growth factor 1 receptor (IGF1R), glucose transporter-1 (GLUT1), glucose transporter-3 (GLUT3), glucose transporter-8 (GLUT8), insulin-like growth factor-binding protein-1 (IGFBP1), insulin-like growth factor-binding protein-2 (IGFBP2), insulin-like growth factor-binding protein-3 (IGFBP3), insulin-like growth factor-binding protein-4 (IGFBP4), insulin-like growth factor-binding protein-5 (IGFBP5), insulin-like growth factor-binding protein-7 (IGFBP7), insulin receptor (IR), insulin receptor substrate-1 (IRS1), MyoD, myogenin (MyoG), cardiac transcription factor mesoderm posterior 1 (Mesp1), myogenic factor-5 (MYF5), myogenic factor-6 (MYF6), GATA-binding protein 4 (GATA4), GATA-binding protein 6 (GATA6), NK2 homeobox 5 (Nkx2.5), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) based on known chicken sequences (Table 1). First, general PCR was performed to confirm the specificity of the primers. Quantitative real-time PCR (qPCR) was then performed with a Roche detection system (Applied Biosystems, Foster City, CA). The reactions were conducted in a 20 μL reaction mixture containing 10 μL of 2x SYBR Green I PCR Master Mix (Roche, Basel, Switzerland), 2 μL of cDNA, 0.4 μL of each primer (10 μM), 0.4 μL of 50x ROX reference Dye II, and 6.8 μL of PCR-grade water. The PCR procedure for IGF1 and AMPK, PI3K, JNK, Akt, FOXO, IGF1R, GLUT1, GLUT3, GLUT8, IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP7, IR, IRS1, MyoD, MyoG, Mesp1, MYF5, MYF6, GATA4, GATA6, Nkx2.5, and GAPDH consisted of 95°C for 30 s followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 60°C for 30 s. For each PCR, Dissociation Curve 1.0 Software (Applied Biosystems) was used to analyze the dissociation curves in order to detect and eliminate possible primer dimers and nonspecific amplification.
Table 1.
The primers used in the present study.
| Target gene | Primer sequence (5′-3′) |
|---|---|
| IGF1 | Forward 5′-GCTTTTGTGATTTCTTGAAGGTGAA-3′ |
| Reverse 5′-CATACCCTGTAGGCTTACTGAAGTA-3′ | |
|
| |
| AMPK | Forward 5′-CCATCTGTCATTAGTCTTCTG-3′ |
| Reverse 5′-AGGCTTCGTCATCAATCAT-3′ | |
|
| |
| PI3K | Forward 5′-GTCCTTGAGCCACTGATG-3′ |
| Reverse 5′-TGTTGCCTTACGGTTGTT-3′ | |
|
| |
| Akt | Forward 5′-AGGAGGAAGAGATGATGGAT-3′ |
| Reverse 5′-GAATGGATGCCGTGAGTT-3′ | |
|
| |
| JNK | Forward 5′-CAGATAAGCAGTTAGATGAGAG-3′ |
| Reverse 5′-GACAGATGACGACGAAGAT-3′ | |
|
| |
| FOXO | Forward 5′-CAGCAATGTCAAGGAGAGCA-3′ |
| Reverse 5′-TGAAGAGGTTGTCCGAGTCC-3′ | |
|
| |
| IGF1R | Forward 5′-GCGTGAGAGGATAGAGTTC-3′ |
| Reverse 5′-TGTTGGCGTTGAGGTATG-3′ | |
|
| |
| GLUT1 | Forward 5′-TAGTACTGGAGCAGGTGGCAGA-3′ |
| Reverse 5′-CGGCACAAGAATGGATGAAA-3′ | |
|
| |
| GLUT3 | Forward 5′-TCCCCAGAGCTTCTTACCTCAC-3′ |
| Reverse 5′-CAGCAAAAGCCAAGACATTCAC-3′ | |
|
| |
| GLUT8 | Forward 5′-CCAAATGGGAACAACTCATCAA-3′ |
| Reverse 5′-GGGCAAAACCAGCAACAAA-3′ | |
|
| |
| IGFBP1 | Forward 5′-TGGCTCGGGCTAGCTGGATG-3′ |
| Reverse 5′-ACCAGCACCCAGCGGAATCT-3′ | |
|
| |
| IGFBP2 | Forward 5′-TGTGACAAGCATGGCTTGTACA-3′ |
| Reverse 5′-TCTCCACGCTGCCCATTC-3′ | |
|
| |
| IGFBP3 | Forward 5′-ATGGTCCCTGTCGTAGAG-3′ |
| Reverse 5′-ATCCAGGAAGCGGTTGT-3′ | |
|
| |
| IGFBP4 | Forward 5′-TGGTGCGTGGACCGCAAGAC′-3′ |
| Reverse 5′-AGCGATGGGGGCGTCCCATA-3′ | |
|
| |
| IGFBP5 | Forward 5′-TGTGCCTCTGGCAGGGGGTA-3′ |
| Reverse 5′-CAACACAGCCCACGCTTCCG-3′ | |
|
| |
| IGFBP7 | Forward 5′-TGTGAAGTCATTGGCATCC-3′ |
| Reverse 5′-CCTCTCCTTTGGCATTTGA-3′ | |
|
| |
| IR | Forward 5′-CAAACGGTGACCAAGCCTCA-3′ |
| Reverse 5′-CATCCTGCCCATCAAACTCC-3′ | |
|
| |
| IRS1 | Forward 5′-TCCACCACCACCACCATCAC-3′ |
| Reverse 5′-ACAGCAGCCGCATCCGAAT-3′ | |
|
| |
| MyoD | Forward 5′-CCGCCGATGACTTCTATG-3′ |
| Reverse 5′-GTTGGTGGTCTTCCTCTTG-3′ | |
|
| |
| MyoG | Forward 5′-AGGCTGAAGAAGGTGAAC-3′ |
| Reverse 5′-GCTCGATGTACTGGATGG-3′ | |
|
| |
| Mesp1 | Forward 5′-GGTCATCACCCTCCTACA-3′ |
| Reverse 5′-CCATCTCTGCATCCACAA-3′ | |
|
| |
| MYF5 | Forward 5′-GAGGAGGAGGCTGAAGAA-3′ |
| Reverse 5′-CGGCAGGTGATAGTAGTTC-3′ | |
|
| |
| MYF6 | Forward 5′-GGAGGAGGCTGAAGAAGA-3′ |
| Reverse 5′-CTCTCGATGTAGCTGATGG-3′ | |
|
| |
| GATA4 | Forward 5′-TCAGACAAGGAAGCGTAAG-3′ |
| Reverse 5′-ATGGCAGAGACCGAGAAT-3′ | |
|
| |
| GATA6 | Forward 5′-CCGACCACTTGCTATGAA-3′ |
| Reverse 5′-TTGCTACAGTCATCTGAGTT-3′ | |
|
| |
| Nkx2.5 | Forward 5′-GACAGAGGAAGAGGAGGAA-3′ |
| Reverse 5′-CGTTCGCTAGATGGTCTC-3′ | |
|
| |
| GAPDH | Forward 5′-AGAACATCATCCCAGCGT-3′ |
| Reverse 5′-AGCCTTCACTACCCTCTTG-3′ | |
2.7. Determination of the Protein Expression of the Proteins Related to IGF1, the PI3K/Akt Pathway, Insulin, and Cardiac Differentiation
For total protein extraction, protein lysates were subjected to 15% SDS-polyacrylamide gel electrophoresis under reducing conditions. The separated proteins were then transferred to a nitrocellulose membrane for 2 h at 100 mA in a transfer apparatus containing Tris-glycine buffer and 20% methanol. The membrane was blocked with 5% skim milk for 24 h and incubated overnight with diluted primary antibodies against IGF1 (1 : 500, Proteintech, China), PI3K (1 : 1000, Santa Cruz Biotechnology, USA), Akt (1 : 500, Santa Cruz Biotechnology, USA), P-Akt (1 : 500, Proteintech, China), FOXO (1 : 1000, Santa Cruz Biotechnology, USA), P-FOXO (1 : 1000, Santa Cruz Biotechnology, USA), JNK (1 : 1000, Santa Cruz Biotechnology, USA), P-JNK (1 : 1000, Santa Cruz Biotechnology, USA), 14-3-3 (1 : 1000, Abcam, Cambridge, UK), P-14-3-3 (1 : 1000, Santa Cruz Biotechnology, USA), GLUT3 (1 : 300, Santa Cruz Biotechnology, USA), IGF1Rβ (1 : 500, Proteintech, China), IGFBP2 (1 : 500, Proteintech, China), MyoG (1 : 1000, Abcam, Cambridge, UK), and MyoD (1 : 1000, Abcam, Cambridge, UK) followed by a horseradish peroxidase- (HRP-) conjugated secondary antibody against rabbit (IGF1, PI3K, Akt, P-Akt, FOXO, P-FOXO, JNK, P-JNK, 14-3-3, P-14-3-3, GLUT, IGF1Rβ, IGFBP2, MyoG, and MyoD) IgG (1 : 5000, Santa Cruz Biotechnology, USA). To verify equal loading of the samples, the membrane was incubated with a monoclonal GAPDH antibody (1 : 1500, Santa Cruz Biotechnology, USA), followed by an HRP-conjugated goat anti-mouse IgG (1 : 3000) secondary antibody. The signal was detected with X-ray films (TransGen Biotech Co., Beijing, China). The optical density (OD) of each band was determined using an Image VCD gel imaging system, and the relative abundance of IGF1, PI3K, Akt, P-Akt, FOXO, P-FOXO, JNK, P-JNK, 14-3-3, P-14-3-3, GLUT3, IGF1Rβ, IGFBP2, MyoG, and MyoD proteins was calculated and presented as the ratios of OD of each of these proteins to that of GAPDH.
2.8. Measurement of ATP
Cardiomyocytes were grown on 6-well plates at a density of 2 × 105 cells/mL, gathered with the lysis solution, and centrifuged at 700 × g. The supernatant was collected, resuspended by salt water and incubated for 35 min at 25°C. The level of adenosine triphosphate (ATP) in the cardiomyocytes was measured by using an ATP detection kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's instructions. The detection was carried out using an ultraviolet spectrophotometer (Synergy NEO, BioTek Instruments) with a detection wavelength of 636 nm.
2.9. Histopathological Examination
Cardiomyocytes were grown at a density of 2 × 105 cells/mL and then washed with PBS three times; 4% paraformaldehyde solution was added in a 24-well plate for cell fixing. After 12 h, the 4% paraformaldehyde solution in the 24-well plate was removed and 0.01 M PBS was added; then, the wells were soaked for 5 min × 3 times. Hematoxylin staining solution was added to the wells and immersed for 1 min. The staining solution was removed, and distilled water was added to soak for 5 min. The distilled water was then removed and placed in 1% hydrochloric acid alcohol. After 1-3 s, it was aspirated. Tap water was added to soak for 5 min to return the cells to blue. Then, the tap water was removed, and Yihong dye solution was added to soak for 1 min. The eosin staining solution was aspirated and soaked in distilled water for 1 min. The climbing pieces were removed, and glycerol ethanol was added dropwise. The staining effect was observed under a microscope and photographed under a 200x microscope [22].
The myocardial tissues were rapidly fixed in 10% formaldehyde for at least 24 h and embedded in paraffin for microscopic examination. From the prepared paraffin blocks, sections (5 μm thick) were cut, obtained, and stained with hematoxylin and eosin (H.E.) for light microscopic observation.
2.10. High-Resolution Respirometry of Mitochondrial Function
Cardiomyocytes were grown on 6-well plates at a density of 2 × 105 cells/mL. All cells were randomly divided into two groups: C (control group) and KD (knockdown group). The cells in the knockdown group were transfected with 3 μL of 20 μM siRNA and 3 μL of Lipofectamine RNAi MAX Reagent (Invitrogen) in 2 mL of Opti-MEM. The cells in the control group were treated with 2 mL of Opti-MEM (Invitrogen) containing the same volume of Lipofectamine RNAi MAX Reagent. Approximately 48 h posttransfection, the cells were harvested for analysis, gathered with the lysis solution, and centrifuged at 700 × g. The supernatant was collected and resuspended by the medium for high-resolution respirometry. The mitochondrial respiratory function was analyzed in a two-channel titration injection respirometer (Oxygraph-2k; Oroboros Instruments, Innsbruck, Austria). The cell suspension was transferred separately to oxygraph chambers at a final density of approximately 2×105 cells/mL. After a short stabilization period, the chambers were closed and data were recorded using DatLab software 5.2 (Oroboros Instruments, Innsbruck, Austria).
2.11. Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0 software, and all data was assessed using the unpaired t-test, where P < 0.05 was considered a statistically significant difference.
3. Results
3.1. Development of an IGF1 Knockdown Model in Cells and Chicken Embryos
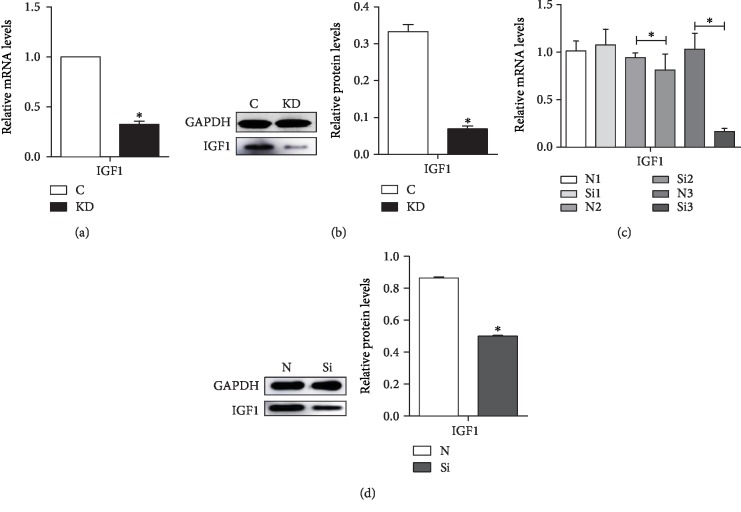
The mRNA and protein levels of IGF1 were significantly decreased (P < 0.05) in the KD group (Figures 1(a) and 1(b)). The results confirmed that we successfully established the model of IGF1 knockdown in vitro.
Figure 1.
The effects of IGF1 knockdown on the mRNA levels (a) and protein levels (b) of the IGF1 gene in cardiomyocytes and the effects of IGF1 silencing on the mRNA levels at 6, 8, and 10 days (c) and on the protein levels (d) of the IGF1 gene in the myocardium. The results were calculated from at least three independent experiments, n = 3. The data are expressed as the means ± SD. C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo. ∗ indicates a significant difference from the corresponding control (P < 0.05).
The mRNA levels of IGF1 were detected at 6, 8, and 10 days, and we found that the expression of IGF1 was significantly decreased at 10 days. For further verification, we took chicken embryos at 10 days to detect the protein level of IGF1. Compared with the N group, the mRNA levels in the Si group decreased (P < 0.05) at 8 days and 10 days (Figure 1(c)). The Si group exhibited significantly decreased protein levels compared with the N group (Figure 1(d)). The results confirmed that we successfully established the IGF1 silence model in vivo.
3.2. Detection of the Antioxidant Capacity in Cells and Chicken Embryos
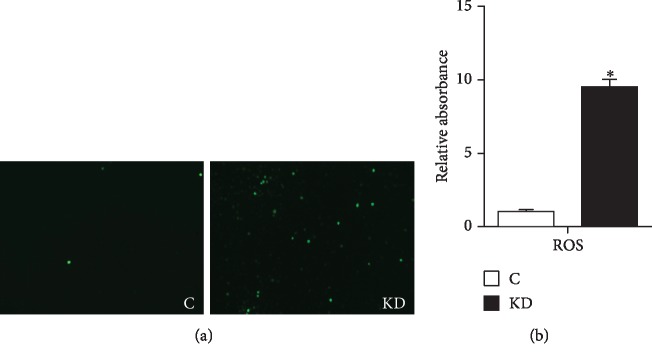
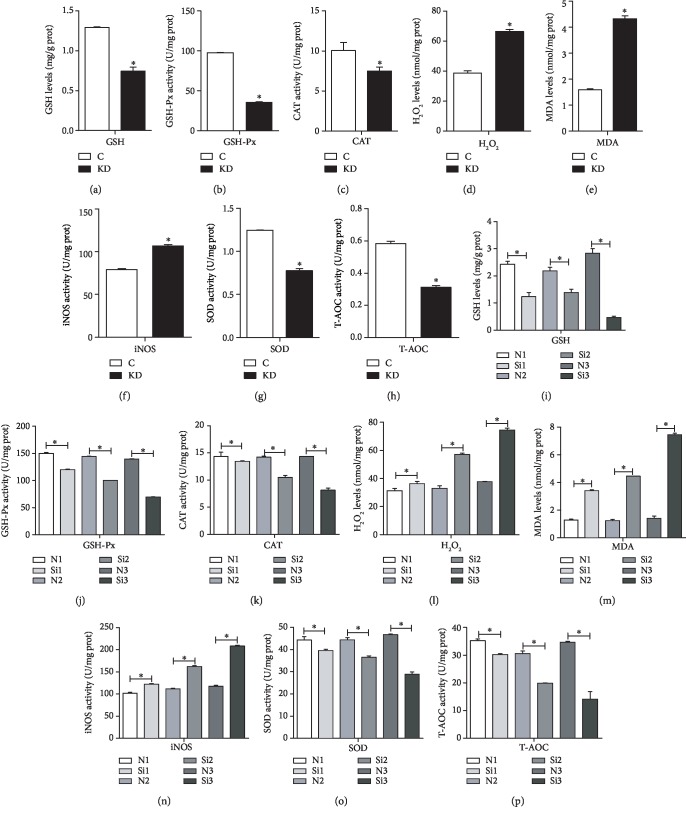
To assess the relationship between oxidative stress and IGF1, the production of ROS, the levels of H2O2, MDA, and T-AOC and the activities of GSH, GSH-Px, SOD, CAT, and iNOS were measured in cardiomyocytes. As presented in Figure 2, the ROS activities were significantly increased (P < 0.05) compared with the C group. The levels of H2O2, MDA, and iNOS were significantly increased in the KD group (P < 0.05) (Figures 3(d)–3(f)). The levels of GSH, GSH-Px, CAT, SOD, and T-AOC in the KD group were significantly lower than those in the C group (P < 0.05) (Figures 3(a)–3(c), 3(g), and 3(h)).
Figure 2.
(a) ROS generation was performed by immunofluorescence using DCFH-DA (green fluorescence, 5 mM) in cells. C indicates the control group; KD indicates the knockdown group. Cardiomyocytes were visualized using fluorescence microscopy. (b) The effects of IGF1 knockdown on the ROS levels in cardiomyocytes were detected by using a fluorescence microplate reader. C indicates the control group; KD indicates the knockdown group. ∗ shows a significant difference from the corresponding control (P < 0.05). n = 3.
Figure 3.
(a–h) Oxidative stress markers of the GSH, GSH-Px, CAT, H2O2, MDA, iNOS, SOD, and T-AOC contents were measured in cardiomyocytes. (i–p) Oxidative stress markers of GSH, GSH-Px, CAT, H2O2, MDA, iNOS, SOD, and T-AOC contents were measured in the myocardium. C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo. ∗ shows a significant difference from the corresponding control (P < 0.05). n = 3.
To further demonstrate the antioxidant capacity of IGF1 silence in vivo, we also studied the antioxidant capacity of the myocardium. As shown in Figure 3, the levels of H2O2, MDA, and iNOS were significantly increased in the Si group (P < 0.05) at 6, 8, and 10 days (Figures 3(l)–3(n)). The levels of GSH, GSH-Px, CAT, SOD, and T-AOC in the Si group were significantly lower than those in the N group (P < 0.05) at 6, 8, and 10 days (Figures 3(i), 3(j), 3(l), 3(o), and 3(p)).
3.3. Protein and mRNA Expression of the PI3K/Akt Pathway-Related Genes in Cells and Chicken Embryos
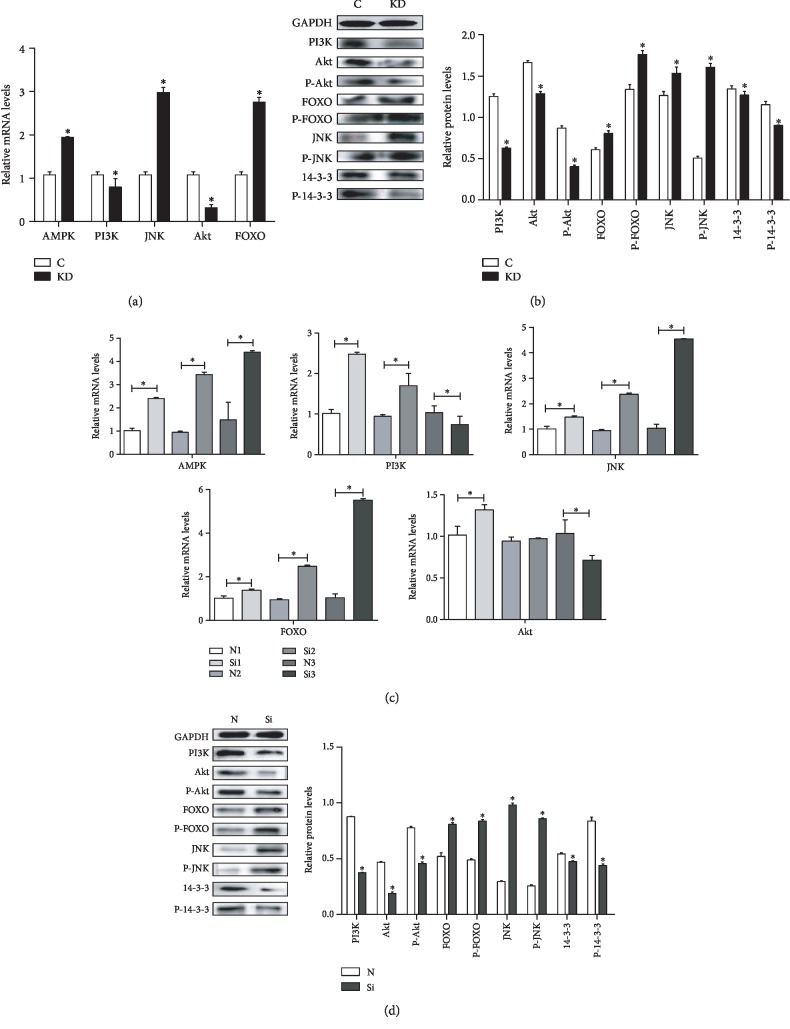
To examine whether IGF1 knockdown changed the expressions of the PI3K/Akt pathway-related genes, we detected the mRNA expression levels of AMPK, PI3K, JNK, Akt, and FOXO as well as the protein expression levels of FOXO, P-FOXO, JNK, P-JNK, PI3K, Akt, P-Akt, 14-3-3, and P-14-3-3. The effects of IGF1 knockdown on the mRNA abundance of PI3K-related genes in chicken cardiomyocytes are shown in Figure 4(a). Compared with the C group, the qPCR results revealed that the mRNA expression of AMPK, JNK, and FOXO was significantly increased (P < 0.05) in the KD group. However, the mRNA expression of PI3K and Akt was decreased (P < 0.05). The results revealed that compared with the C group, the protein expression of FOXO, P-FOXO, JNK, and P-JNK in the KD group was significantly increased (P < 0.05). However, the protein expression of PI3K, Akt, P-Akt, 14-3-3, and P-14-3-3 decreased in the KD group (P < 0.05) (Figure 4(b)).
Figure 4.
The effects of IGF1 knockdown on the mRNA levels (a) and protein levels (b) of PI3K-related genes in cardiomyocytes and the effects of IGF1 silencing on the mRNA levels at 6, 8, and 10 days (c) and on the protein levels (d) of PI3K-related genes in the myocardium. C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo. GAPDH was selected as the reference. ∗ shows a significant difference from the corresponding control (P < 0.05). n = 3.
Moreover, we detected the effects of IGF1 silencing on the mRNA abundance of PI3K-related genes (AMPK, PI3K, JNK, Akt, and FOXO) in the myocardium of chicken embryos as shown in Figure 4(c). The qPCR results revealed that the mRNA expression of AMPK, JNK, and FOXO was significantly increased (P < 0.05) in the Si group at 6, 8, and 10 days. The mRNA expression of PI3K increased at 6 and 8 days but significantly decreased (P < 0.05) at 10 days. The mRNA expression of Akt increased at 6 days, showed no significant change at 8 days, and significantly decreased (P < 0.05) at 10 days. A western blot analysis was performed to determine the protein expression of PI3K-related genes. The results revealed that compared with the N group, the protein expression of FOXO, P-FOXO, JNK, and P-JNK in the Si group was significantly increased (P < 0.05). However, the protein expression of PI3K, Akt, P-Akt, 14-3-3, and P-14-3-3 decreased in the Si group (P < 0.05) (Figure 4(d)).
3.4. Protein and mRNA Expression of Insulin-Related Genes in Cells and Chicken Embryos
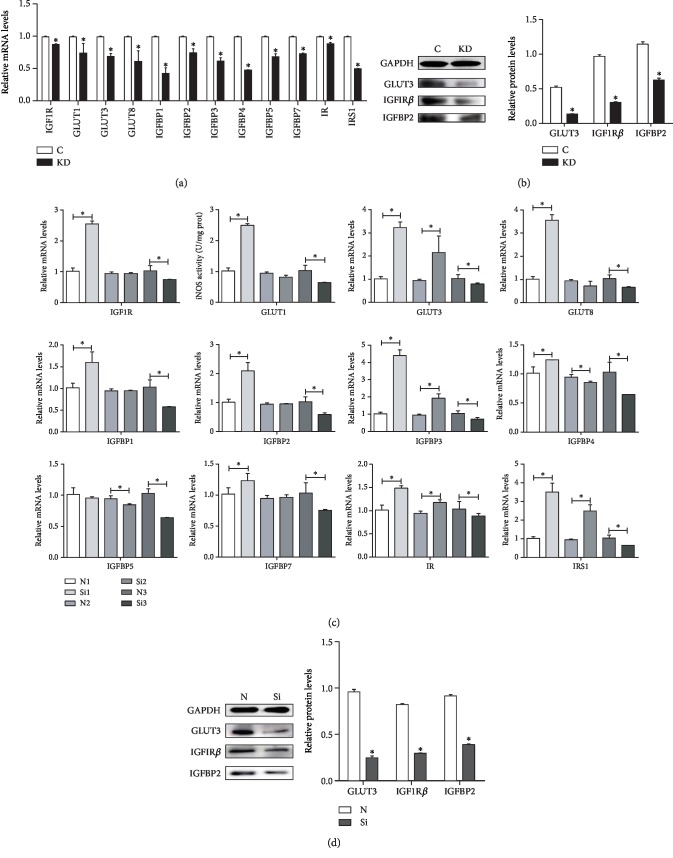
We also examined the effects of IGF1 knockdown on the mRNA abundance of insulin-related genes (IGF1R, GLUT1, GLUT3, GLUT8, IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP7, IR, and IRS1) in chicken cardiomyocytes (Figure 5(a)); the qPCR results revealed that the mRNA expression of IGF1R, GLUT1, GLUT3, GLUT8, IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP7, IR, and IRS1 was significantly decreased (P < 0.05) in the KD group. A western blot analysis was performed to determine the protein expression of insulin-related genes. The results revealed that compared with the C group, the protein expression of GLUT3, IGF1Rβ, and IGFBP2 in the KD group was significantly decreased (P < 0.05) (Figure 5(b)).
Figure 5.
The effects of IGF1 knockdown on the mRNA levels (a) and protein levels (b) of insulin-related genes in cardiomyocytes and the effects of IGF1 silencing on the mRNA levels at 6, 8, and 10 days (c) and on the protein levels (d) of insulin-related genes in the myocardium. C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo. GAPDH was selected as the reference. ∗ shows a significant difference from the corresponding control (P < 0.05). n = 3.
Furthermore, we examined the effects of IGF1 knockdown on the mRNA abundance of insulin-related genes (IGF1R, GLUT1, GLUT3, GLUT8, IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP7, IR, and IRS1) in the myocardium of chicken embryos as shown in Figure 5(c). The qPCR results revealed that the mRNA expression of IGF1R, GLUT1, GLUT8, IGFBP1, IGFBP2, and IGFBP7 increased in the Si group at 6 days, showed no significant change at 8 days, and significantly decreased (P < 0.05) at 10 days. The mRNA expression of GLUT3, IGFBP3, IR, and IRS1 increased at 6 and 8 days, but significantly decreased (P < 0.05) at 10 days. The mRNA expression of IGFBP4 increased at 6 days but significantly decreased at 8 and 10 days. The mRNA expression of IGFBP5 significantly decreased (P < 0.05) at 8 and 10 days. A western blot analysis was performed to determine the protein expression of insulin-related genes. The results revealed that compared with the N group, the protein expression of GLUT3, IGF1Rβ, and IGFBP2 in the Si group was significantly decreased (P < 0.05) (Figure 5(d)).
3.5. The Oxygen Consumption Rate and ATP Content in Myocardial Cells
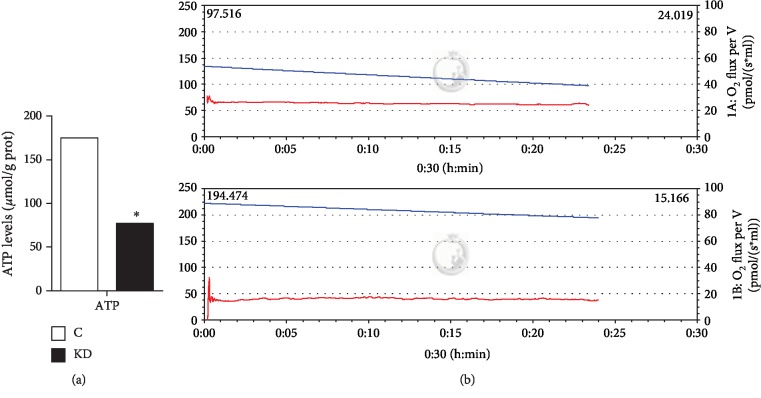
To assess the effect of IGF1 knockdown on energy metabolism, the oxygen consumption rate and ATP content were detected. The results revealed that IGF1 knockdown significantly decreased the ATP content (Figure 6(a)). The results of the oxygen consumption rate revealed that the oxygen consumption rate of the IGF1 knockdown group was 15.166 while the oxygen consumption rate of the control group was 24.019, indicating that IGF1 knockdown significantly decreased the myocardial oxygen consumption rate under the same conditions and the same initial oxygen concentration (Figure 6(b)).
Figure 6.
(a) The level of ATP was determined to investigate the function of energy metabolism. The data are represented as the means ± SD. Samples with an asterisk (∗) represent significant differences (P < 0.05), n = 3. The myocardial oxygen consumption rate results are shown in (b). The red curve indicates the oxygen consumption, and the blue curve indicates the oxygen concentration. C indicates the control group; KD indicates the knockdown group.
The instantaneous oxygen consumption rates of the cardiomyocytes for different groups are provided in Table 2.
Table 2.
Oxygen concentration and the oxygen consumption rate.
| Time (min) | 1A: O2 concentration (nmol/mL) | 1A: O2 flux per V (pmol/(s∗mL)) | 1B: O2 concentration (nmol/mL) | 1B: O2 flux per V (pmol/(s∗mL)) |
|---|---|---|---|---|
| 0.03 | 134.4705 | 222.2083 | ||
| 0.07 | 134.361 | 222.1058 | ||
| 0.1 | 134.3131 | 222.0585 | ||
| 0.13 | 134.2618 | 222.1088 | ||
| 0.17 | 134.1943 | 31.2253 | 222.157 | 1.3718 |
| 0.2 | 134.1481 | 25.9247 | 222.0753 | -5.4236 |
| 0.23 | 134.0865 | 26.9524 | 221.9699 | 6.9917 |
| 0.27 | 134.0053 | 29.6908 | 221.9236 | 24.3313 |
| 0.3 | 133.9386 | 31.3172 | 221.8753 | 32.2136 |
| 0.33 | 133.8958 | 29.6935 | 221.8517 | 14.5802 |
| 0.37 | 133.8531 | 28.1554 | 221.8585 | 13.5949 |
| 0.4 | 133.8078 | 27.8145 | 221.8034 | 15.271 |
| 0.43 | 133.7513 | 27.2173 | 221.7768 | 17.5375 |
| 0.47 | 133.694 | 26.4491 | 221.7354 | 17.44 |
| 0.5 | 133.6496 | 27.3053 | 221.6911 | 14.1902 |
| 0.53 | 133.5888 | 26.5373 | 221.6527 | 14.0926 |
| 0.57 | 133.5316 | 26.3677 | 221.6044 | 15.079 |
| 0.6 | 133.4914 | 26.0266 | 221.5699 | 16.3605 |
| 0.63 | 133.4358 | 25.6004 | 221.5187 | 16.5588 |
| 0.67 | 133.3802 | 26.457 | 221.499 | 14.8846 |
| 0.7 | 133.3238 | 25.9453 | 221.4684 | 14.7868 |
| 0.73 | 133.2365 | 26.033 | 221.431 | 15.2803 |
| 0.77 | 133.1827 | 26.1198 | 221.3995 | 15.7737 |
| 0.8 | 133.1467 | 25.9497 | 221.365 | 15.676 |
| 0.83 | 133.1014 | 26.4639 | 221.3246 | 14.7904 |
| 0.87 | 133.0279 | 26.2092 | 221.2931 | 14.6927 |
| 0.9 | 132.9774 | 26.2105 | 221.2497 | 14.9893 |
| 0.93 | 132.9338 | 26.1261 | 221.2143 | 15.2857 |
| 0.97 | 132.8663 | 26.0422 | 221.1739 | 15.2868 |
| 1 | 132.8064 | 26.4713 | 221.1473 | 14.6963 |
| 1.03 | 132.7414 | 26.3019 | 221.1256 | 14.5984 |
| 1.07 | 132.6687 | 26.3892 | 221.0951 | 14.6976 |
| 1.1 | 132.6345 | 26.3901 | 221.0586 | 14.8956 |
| 1.13 | 132.5969 | 26.3055 | 221.0182 | 14.7981 |
| 1.17 | 132.5524 | 26.4776 | 220.9788 | 14.5035 |
| 1.2 | 132.4772 | 26.3085 | 220.9493 | 14.4057 |
| 1.23 | 132.4361 | 26.3095 | 220.9168 | 14.5051 |
| 1.27 | 132.3746 | 26.3111 | 220.8714 | 14.6047 |
| 1.3 | 132.3147 | 26.2271 | 220.836 | 14.6056 |
| 1.33 | 132.2617 | 26.3994 | 220.7926 | 14.4097 |
| 1.37 | 132.189 | 26.3157 | 220.7394 | 14.411 |
| 1.4 | 132.124 | 26.4028 | 220.7089 | 14.5103 |
| 1.43 | 132.089 | 26.4037 | 220.6685 | 14.7083 |
| 1.47 | 132.024 | 26.3198 | 220.6439 | 14.6104 |
| 1.5 | 131.9581 | 26.407 | 220.6064 | 14.5128 |
| 1.53 | 131.9094 | 26.4082 | 220.5749 | 14.3166 |
| 1.57 | 131.8777 | 26.409 | 220.5385 | 14.3175 |
| 1.6 | 131.8495 | 26.3242 | 220.4872 | 14.4173 |
| 1.63 | 131.7649 | 26.4118 | 220.4695 | 14.4177 |
| 1.67 | 131.6828 | 26.4994 | 220.4242 | 14.5174 |
| 1.7 | 131.6366 | 26.5005 | 220.3641 | 14.5189 |
| 1.73 | 131.5896 | 26.5017 | 220.3326 | 14.5197 |
| 1.77 | 131.5374 | 26.503 | 220.3099 | 14.5202 |
| 1.8 | 131.5041 | 26.5039 | 220.2744 | 14.5211 |
| 1.83 | 131.4545 | 26.4196 | 220.2311 | 14.5222 |
| 1.87 | 131.3698 | 26.4217 | 220.2134 | 14.5227 |
| 1.9 | 131.2988 | 26.4235 | 220.1651 | 14.6224 |
| 1.93 | 131.2518 | 26.4247 | 220.1099 | 14.6238 |
| 1.97 | 131.1979 | 26.426 | 220.0469 | 14.8224 |
| 2 | 131.1304 | 26.4277 | 220.0232 | 14.9215 |
| 2.03 | 131.0833 | 26.4289 | 219.9888 | 14.9223 |
| 2.07 | 131.0269 | 26.4303 | 219.9523 | 15.0217 |
| 2.1 | 130.9645 | 26.5173 | 219.909 | 15.1213 |
| 2.13 | 130.9021 | 26.5189 | 219.8597 | 15.2211 |
| 2.17 | 130.8559 | 26.5201 | 219.8272 | 15.3204 |
| 2.2 | 130.802 | 26.6069 | 219.7829 | 15.3215 |
| 2.23 | 130.7396 | 26.6085 | 219.7464 | 15.421 |
| 2.27 | 130.6797 | 26.61 | 219.7228 | 15.5201 |
| 2.3 | 130.6318 | 26.6112 | 219.6735 | 15.6198 |
| 2.33 | 130.5899 | 26.6122 | 219.639 | 15.7192 |
| 2.37 | 130.5532 | 26.6131 | 219.71 | 15.5204 |
| 2.4 | 130.5147 | 26.6141 | 219.71 | 15.3233 |
| 2.43 | 130.4591 | 26.53 | 219.6095 | 15.1288 |
| 2.47 | 130.3608 | 26.5324 | 219.5297 | 15.1308 |
| 2.5 | 130.3129 | 26.5336 | 219.4824 | 15.132 |
| 2.53 | 130.2658 | 26.4493 | 219.441 | 15.133 |
| 2.57 | 130.2085 | 26.4507 | 219.3996 | 15.1341 |
| 2.6 | 130.1632 | 26.3664 | 219.3977 | 15.0356 |
| 2.63 | 130.1119 | 26.3676 | 219.3455 | 15.0369 |
| 2.67 | 130.0546 | 26.2836 | 219.2676 | 15.0389 |
| 2.7 | 129.9785 | 26.371 | 219.2115 | 15.1388 |
| 2.73 | 129.9281 | 26.3722 | 219.1632 | 15.2385 |
| 2.77 | 129.887 | 26.3733 | 219.114 | 15.4368 |
| 2.8 | 129.8391 | 26.2889 | 219.0834 | 15.536 |
| 2.83 | 129.781 | 26.2904 | 219.048 | 15.6354 |
| 2.87 | 129.7194 | 26.2919 | 219.0145 | 15.6363 |
| 2.9 | 129.6715 | 26.2931 | 218.979 | 15.7357 |
| 2.93 | 129.6159 | 26.209 | 218.9317 | 15.7369 |
| 2.97 | 129.5441 | 26.2108 | 218.8736 | 15.8368 |
| 3 | 129.4774 | 26.2125 | 218.8726 | 15.8369 |
| 3.03 | 129.4099 | 26.2997 | 218.8539 | 15.9358 |
| 3.07 | 129.368 | 26.3007 | 218.8224 | 15.9366 |
| 3.1 | 129.3235 | 26.3018 | 218.7642 | 15.9381 |
| 3.13 | 129.2585 | 26.3035 | 218.7248 | 15.9391 |
| 3.17 | 129.2055 | 26.2193 | 218.6795 | 15.9402 |
| 3.2 | 129.1627 | 26.2203 | 218.6342 | 15.9413 |
| 3.23 | 129.1157 | 26.2215 | 218.5977 | 15.9422 |
| 3.27 | 129.0456 | 26.2233 | 218.5652 | 15.943 |
| 3.3 | 128.9874 | 26.2247 | 218.5278 | 16.0425 |
| 3.33 | 128.9421 | 26.2259 | 218.4579 | 16.1428 |
| 3.37 | 128.9062 | 26.2268 | 218.4214 | 16.2422 |
| 3.4 | 128.8455 | 26.2283 | 218.3987 | 16.2428 |
| 3.43 | 128.7796 | 26.2299 | 218.3524 | 16.3424 |
| 3.47 | 128.7463 | 26.2308 | 218.316 | 16.4418 |
| 3.5 | 128.6693 | 26.2327 | 218.2786 | 16.5413 |
| 3.53 | 128.5992 | 26.3199 | 218.2559 | 16.5419 |
| 3.57 | 128.5308 | 26.4072 | 218.2234 | 16.6412 |
| 3.6 | 128.4803 | 26.4939 | 218.18 | 16.6423 |
| 3.63 | 128.4231 | 26.5809 | 218.1288 | 16.7421 |
| 3.67 | 128.3837 | 26.5819 | 218.0855 | 16.8417 |
| 3.7 | 128.3162 | 26.5836 | 218.0559 | 16.6454 |
| 3.73 | 128.2469 | 26.5853 | 218.0185 | 16.3508 |
| 3.77 | 128.1947 | 26.5866 | 217.9643 | 16.2536 |
| 3.8 | 128.1494 | 26.6732 | 217.9436 | 16.2541 |
| 3.83 | 128.1032 | 26.6744 | 217.9298 | 16.0574 |
| 3.87 | 128.0656 | 26.6753 | 217.8875 | 15.96 |
| 3.9 | 128.0075 | 26.5913 | 217.8421 | 15.9611 |
| 3.93 | 127.9288 | 26.5932 | 217.7978 | 15.7652 |
| 3.97 | 127.8724 | 26.5947 | 217.7387 | 15.6682 |
| 4 | 127.845 | 26.5953 | 217.6865 | 15.768 |
| 4.03 | 127.7877 | 26.5968 | 217.6372 | 15.7692 |
| 4.07 | 127.7099 | 26.5987 | 217.6225 | 15.8681 |
| 4.1 | 127.6654 | 26.5998 | 217.6008 | 15.8686 |
| 4.13 | 127.6081 | 26.6013 | 217.5614 | 15.8696 |
| 4.17 | 127.5363 | 26.6031 | 217.5259 | 15.8705 |
| 4.2 | 127.4713 | 26.6047 | 217.4767 | 15.8717 |
| 4.23 | 127.408 | 26.6063 | 217.4402 | 15.8727 |
| 4.27 | 127.3618 | 26.6074 | 217.3969 | 15.8737 |
| 4.3 | 127.3105 | 26.6942 | 217.3565 | 15.9733 |
| 4.33 | 127.2763 | 26.6951 | 217.325 | 15.9741 |
| 4.37 | 127.2233 | 26.6964 | 217.2836 | 15.9751 |
| 4.4 | 127.1626 | 26.6979 | 217.2422 | 15.8776 |
| 4.43 | 127.1216 | 26.6989 | 217.1742 | 15.8793 |
| 4.47 | 127.054 | 26.7006 | 217.1378 | 15.9787 |
| 4.5 | 126.9813 | 26.7024 | 217.0875 | 15.98 |
| 4.53 | 126.942 | 26.7034 | 217.0294 | 16.08 |
| 4.57 | 126.8941 | 26.7046 | 217.0038 | 16.1791 |
| 4.6 | 126.8342 | 26.6206 | 216.9585 | 16.2788 |
| 4.63 | 126.7932 | 26.6216 | 216.8984 | 16.3788 |
| 4.67 | 126.7145 | 26.6236 | 216.8846 | 16.4776 |
| 4.7 | 126.6461 | 26.6253 | 216.858 | 16.4783 |
| 4.73 | 126.5948 | 26.5411 | 216.8235 | 16.5777 |
| 4.77 | 126.5401 | 26.5424 | 216.7762 | 16.5789 |
| 4.8 | 126.4862 | 26.5438 | 216.7457 | 16.6781 |
| 4.83 | 126.4529 | 26.4591 | 216.7033 | 16.6792 |
| 4.87 | 126.3981 | 26.4605 | 216.6708 | 16.68 |
| 4.9 | 126.3494 | 26.4617 | 216.6294 | 16.681 |
| 4.93 | 126.2955 | 26.3775 | 216.5881 | 16.6821 |
| 4.97 | 126.2314 | 26.3791 | 216.5703 | 16.6825 |
| 5 | 126.1527 | 26.3811 | 216.5102 | 16.684 |
| 5.03 | 126.0963 | 26.468 | 216.4748 | 16.6849 |
| 5.07 | 126.0638 | 26.4688 | 216.4482 | 16.6856 |
| 5.1 | 126.0159 | 26.47 | 216.4009 | 16.6868 |
| 5.13 | 125.9492 | 26.4717 | 216.3457 | 16.6881 |
| 5.17 | 125.9039 | 26.4728 | 216.3388 | 16.5898 |
| 5.2 | 125.844 | 26.3888 | 216.4088 | 16.2925 |
| 5.23 | 125.7901 | 26.3902 | 216.3132 | 16.0979 |
| 5.27 | 125.726 | 26.3918 | 216.2078 | 16.1005 |
| 5.3 | 125.6944 | 26.3071 | 216.1861 | 16.0025 |
| 5.33 | 125.6525 | 26.2226 | 216.1142 | 16.1028 |
| 5.37 | 125.5875 | 26.1387 | 216.0659 | 16.2026 |
| 5.4 | 125.5413 | 26.1399 | 215.997 | 16.3028 |
| 5.43 | 125.4806 | 26.0559 | 216.0039 | 16.2041 |
| 5.47 | 125.4224 | 25.9718 | 215.9891 | 16.106 |
| 5.5 | 125.3745 | 25.973 | 215.9359 | 16.1073 |
| 5.53 | 125.3335 | 25.974 | 215.8955 | 16.0098 |
| 5.57 | 125.2753 | 25.9755 | 215.8601 | 16.0107 |
| 5.6 | 125.2163 | 25.977 | 215.8039 | 16.0121 |
| 5.63 | 125.1462 | 25.9787 | 215.7743 | 15.9143 |
| 5.67 | 125.0855 | 25.9802 | 215.7409 | 15.9151 |
| 5.7 | 125.0393 | 25.8959 | 215.6946 | 15.8178 |
| 5.73 | 124.9889 | 25.8971 | 215.6443 | 15.819 |
| 5.77 | 124.9119 | 25.8991 | 215.5921 | 15.9189 |
| 5.8 | 124.8709 | 25.9001 | 215.5665 | 15.9195 |
| 5.83 | 124.8264 | 25.9012 | 215.5192 | 15.9207 |
| 5.87 | 124.7777 | 25.9024 | 215.4818 | 16.0201 |
| 5.9 | 124.7204 | 25.8183 | 215.464 | 16.1191 |
| 5.93 | 124.6631 | 25.8198 | 215.4227 | 16.1201 |
| 5.97 | 124.5827 | 25.8218 | 215.3586 | 16.2202 |
| 6 | 124.5117 | 25.9091 | 215.3468 | 16.3191 |
| 6.03 | 124.4827 | 25.9953 | 215.3212 | 16.3197 |
| 6.07 | 124.4408 | 25.9963 | 215.2867 | 16.3206 |
| 6.1 | 124.4091 | 25.9971 | 215.2256 | 16.3221 |
| 6.13 | 124.3552 | 25.9985 | 215.1675 | 16.422 |
| 6.17 | 124.3014 | 25.9143 | 215.1261 | 16.4231 |
| 6.2 | 124.2501 | 25.9156 | 215.0995 | 16.4237 |
| 6.23 | 124.1782 | 25.8319 | 215.0641 | 16.5231 |
| 6.27 | 124.1115 | 25.8335 | 215.001 | 16.5247 |
| 6.3 | 124.0619 | 25.8348 | 214.9695 | 16.624 |
| 6.33 | 124.014 | 25.9215 | 214.9439 | 16.6247 |
| 6.37 | 123.9456 | 26.0087 | 214.9252 | 16.6251 |
| 6.4 | 123.8875 | 26.0102 | 214.8729 | 16.6264 |
| 6.43 | 123.8302 | 26.0116 | 214.8158 | 16.6279 |
| 6.47 | 123.7609 | 26.0989 | 214.7577 | 16.7278 |
| 6.5 | 123.7301 | 26.1851 | 214.7321 | 16.7285 |
| 6.53 | 123.684 | 26.1863 | 214.7045 | 16.5321 |
| 6.57 | 123.6215 | 26.1878 | 214.672 | 16.3359 |
| 6.6 | 123.5685 | 26.2747 | 214.6276 | 16.337 |
| 6.63 | 123.5138 | 26.2761 | 214.5715 | 16.3384 |
| 6.67 | 123.4497 | 26.2777 | 214.535 | 16.3393 |
| 6.7 | 123.4009 | 26.2789 | 214.4897 | 16.439 |
| 6.73 | 123.3778 | 26.1939 | 214.4395 | 16.5388 |
| 6.77 | 123.3077 | 26.1957 | 214.3794 | 16.6388 |
| 6.8 | 123.2479 | 26.1972 | 214.339 | 16.7383 |
| 6.83 | 123.2119 | 26.1126 | 214.3065 | 16.7391 |
| 6.87 | 123.1401 | 26.1144 | 214.2681 | 16.8386 |
| 6.9 | 123.1025 | 26.0298 | 214.2287 | 16.8396 |
| 6.93 | 123.0332 | 26.0315 | 214.1705 | 16.9395 |
| 6.97 | 122.987 | 25.9472 | 214.1252 | 17.0392 |
| 7 | 122.9255 | 25.9487 | 214.0888 | 17.0401 |
| 7.03 | 122.875 | 25.95 | 214.0799 | 17.0403 |
| 7.07 | 122.8015 | 25.9518 | 214.0504 | 17.0411 |
| 7.1 | 122.7596 | 26.0384 | 214.01 | 17.0421 |
| 7.13 | 122.7339 | 25.9535 | 213.9587 | 17.1419 |
| 7.17 | 122.663 | 25.9553 | 213.8967 | 17.2419 |
| 7.2 | 122.6134 | 25.9565 | 213.8632 | 17.2428 |
| 7.23 | 122.5544 | 25.8725 | 213.8346 | 17.2435 |
| 7.27 | 122.5013 | 25.8738 | 213.8248 | 17.1452 |
| 7.3 | 122.4304 | 25.9611 | 213.7834 | 17.1462 |
| 7.33 | 122.3731 | 26.048 | 213.7528 | 17.0485 |
| 7.37 | 122.3132 | 26.0495 | 213.7213 | 16.9508 |
| 7.4 | 122.279 | 26.0504 | 213.6662 | 16.8536 |
| 7.43 | 122.2243 | 26.0518 | 213.6041 | 16.8552 |
| 7.47 | 122.1619 | 25.9678 | 213.5509 | 16.8565 |
| 7.5 | 122.0986 | 25.9694 | 213.4987 | 16.9563 |
| 7.53 | 122.0456 | 25.9707 | 213.4484 | 16.9576 |
| 7.57 | 122.0139 | 25.9715 | 213.4031 | 16.9587 |
| 7.6 | 121.9806 | 25.8868 | 213.3824 | 17.0578 |
| 7.63 | 121.9173 | 25.8029 | 213.3529 | 17.0585 |
| 7.67 | 121.8626 | 25.8043 | 213.3036 | 17.0597 |
| 7.7 | 121.8078 | 25.8056 | 213.2652 | 16.9622 |
| 7.73 | 121.7326 | 25.8075 | 213.2327 | 16.963 |
| 7.77 | 121.6873 | 25.8086 | 213.213 | 16.865 |
| 7.8 | 121.6248 | 25.8957 | 213.1834 | 16.7672 |
| 7.83 | 121.5855 | 25.8967 | 213.1174 | 16.7688 |
| 7.87 | 121.547 | 25.8122 | 213.0642 | 16.7702 |
| 7.9 | 121.4957 | 25.8134 | 213.0258 | 16.7711 |
| 7.93 | 121.4495 | 25.8146 | 212.9894 | 16.6735 |
| 7.97 | 121.3803 | 25.7308 | 212.949 | 16.6745 |
| 8 | 121.3161 | 25.8179 | 212.9175 | 16.5768 |
| 8.03 | 121.2648 | 25.8192 | 212.9027 | 16.5772 |
| 8.07 | 121.2092 | 25.8206 | 212.8455 | 16.4801 |
| 8.1 | 121.1742 | 25.736 | 212.8081 | 16.481 |
| 8.13 | 121.1349 | 25.6514 | 212.7677 | 16.5806 |
| 8.17 | 121.0835 | 25.5672 | 212.7234 | 16.5817 |
| 8.2 | 121.0117 | 25.569 | 212.6643 | 16.5831 |
| 8.23 | 120.9604 | 25.4848 | 212.614 | 16.6829 |
| 8.27 | 120.8852 | 25.5722 | 212.5707 | 16.7825 |
| 8.3 | 120.8227 | 25.5737 | 212.5559 | 16.7829 |
| 8.33 | 120.7919 | 25.5745 | 212.5165 | 16.8824 |
| 8.37 | 120.7458 | 25.4901 | 212.4958 | 16.7844 |
| 8.4 | 120.6979 | 25.4913 | 212.4791 | 16.6863 |
| 8.43 | 120.6346 | 25.4929 | 212.4111 | 16.688 |
| 8.47 | 120.5679 | 25.4946 | 212.4022 | 16.5897 |
| 8.5 | 120.5183 | 25.4958 | 212.4633 | 16.3911 |
| 8.53 | 120.4773 | 25.4968 | 212.3402 | 16.2957 |
| 8.57 | 120.4277 | 25.4126 | 212.2249 | 16.3971 |
| 8.6 | 120.355 | 25.4144 | 212.1825 | 16.3982 |
| 8.63 | 120.3105 | 25.4155 | 212.1116 | 16.4984 |
| 8.67 | 120.2857 | 25.4161 | 212.0653 | 16.4996 |
| 8.7 | 120.237 | 25.4173 | 212.0722 | 16.4009 |
| 8.73 | 120.166 | 25.4191 | 212.0702 | 16.3025 |
| 8.77 | 120.1292 | 25.3345 | 212.018 | 16.3038 |
| 8.8 | 120.0754 | 25.3359 | 211.955 | 16.3053 |
| 8.83 | 119.9967 | 25.4233 | 211.9087 | 16.405 |
| 8.87 | 119.9497 | 25.4245 | 211.8781 | 16.5043 |
| 8.9 | 119.9009 | 25.4257 | 211.8998 | 16.4052 |
| 8.93 | 119.8265 | 25.4276 | 211.8703 | 16.3075 |
| 8.97 | 119.7752 | 25.4289 | 211.7816 | 16.3097 |
| 9 | 119.7059 | 25.5161 | 211.7107 | 16.41 |
| 9.03 | 119.6717 | 25.517 | 211.6584 | 16.5098 |
| 9.07 | 119.617 | 25.6039 | 211.5944 | 16.6099 |
| 9.1 | 119.5503 | 25.6055 | 211.5461 | 16.7096 |
| 9.13 | 119.4981 | 25.6923 | 211.5245 | 16.8087 |
| 9.17 | 119.434 | 25.7795 | 211.4782 | 16.8098 |
| 9.2 | 119.3853 | 25.7807 | 211.4289 | 17.0081 |
| 9.23 | 119.3348 | 25.8675 | 211.3816 | 17.1078 |
| 9.27 | 119.2698 | 25.8691 | 211.3787 | 17.2064 |
| 9.3 | 119.2356 | 25.8699 | 211.3639 | 17.2067 |
| 9.33 | 119.2006 | 25.8708 | 211.3087 | 17.2081 |
| 9.37 | 119.1458 | 25.8722 | 211.354 | 17.01 |
| 9.4 | 119.1022 | 25.8733 | 211.3797 | 16.8123 |
| 9.43 | 119.0672 | 25.7886 | 211.2358 | 16.7174 |
| 9.47 | 119.0167 | 25.7044 | 211.1373 | 16.7198 |
| 9.5 | 118.9432 | 25.6207 | 211.0743 | 16.8199 |
| 9.53 | 118.8842 | 25.6222 | 211.0181 | 17.0183 |
| 9.57 | 118.8551 | 25.5374 | 211.0181 | 17.1169 |
| 9.6 | 118.8012 | 25.5387 | 210.9698 | 17.2166 |
| 9.63 | 118.726 | 25.5406 | 210.9294 | 17.3161 |
| 9.67 | 118.6618 | 25.6277 | 210.8605 | 17.4163 |
| 9.7 | 118.6148 | 25.5434 | 210.8221 | 17.5158 |
| 9.73 | 118.5567 | 25.5449 | 210.7679 | 17.5172 |
| 9.77 | 118.5036 | 25.5462 | 210.7699 | 17.5171 |
| 9.8 | 118.4224 | 25.6337 | 210.8802 | 17.2188 |
| 9.83 | 118.3933 | 25.72 | 210.7452 | 16.9267 |
| 9.87 | 118.3591 | 25.6353 | 210.6369 | 16.8309 |
| 9.9 | 118.2958 | 25.6369 | 210.5807 | 16.9308 |
| 9.93 | 118.2266 | 25.7241 | 210.5226 | 17.0307 |
| 9.97 | 118.1958 | 25.7249 | 210.4714 | 17.229 |
| 10 | 118.1376 | 25.6408 | 210.4832 | 17.4258 |
| 10.03 | 118.0855 | 25.6422 | 210.428 | 17.4272 |
| 10.07 | 118.0299 | 25.558 | 210.3935 | 17.428 |
| 10.1 | 117.994 | 25.4734 | 210.3591 | 17.4289 |
| 10.13 | 117.9307 | 25.3895 | 210.3059 | 17.4302 |
| 10.17 | 117.8666 | 25.3911 | 210.294 | 17.4305 |
| 10.2 | 117.8016 | 25.3927 | 210.3728 | 17.2315 |
| 10.23 | 117.7708 | 25.3935 | 210.2822 | 16.9382 |
| 10.27 | 117.7272 | 25.3946 | 210.166 | 16.8426 |
| 10.3 | 117.6562 | 25.3963 | 210.1246 | 16.7451 |
| 10.33 | 117.5972 | 25.3978 | 210.0576 | 16.8453 |
| 10.37 | 117.5716 | 25.3985 | 210.0162 | 16.9449 |
| 10.4 | 117.51 | 25.4 | 210.0113 | 16.945 |
| 10.43 | 117.4536 | 25.4014 | 209.968 | 17.0446 |
| 10.47 | 117.4193 | 25.4023 | 209.9088 | 17.1446 |
| 10.5 | 117.3783 | 25.4033 | 209.8714 | 17.244 |
| 10.53 | 117.2954 | 25.4054 | 209.8419 | 17.2448 |
| 10.57 | 117.2355 | 25.4924 | 209.7877 | 17.4432 |
| 10.6 | 117.1962 | 25.5789 | 209.7512 | 17.4441 |
| 10.63 | 117.1551 | 25.5799 | 209.7059 | 17.4452 |
| 10.67 | 117.1004 | 25.4958 | 209.6665 | 17.4462 |
| 10.7 | 117.0474 | 25.4971 | 209.6143 | 17.349 |
| 10.73 | 116.9721 | 25.499 | 209.6833 | 16.9532 |
| 10.77 | 116.9345 | 25.4999 | 209.6232 | 16.6592 |
| 10.8 | 116.9046 | 25.3296 | 209.4882 | 16.7611 |
| 10.83 | 116.8396 | 25.3313 | 209.4616 | 16.8602 |
| 10.87 | 116.7891 | 25.247 | 209.3897 | 16.9605 |
| 10.9 | 116.7464 | 25.1626 | 209.3463 | 17.0601 |
| 10.93 | 116.6891 | 25.0785 | 209.3286 | 17.1591 |
| 10.97 | 116.6395 | 24.9942 | 209.3444 | 17.0602 |
| 11 | 116.5856 | 24.9956 | 209.2793 | 17.1603 |
| 11.03 | 116.5377 | 24.9967 | 209.2153 | 17.2604 |
| 11.07 | 116.4676 | 24.9985 | 209.1789 | 17.3599 |
| 11.1 | 116.4163 | 24.9998 | 209.1345 | 17.361 |
| 11.13 | 116.3522 | 25.0869 | 209.0971 | 17.1649 |
| 11.17 | 116.3248 | 25.0021 | 209.1651 | 16.8676 |
| 11.2 | 116.2735 | 25.0034 | 209.0853 | 16.7711 |
| 11.23 | 116.2059 | 25.005 | 208.969 | 16.8725 |
| 11.27 | 116.1444 | 25.0921 | 208.9582 | 16.8728 |
| 11.3 | 116.0871 | 25.0935 | 208.9119 | 17.071 |
| 11.33 | 116.0229 | 25.1806 | 208.8528 | 17.0725 |
| 11.37 | 115.9836 | 25.1816 | 208.837 | 17.0729 |
| 11.4 | 115.946 | 25.1826 | 208.7878 | 17.1726 |
| 11.43 | 115.8827 | 25.1841 | 208.7444 | 17.1737 |
| 11.47 | 115.8254 | 25.1856 | 208.6873 | 17.2736 |
| 11.5 | 115.7809 | 25.2722 | 208.6548 | 17.2745 |
| 11.53 | 115.7219 | 25.3592 | 208.6193 | 17.0783 |
| 11.57 | 115.6792 | 25.3603 | 208.5819 | 16.9807 |
| 11.6 | 115.6125 | 25.3619 | 208.6223 | 16.7827 |
| 11.63 | 115.5646 | 25.4486 | 208.637 | 16.5853 |
| 11.67 | 115.5295 | 25.4495 | 208.5366 | 16.4893 |
| 11.7 | 115.4825 | 25.4507 | 208.4577 | 16.4913 |
| 11.73 | 115.4235 | 25.4522 | 208.442 | 16.3932 |
| 11.77 | 115.3713 | 25.4535 | 208.3848 | 16.2961 |
| 11.8 | 115.3294 | 25.369 | 208.311 | 16.3964 |
| 11.83 | 115.2867 | 25.2846 | 208.3159 | 16.2978 |
| 11.87 | 115.2268 | 25.2861 | 208.2676 | 16.299 |
| 11.9 | 115.1704 | 25.2875 | 208.2243 | 16.3001 |
| 11.93 | 115.1088 | 25.3745 | 208.1819 | 16.3011 |
| 11.97 | 115.0558 | 25.2903 | 208.1504 | 16.3019 |
| 12 | 114.9959 | 25.2918 | 208.0982 | 16.3032 |
| 12.03 | 114.9549 | 25.2929 | 208.0578 | 16.3042 |
| 12.07 | 114.8967 | 25.3798 | 208.0282 | 16.108 |
| 12.1 | 114.8258 | 25.3816 | 208.1248 | 15.7115 |
| 12.13 | 114.7727 | 25.3829 | 208.0134 | 15.7143 |
| 12.17 | 114.7326 | 25.4694 | 207.8992 | 15.7171 |
| 12.2 | 114.6787 | 25.4708 | 207.8736 | 15.8163 |
| 12.23 | 114.6334 | 25.3864 | 207.8223 | 15.9161 |
| 12.27 | 114.6043 | 25.3016 | 207.7583 | 16.0162 |
| 12.3 | 114.553 | 25.3029 | 207.7741 | 16.0158 |
| 12.33 | 114.4846 | 25.2191 | 207.7534 | 15.9178 |
| 12.37 | 114.4127 | 25.2209 | 207.6982 | 15.9192 |
| 12.4 | 114.3537 | 25.2224 | 207.6588 | 15.9202 |
| 12.43 | 114.3093 | 25.309 | 207.6095 | 16.0199 |
| 12.47 | 114.2426 | 25.3107 | 207.5268 | 16.1205 |
| 12.5 | 114.2015 | 25.3117 | 207.5051 | 16.0225 |
| 12.53 | 114.1733 | 25.3124 | 207.5603 | 15.8241 |
| 12.57 | 114.1314 | 25.2279 | 207.5573 | 15.7257 |
| 12.6 | 114.0604 | 25.2297 | 207.4293 | 15.7289 |
| 12.63 | 113.9963 | 25.2313 | 207.3593 | 15.7306 |
| 12.67 | 113.9535 | 25.3179 | 207.2973 | 15.9292 |
| 12.7 | 113.9056 | 25.3191 | 207.247 | 16.029 |
| 12.73 | 113.8509 | 25.3205 | 207.2224 | 16.1281 |
| 12.77 | 113.8184 | 25.3213 | 207.246 | 16.226 |
| 12.8 | 113.7663 | 25.2371 | 207.1948 | 16.2273 |
| 12.83 | 113.7141 | 25.2384 | 207.1377 | 16.3273 |
| 12.87 | 113.6551 | 25.2398 | 207.1002 | 16.4267 |
| 12.9 | 113.5833 | 25.2416 | 207.0451 | 16.6251 |
| 12.93 | 113.5285 | 25.3285 | 206.9948 | 16.6264 |
| 12.97 | 113.4841 | 25.3296 | 206.9603 | 16.4302 |
| 13 | 113.4413 | 25.3307 | 206.9298 | 16.431 |
| 13.03 | 113.3883 | 25.332 | 206.8579 | 16.5313 |
| 13.07 | 113.3327 | 25.3334 | 206.8756 | 16.5308 |
| 13.1 | 113.2848 | 25.3346 | 206.9396 | 16.3322 |
| 13.13 | 113.2258 | 25.2506 | 206.8372 | 16.3348 |
| 13.17 | 113.1839 | 25.2516 | 206.7466 | 16.337 |
| 13.2 | 113.1506 | 25.1669 | 206.716 | 16.3378 |
| 13.23 | 113.0719 | 25.0834 | 206.651 | 16.3394 |
| 13.27 | 113.0155 | 25.0848 | 206.5899 | 16.538 |
| 13.3 | 112.9573 | 25.0863 | 206.5939 | 16.5379 |
| 13.33 | 112.918 | 25.0872 | 206.5535 | 16.5389 |
| 13.37 | 112.8684 | 25.0885 | 206.517 | 16.6383 |
| 13.4 | 112.8025 | 25.0901 | 206.4658 | 16.6396 |
| 13.43 | 112.7624 | 25.1766 | 206.4106 | 16.4439 |
| 13.47 | 112.7145 | 25.1778 | 206.3643 | 16.4451 |
| 13.5 | 112.6572 | 25.1793 | 206.3121 | 16.5449 |
| 13.53 | 112.5965 | 25.1808 | 206.2688 | 16.6445 |
| 13.57 | 112.5306 | 25.268 | 206.2363 | 16.7438 |
| 13.6 | 112.493 | 25.1834 | 206.2116 | 16.843 |
| 13.63 | 112.4554 | 25.1843 | 206.1821 | 16.8437 |
| 13.67 | 112.3998 | 25.1002 | 206.1545 | 16.8444 |
| 13.7 | 112.3425 | 25.1871 | 206.122 | 16.7467 |
| 13.73 | 112.2963 | 25.1883 | 206.0875 | 16.7476 |
| 13.77 | 112.2279 | 25.2755 | 206.053 | 16.7484 |
| 13.8 | 112.1638 | 25.3626 | 206.0176 | 16.7493 |
| 13.83 | 112.1219 | 25.3637 | 205.988 | 16.75 |
| 13.87 | 112.0791 | 25.3648 | 205.925 | 16.6531 |
| 13.9 | 112.0124 | 25.3664 | 205.9171 | 16.3578 |
| 13.93 | 111.962 | 25.4532 | 205.986 | 16.0605 |
| 13.97 | 111.9286 | 25.454 | 205.8836 | 15.866 |
| 14 | 111.8696 | 25.4555 | 205.7969 | 15.9667 |
| 14.03 | 111.826 | 25.4566 | 205.7732 | 15.9673 |
| 14.07 | 111.7644 | 25.4581 | 205.7102 | 15.9689 |
| 14.1 | 111.7251 | 25.3736 | 205.659 | 15.9702 |
| 14.13 | 111.6738 | 25.3749 | 205.6639 | 15.8715 |
| 14.17 | 111.6105 | 25.291 | 205.5861 | 15.8735 |
| 14.2 | 111.5566 | 25.2923 | 205.522 | 15.8751 |
| 14.23 | 111.4865 | 25.3796 | 205.4826 | 15.9746 |
| 14.27 | 111.4455 | 25.3806 | 205.4521 | 16.0739 |
| 14.3 | 111.4164 | 25.3813 | 205.4186 | 16.0747 |
| 14.33 | 111.36 | 25.3827 | 205.4018 | 16.0751 |
| 14.37 | 111.3009 | 25.3842 | 205.3506 | 16.1749 |
| 14.4 | 111.2325 | 25.3859 | 205.3201 | 16.1757 |
| 14.43 | 111.1906 | 25.387 | 205.2836 | 15.9795 |
| 14.47 | 111.1479 | 25.388 | 205.3152 | 15.6832 |
| 14.5 | 111.0923 | 25.3039 | 205.324 | 15.486 |
| 14.53 | 111.0324 | 25.3054 | 205.2265 | 15.3899 |
| 14.57 | 110.9657 | 25.3071 | 205.1408 | 15.392 |
| 14.6 | 110.9332 | 25.3079 | 205.0945 | 15.4917 |
| 14.63 | 110.876 | 25.3093 | 205.0373 | 15.4931 |
| 14.67 | 110.811 | 25.3965 | 204.992 | 15.4943 |
| 14.7 | 110.7622 | 25.3977 | 204.9782 | 15.4946 |
| 14.73 | 110.7118 | 25.3989 | 204.9142 | 15.5947 |
| 14.77 | 110.6485 | 25.4005 | 204.8728 | 15.6943 |
| 14.8 | 110.6066 | 25.4016 | 204.8482 | 15.7934 |
| 14.83 | 110.5844 | 25.4021 | 204.8029 | 15.8931 |
| 14.87 | 110.5262 | 25.3181 | 204.7418 | 16.0916 |
| 14.9 | 110.4715 | 25.3194 | 204.7162 | 16.2893 |
| 14.93 | 110.4108 | 25.3209 | 204.6926 | 16.3884 |
| 14.97 | 110.3586 | 25.3223 | 204.6591 | 16.4877 |
| 15 | 110.297 | 25.3238 | 204.6118 | 16.5874 |
| 15.03 | 110.2474 | 25.325 | 204.5763 | 16.5883 |
| 15.07 | 110.203 | 25.3261 | 204.5605 | 16.5887 |
| 15.1 | 110.1371 | 25.3278 | 204.5162 | 16.5898 |
| 15.13 | 110.079 | 25.4148 | 204.4719 | 16.6894 |
| 15.17 | 110.0328 | 25.4159 | 204.4098 | 16.691 |
| 15.2 | 109.9926 | 25.4169 | 204.3803 | 16.7902 |
| 15.23 | 109.9302 | 25.4185 | 204.3744 | 16.6919 |
| 15.27 | 109.8883 | 25.505 | 204.4561 | 16.2958 |
| 15.3 | 109.8609 | 25.4202 | 204.3576 | 16.0027 |
| 15.33 | 109.8122 | 25.3359 | 204.2778 | 15.9062 |
| 15.37 | 109.7386 | 25.3378 | 204.2266 | 15.9075 |
| 15.4 | 109.6762 | 25.3393 | 204.1566 | 15.9092 |
| 15.43 | 109.63 | 25.255 | 204.1133 | 16.0088 |
| 15.47 | 109.5796 | 25.2562 | 204.1172 | 15.8117 |
| 15.5 | 109.5377 | 25.2573 | 204.0798 | 15.8126 |
| 15.53 | 109.4975 | 25.1728 | 204.0148 | 15.9128 |
| 15.57 | 109.4633 | 25.1736 | 203.9606 | 16.0126 |
| 15.6 | 109.3966 | 25.0898 | 203.933 | 16.1118 |
| 15.63 | 109.3188 | 25.0917 | 203.8966 | 16.1127 |
| 15.67 | 109.264 | 25.0931 | 203.867 | 16.1135 |
| 15.7 | 109.2221 | 25.0086 | 203.8246 | 16.2131 |
| 15.73 | 109.1512 | 25.0959 | 203.7872 | 16.214 |
| 15.77 | 109.1204 | 25.0967 | 203.7557 | 16.2148 |
| 15.8 | 109.0853 | 25.012 | 203.7251 | 16.117 |
| 15.83 | 109.0297 | 24.9279 | 203.6769 | 15.8227 |
| 15.87 | 108.9853 | 24.929 | 203.6355 | 15.7252 |
| 15.9 | 108.9297 | 24.9304 | 203.5941 | 15.7262 |
| 15.93 | 108.8775 | 24.8462 | 203.5567 | 15.7272 |
| 15.97 | 108.8151 | 24.8478 | 203.5193 | 15.7281 |
| 16 | 108.7647 | 24.849 | 203.4917 | 15.8273 |
| 16.03 | 108.7245 | 24.85 | 203.4651 | 15.7295 |
| 16.07 | 108.6757 | 24.8512 | 203.4207 | 15.7306 |
| 16.1 | 108.6364 | 24.8522 | 203.3498 | 15.8309 |
| 16.13 | 108.5765 | 24.8537 | 203.2897 | 15.9309 |
| 16.17 | 108.5167 | 24.7697 | 203.272 | 16.0298 |
| 16.2 | 108.4594 | 24.7711 | 203.2375 | 16.1292 |
| 16.23 | 108.3978 | 24.7727 | 203.1922 | 16.2289 |
| 16.27 | 108.3499 | 24.7739 | 203.1518 | 16.3284 |
| 16.3 | 108.3037 | 24.775 | 203.137 | 16.3288 |
| 16.33 | 108.2576 | 24.7762 | 203.1055 | 16.4281 |
| 16.37 | 108.1986 | 24.7777 | 203.0612 | 16.4292 |
| 16.4 | 108.1558 | 24.7787 | 203.0011 | 16.4307 |
| 16.43 | 108.1131 | 24.7798 | 202.9705 | 16.5299 |
| 16.47 | 108.0446 | 24.867 | 202.9508 | 16.5304 |
| 16.5 | 107.9856 | 24.8685 | 202.9173 | 16.5313 |
| 16.53 | 107.9352 | 24.9553 | 202.8809 | 16.6307 |
| 16.57 | 107.8805 | 25.0422 | 202.8375 | 16.5333 |
| 16.6 | 107.8497 | 25.0429 | 202.7804 | 16.3377 |
| 16.63 | 107.8086 | 24.9584 | 202.7489 | 16.2399 |
| 16.67 | 107.7633 | 24.8741 | 202.7193 | 16.1422 |
| 16.7 | 107.7094 | 24.8754 | 202.6908 | 16.0444 |
| 16.73 | 107.6436 | 24.8771 | 202.6484 | 16.1439 |
| 16.77 | 107.6111 | 24.8779 | 202.6307 | 16.0459 |
| 16.8 | 107.5435 | 24.8796 | 202.6031 | 15.948 |
| 16.83 | 107.4939 | 24.8808 | 202.5489 | 15.8509 |
| 16.87 | 107.4486 | 24.8819 | 202.5046 | 15.852 |
| 16.9 | 107.3905 | 24.7979 | 202.4721 | 15.8528 |
| 16.93 | 107.3152 | 24.7998 | 202.409 | 15.8544 |
| 16.97 | 107.2827 | 24.8861 | 202.3755 | 15.8552 |
| 17 | 107.2391 | 24.8872 | 202.3469 | 15.8559 |
| 17.03 | 107.1724 | 24.8888 | 202.3184 | 15.8567 |
| 17.07 | 107.1126 | 24.9758 | 202.275 | 15.7592 |
| 17.1 | 107.0561 | 25.0628 | 202.2445 | 15.76 |
| 17.13 | 106.9997 | 25.0642 | 202.1962 | 15.7612 |
| 17.17 | 106.9544 | 25.1508 | 202.1489 | 15.7624 |
| 17.2 | 106.9202 | 25.1517 | 202.1135 | 15.6647 |
| 17.23 | 106.8783 | 25.0672 | 202.1834 | 15.466 |
| 17.27 | 106.8158 | 25.0688 | 202.1263 | 15.3689 |
| 17.3 | 106.7577 | 25.1557 | 202.0248 | 15.3714 |
| 17.33 | 106.7184 | 25.1567 | 201.9992 | 15.2736 |
| 17.37 | 106.6611 | 25.1582 | 201.946 | 15.1764 |
| 17.4 | 106.5995 | 25.1597 | 201.8967 | 15.1776 |
| 17.43 | 106.5499 | 25.0754 | 201.8958 | 15.1776 |
| 17.47 | 106.5011 | 25.0766 | 201.8524 | 15.1787 |
| 17.5 | 106.4687 | 25.0775 | 201.8022 | 15.18 |
| 17.53 | 106.4208 | 25.0786 | 201.7657 | 15.1809 |
| 17.57 | 106.3541 | 25.0803 | 201.7174 | 15.2806 |
| 17.6 | 106.3173 | 25.0812 | 201.6593 | 15.3806 |
| 17.63 | 106.2609 | 25.0826 | 201.617 | 15.4801 |
| 17.67 | 106.1907 | 25.0844 | 201.5825 | 15.481 |
| 17.7 | 106.1454 | 25.0855 | 201.547 | 15.5804 |
| 17.73 | 106.0924 | 25.0869 | 201.5155 | 15.5812 |
| 17.77 | 106.0445 | 25.0881 | 201.4692 | 15.6809 |
| 17.8 | 105.9975 | 25.0892 | 201.4396 | 15.7801 |
| 17.83 | 105.9376 | 25.1762 | 201.3845 | 15.7815 |
| 17.87 | 105.8923 | 25.1774 | 201.4436 | 15.6815 |
| 17.9 | 105.8376 | 25.1787 | 201.4495 | 15.4843 |
| 17.93 | 105.788 | 25.18 | 201.3254 | 15.4874 |
| 17.97 | 105.7521 | 25.1809 | 201.2899 | 15.4883 |
| 18 | 105.6973 | 25.0967 | 201.219 | 15.5886 |
| 18.03 | 105.6221 | 25.0986 | 201.151 | 15.6888 |
| 18.07 | 105.5785 | 25.0997 | 201.148 | 15.7874 |
| 18.1 | 105.5417 | 25.0151 | 201.1106 | 15.7883 |
| 18.13 | 105.475 | 25.0168 | 201.0584 | 15.7896 |
| 18.17 | 105.4186 | 25.0182 | 200.9953 | 15.8897 |
| 18.2 | 105.3724 | 25.0193 | 200.9648 | 15.8905 |
| 18.23 | 105.3262 | 25.0205 | 200.9313 | 15.9898 |
| 18.27 | 105.2826 | 25.0216 | 200.9047 | 16.089 |
| 18.3 | 105.2287 | 24.9374 | 200.8781 | 16.0897 |
| 18.33 | 105.168 | 24.9389 | 200.9707 | 15.8903 |
| 18.37 | 105.1201 | 24.9401 | 200.8722 | 15.8928 |
| 18.4 | 105.074 | 24.9413 | 200.7747 | 15.8952 |
| 18.43 | 105.0363 | 24.9422 | 200.7491 | 15.8959 |
| 18.47 | 104.973 | 25.0293 | 200.6919 | 15.8973 |
| 18.5 | 104.9235 | 25.0306 | 200.6338 | 16.0958 |
| 18.53 | 104.8781 | 25.0317 | 200.6141 | 16.0963 |
| 18.57 | 104.8123 | 24.9478 | 200.5757 | 15.9987 |
| 18.6 | 104.7413 | 25.0351 | 200.5382 | 15.9012 |
| 18.63 | 104.708 | 25.036 | 200.5028 | 15.902 |
| 18.67 | 104.6703 | 25.0369 | 200.4446 | 15.9035 |
| 18.7 | 104.631 | 24.9524 | 200.4141 | 16.0028 |
| 18.73 | 104.5805 | 24.9536 | 200.3727 | 16.0038 |
| 18.77 | 104.5181 | 24.9552 | 200.3323 | 16.0048 |
| 18.8 | 104.4651 | 24.9565 | 200.3195 | 16.0051 |
| 18.83 | 104.4044 | 24.958 | 200.3796 | 15.8066 |
| 18.87 | 104.3625 | 24.9591 | 200.2663 | 15.7109 |
| 18.9 | 104.3197 | 24.8746 | 200.1777 | 15.8117 |
| 18.93 | 104.2607 | 24.8761 | 200.1461 | 15.8124 |
| 18.97 | 104.2154 | 24.8772 | 200.1077 | 15.9119 |
| 19 | 104.1701 | 24.7929 | 200.0664 | 16.0115 |
| 19.03 | 104.1111 | 24.7943 | 200.0752 | 16.0112 |
| 19.07 | 104.0623 | 24.7956 | 200.0181 | 16.1112 |
| 19.1 | 104.0222 | 24.7966 | 199.9629 | 16.1126 |
| 19.13 | 103.9862 | 24.712 | 199.9215 | 16.2121 |
| 19.17 | 103.9221 | 24.7136 | 199.8821 | 16.3116 |
| 19.2 | 103.8853 | 24.629 | 199.8792 | 16.2132 |
| 19.23 | 103.8263 | 24.6304 | 199.823 | 16.0175 |
| 19.27 | 103.763 | 24.632 | 199.7767 | 15.9202 |
| 19.3 | 103.7194 | 24.5476 | 199.8575 | 15.7211 |
| 19.33 | 103.6767 | 24.4632 | 199.7807 | 15.526 |
| 19.37 | 103.6245 | 24.4645 | 199.6605 | 15.6276 |
| 19.4 | 103.5476 | 24.5519 | 199.6437 | 15.628 |
| 19.43 | 103.5022 | 24.553 | 199.5817 | 15.728 |
| 19.47 | 103.4595 | 24.5541 | 199.5324 | 15.8278 |
| 19.5 | 103.4107 | 24.5553 | 199.5166 | 15.9267 |
| 19.53 | 103.3654 | 24.5564 | 199.4851 | 16.026 |
| 19.57 | 103.3218 | 24.5575 | 199.4585 | 16.0267 |
| 19.6 | 103.2722 | 24.4733 | 199.4103 | 16.1264 |
| 19.63 | 103.2064 | 24.4749 | 199.3679 | 16.1274 |
| 19.67 | 103.1653 | 24.4759 | 199.3068 | 15.9319 |
| 19.7 | 103.12 | 24.4771 | 199.2645 | 15.933 |
| 19.73 | 103.0618 | 24.4785 | 199.2507 | 15.9333 |
| 19.77 | 103.0063 | 24.4799 | 199.3393 | 15.6356 |
| 19.8 | 102.9524 | 24.4813 | 199.2241 | 15.6385 |
| 19.83 | 102.9028 | 24.4825 | 199.1285 | 15.6409 |
| 19.87 | 102.8412 | 24.5696 | 199.0999 | 15.7401 |
| 19.9 | 102.8045 | 24.5705 | 199.0379 | 15.8402 |
| 19.93 | 102.7514 | 24.6573 | 198.9975 | 15.8412 |
| 19.97 | 102.6916 | 24.6588 | 198.9935 | 15.9398 |
| 20 | 102.6463 | 24.7455 | 198.9512 | 15.9408 |
| 20.03 | 102.5941 | 24.7468 | 198.8931 | 16.0408 |
| 20.07 | 102.5342 | 24.7483 | 198.8428 | 16.1406 |
| 20.1 | 102.4821 | 24.7496 | 198.8133 | 16.2398 |
| 20.13 | 102.4573 | 24.7502 | 198.7857 | 16.339 |
| 20.17 | 102.4111 | 24.7513 | 198.7561 | 16.1427 |
| 20.2 | 102.3624 | 24.7525 | 198.697 | 16.1442 |
| 20.23 | 102.2982 | 24.7542 | 198.6813 | 16.1446 |
| 20.27 | 102.2452 | 24.841 | 198.7532 | 16.0443 |
| 20.3 | 102.2127 | 24.8418 | 198.6172 | 16.0477 |
| 20.33 | 102.1597 | 24.7576 | 198.5364 | 16.1482 |
| 20.37 | 102.087 | 24.8449 | 198.5 | 16.2477 |
| 20.4 | 102.0477 | 24.8459 | 198.4488 | 16.3474 |
| 20.43 | 101.9964 | 24.8472 | 198.4094 | 16.4469 |
| 20.47 | 101.9553 | 24.7627 | 198.4143 | 16.4468 |
| 20.5 | 101.9134 | 24.7638 | 198.3493 | 16.547 |
| 20.53 | 101.8647 | 24.6795 | 198.3089 | 16.548 |
| 20.57 | 101.798 | 24.6811 | 198.2586 | 16.6477 |
| 20.6 | 101.7338 | 24.6827 | 198.2271 | 16.747 |
| 20.63 | 101.6817 | 24.6841 | 198.1926 | 16.6494 |
| 20.67 | 101.6372 | 24.6852 | 198.1808 | 16.4527 |
| 20.7 | 101.5928 | 24.6863 | 198.1335 | 16.4538 |
| 20.73 | 101.5508 | 24.6873 | 198.1197 | 16.4542 |
| 20.77 | 101.4953 | 24.6887 | 198.1936 | 16.2553 |
| 20.8 | 101.4431 | 24.69 | 198.0685 | 16.1599 |
| 20.83 | 101.3952 | 24.6912 | 197.9907 | 16.1619 |
| 20.87 | 101.3499 | 24.6923 | 197.968 | 16.1624 |
| 20.9 | 101.296 | 24.6937 | 197.901 | 16.1641 |
| 20.93 | 101.2285 | 24.6954 | 197.8577 | 16.1652 |
| 20.97 | 101.1797 | 24.6966 | 197.8616 | 16.1651 |
| 21 | 101.131 | 24.6978 | 197.8104 | 16.1664 |
| 21.03 | 101.0771 | 24.6992 | 197.7444 | 16.2665 |
| 21.07 | 101.0275 | 24.7004 | 197.6823 | 16.3666 |
| 21.1 | 100.9796 | 24.7016 | 197.6518 | 16.1703 |
| 21.13 | 100.9437 | 24.7025 | 197.64 | 16.0721 |
| 21.17 | 100.9027 | 24.618 | 197.6114 | 16.0728 |
| 21.2 | 100.8556 | 24.6192 | 197.571 | 16.0739 |
| 21.23 | 100.8095 | 24.6203 | 197.5286 | 16.0749 |
| 21.27 | 100.759 | 24.5361 | 197.5996 | 15.9746 |
| 21.3 | 100.7017 | 24.5375 | 197.505 | 15.78 |
| 21.33 | 100.6427 | 24.539 | 197.4085 | 15.8809 |
| 21.37 | 100.5948 | 24.5402 | 197.373 | 15.8818 |
| 21.4 | 100.5392 | 24.6271 | 197.3257 | 15.9815 |
| 21.43 | 100.4896 | 24.6283 | 197.2981 | 15.9822 |
| 21.47 | 100.4452 | 24.6295 | 197.2804 | 15.9826 |
| 21.5 | 100.3879 | 24.6309 | 197.2311 | 15.9838 |
| 21.53 | 100.3468 | 24.5464 | 197.1917 | 15.9848 |
| 21.57 | 100.2964 | 24.5477 | 197.1474 | 16.0844 |
| 21.6 | 100.2357 | 24.6347 | 197.1178 | 15.7896 |
| 21.63 | 100.1818 | 24.5505 | 197.0804 | 15.7906 |
| 21.67 | 100.1219 | 24.6375 | 197.0607 | 15.7911 |
| 21.7 | 100.0886 | 24.6384 | 197.1238 | 15.5925 |
| 21.73 | 100.039 | 24.6396 | 197.0646 | 15.4954 |
| 21.77 | 99.9937 | 24.6407 | 196.9642 | 15.4979 |
| 21.8 | 99.9338 | 24.6422 | 196.9326 | 15.4987 |
| 21.83 | 99.8817 | 24.558 | 196.8568 | 15.5006 |
| 21.87 | 99.8415 | 24.559 | 196.8016 | 15.6005 |
| 21.9 | 99.7902 | 24.5603 | 196.8026 | 15.699 |
| 21.93 | 99.7397 | 24.5616 | 196.7612 | 15.7986 |
| 21.97 | 99.6961 | 24.5627 | 196.7208 | 15.7996 |
| 22 | 99.6294 | 24.5643 | 196.6834 | 15.8005 |
| 22.03 | 99.5918 | 24.5653 | 196.6351 | 15.9002 |
| 22.07 | 99.5405 | 24.5666 | 196.5928 | 15.9013 |
| 22.1 | 99.4909 | 24.5678 | 196.5691 | 15.7048 |
| 22.13 | 99.4396 | 24.5691 | 196.5238 | 15.6075 |
| 22.17 | 99.3865 | 24.5704 | 196.5021 | 15.608 |
3.6. Protein and mRNA Expression of Cardiac Differentiation-Related Genes in Cells and Chicken Embryos
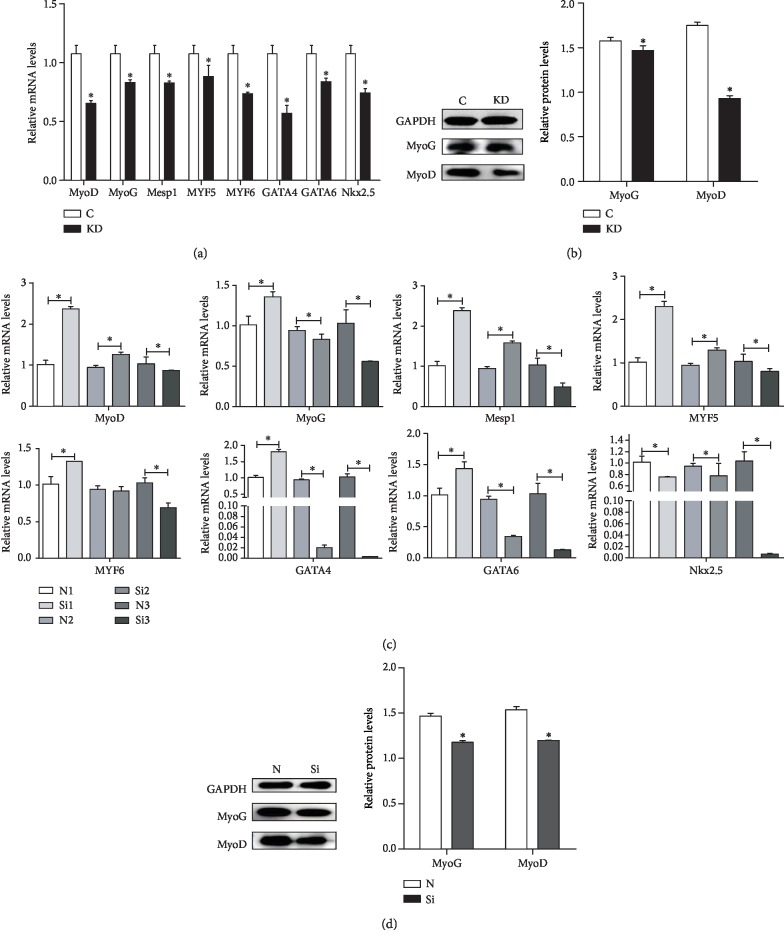
The effects of IGF1 knockdown on the mRNA abundance of cardiac differentiation-related genes (MyoD, MyoG, Mesp1, MYF5, MYF6, GATA4, GATA6, and Nkx2.5) in chicken cardiomyocytes are shown (Figure 7(a)), and the qPCR results revealed that the mRNA expression of MyoD, MyoG, Mesp1, MYF5, MYF6, GATA4, GATA6, and Nkx2.5 was significantly decreased (P < 0.05) in the KD group. A western blot analysis was performed to determine the protein expression of cardiac differentiation-related genes. The results revealed that compared with the C group, the protein expression of MyoG and MyoD in the KD group was significantly decreased (P < 0.05) (Figure 7(b)).
Figure 7.
The effects of IGF1 knockdown on the mRNA levels (a) and protein levels (b) of cardiac differentiation-related genes in cardiomyocytes and the effects of IGF1 silencing on the mRNA levels at 6, 8, and 10 days (c) and on the protein levels (d) of cardiac differentiation-related genes in the myocardium. C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo. GAPDH was selected as the reference. ∗ shows a significant difference from the corresponding control (P < 0.05). n = 3.
The effects of IGF1 knockdown on the mRNA abundance of cardiac differentiation-related genes (MyoD, MyoG, Mesp1, MYF5, MYF6, GATA4, GATA6, and Nkx2.5) in the myocardium of chicken embryos are shown in Figure 7(c). The qPCR results revealed that the mRNA expression of MyoD, Mesp1, and MYF5 increased in the Si group at 6 and 8 days, but significantly decreased (P < 0.05) at 10 days. The mRNA expression of MyoG, GATA4, and GATA6 increased at 6 days, but significantly decreased at 8 and 10 days. The mRNA expression of MYF6 increased at 6 days, showed no significant change at 8 days, and significantly decreased (P < 0.05) at 10 days. The mRNA expression of Nkx2.5 decreased (P < 0.05) at 6, 8, and 10 days. A western blot analysis was performed to determine the protein expression of cardiac differentiation-related genes. The results revealed that compared with the N group, the protein expression of MyoG and MyoD in the Si group was significantly decreased (P < 0.05) (Figure 7(d)).
3.7. Intracellular Morphological Observation and H.E. Stain in Cells and Chicken Embryos' Myocardium
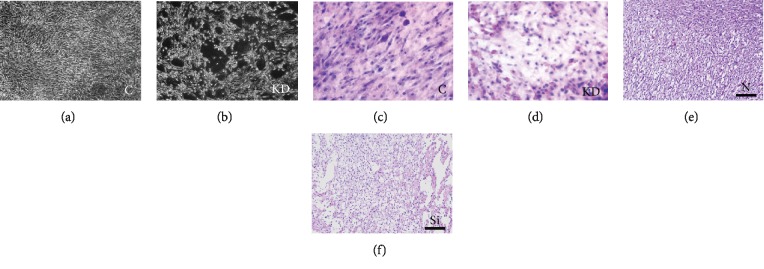
As observed under a microscope, normal cardiomyocytes were fusiform and tightly connected and the whole looked similar to paving stones accompanied by protruding pseudopodia that stretched out between cells, interweaving into a mesh in the control group (Figure 8(a)). In the KD group, as the density of cell growth decreased, the volume of the cardiomyocytes and intercellular junctions was evidently reduced. We observed that myocardial fibers and muscle fiber bundles were disintegrated, and the pseudopodia between cells did not interweave into a mesh in the KD group (Figure 8(b)). We observed myocardial cells stained by hematoxylin and eosin (H.E.). The cardiomyocytes in the control group displayed normal morphologies (Figure 8(c)). However, many slender cardiomyocytes appeared in the IGF1 knockdown group (Figure 8(d)), which indicated that cardiomyocyte development was blocked.
Figure 8.
Morphological observation was performed in cardiomyocytes transfected with Opti-MEM (C group, a) and siRNA (KD group, b) for 24 h. Cardiomyocytes were stained by H.E., and the results are shown in (c) and (d). H.E. staining for myocardial tissues in the N group (e) and the Si group (f). C indicates the control group; KD indicates the knockdown group in vitro; N indicates the normal group; Si indicates the knockdown group in vivo.
Myocardial injury and ultrastructural damage are shown in Figure 8(f). Blood vessel rupturing and increased tissue gaps were observed in the IGF1-deficient chicken heart group more than in the normal group (Figure 8(e)).
4. Discussion
IGF1 is a polypeptide neurotropic factor with a structure and function similar to insulin. IGF1 is a single-chain protein that promotes cell differentiation and proliferation, and it has a wide range of biological functions and participates in the regulation of various organs. IGF1 plays an important role in the development of human and vertebrate embryos [23]. In the present study, we established an IGF1 knockdown model in vivo and in vitro through transfection of small interfering RNA (siRNA) and the mRNA and protein levels of IGF1 were detected to support this point. Significant damage to myocardial tissue, rupturing of blood vessels, and increased tissue gaps were observed in IGF1-deficient chicken heart through histopathological observation, demonstrating that IGF1 suppression leads to dysplasia of cardiomyocytes and myocardial tissue.
The concentration of free radicals is low under physiological conditions, which is important to cell signal regulation, metabolism, survival, and apoptosis [24, 25]. However, a large number of free radicals are induced when organisms are stimulated by physical factors or exogenous chemical substances; free radicals can covalently bind to biomacromolecules and peroxidize biofilm lipids to produce various toxic effects [26], which can lead to the occurrence of many diseases such as myocardial infarction [27, 28] and various cancers [29]. Oxygen free radicals can make lipid fatty acids into lipid peroxides and further decompose into a series of complex compounds, including MDA; therefore, the level of lipid oxidation can be detected by the level of MDA [30]. iNOS is an oxidative stress (free radical) that utilizes nitric oxide, which can be produced when cells are stimulated and activated. The cells can form a complex antioxidant enzyme defense system, which mainly includes SOD, CAT, GSH, and so on, to protect the body from peroxidative damage [26, 31]. SOD can eliminate free radicals in the body and protect cells from free radical damage. The level of SOD activity reflects the ability of antifree radicals. SOD can convert superoxide anion (O2−) into H2O2, and CAT converts H2O2 into water such that toxic O2− and H2O2 are converted into harmless water molecules [32, 33]. GSH is catalyzed to convert to oxidized glutathione (GSSH) with the assistance of GSH-Px, which can reduce oxidized substances and relieve their toxicity [34, 35]. In recent years, the antioxidant functions of IGF1 have been gradually discovered; Tumati et al. found that low IGF1 can induced excessive ROS, which will be further moderated by JNK-induced epithelial cytoprotection [36]. ROS, as an important endogenous stimulator in the body, can stimulate multiple pathways including myocardial development. Huk et al. found that ROS serves as secondary messengers to influence cardiac valve development [37]. ROS may be involved in adverse cardiac remodeling [38]. In our present study, we found that decreasing expression of IGF1 results in increasing ROS generation, suggesting that IGF1 may be involved in the regulation of ROS and the occurrence of oxidative stress. IGF1 significantly decreased the CAT, SOD, GSH, and GSH-Px activities in vivo and in vitro. These changes were accompanied by reduced T-AOC, which is an important indicator for determining the body's antioxidant capacity; meanwhile, the expression of iNOS was significantly increased. All of these results demonstrate that IGF1 suppression can significantly reduce the body's antioxidant capacity and enrich many oxygen free radicals in the body; this may be one of the important causes of myocardial damage and dysplasia.
FOXO, a highly conserved transcriptional regulatory protein, is a downstream protein of the PI3K/Akt and AMPK/14-3-3 signaling pathways [39, 40]. The PI3K/Akt/FOXO signaling pathway is involved in the regulation of various physiological processes such as proliferation, apoptosis, and insulin resistance. The PI3K/Akt pathway can be activated by several of these stimuli, and ROS is one of the most important factors. Adipogenic differentiation can be mediated through activation of the PI3K/Akt pathway by oxidative stress in primary rat osteoblasts [41]. Stitt et al. found that the IGF1/PI3K/Akt pathway plays an important role in preventing the expression of ubiquitin ligases by inhibiting FOXO transcription factors [42]. AMPK is an important protein kinase in eukaryotic cells. The energy regulation of AMPK can maintain normal ATP levels in cardiomyocytes [43]. AMPK regulates the utilization of energy throughout the body, and it is considered the energy regulator [44]. FOXO is an important downstream molecule of AMPK involved in the signal transduction and regulation of various cellular biological processes such as inhibiting cell proliferation and promoting apoptosis [45, 46]. IGF1 stimulation can decrease the level of AMPK phosphorylation in F1 and F3/4 granulosa cells [47]. Hinchy proved that the ROS released from mitochondria can activate AMPK indirectly [48]. Furthermore, FOXO has been proven to be required in endothelial but not myocardial cell lineages during cardiovascular development [49]. Evans-Anderson et al. demonstrated that a FOXO transcription factor is a negative regulator of cardiomyocyte proliferation during heart development [50]. In our present study, we found that IGF1 depletion inhibited the expression of IRS1, PI3K, and Akt and upregulated both the mRNA and protein expression of AMPK and FOXO; meanwhile, the expression of JNK significantly increased in the IGF1 suppression group, which is important upstream of IRS1. All of these results suggest that IGF1 knockdown can activate FOXO through inhibiting the expression of IRS1. Combined with the results of ROS, we concluded that IGF1 suppression triggers ROS release to activate FOXO, which further inhibits myocardial development.
Mitochondria, as an energy converter in animal cells, are a main site for intracellular oxidative phosphorylation and ATP formation. Pawlikowska et al. showed that mitochondria are essential organelles for insulin-mediated muscle formation and that insulin stimulates mitochondria and promotes mitochondrial function [51]. Insulin-like growth factor-binding proteins (IGFBPs) are important parts of the IGF family; IGFBPs cannot bind with insulin but rather form a complex with IGFs. They can regulate the normal growth of embryonic and postnatal organs, which is also a very important tool for IGF1 transport and storage. The IGF1-IGFBP complex will decompose and release IGF1, which will further combine with IGF1R in the cell membrane under the catalysis of IR [52]. IGF1 is protected from decomposition by its high affinity with IGFBP, prolonging the half-life of IGF1 in the body's circulation cells. IGFBPs avoid the body's negative effects for insulin overdose by reducing the concentration of free IGF1 in the blood. In addition, studies have shown that IGFBP can help IGF1 recognize target cell and regulate the activity of IGF1 [53]. The IGF1-IGFBP complex can cause a cascade of various phosphorylation processes in the cell, ultimately leading to the entry of the glucose transporter molecules (GLUT1, 3, and 8) into the cell membrane and increasing the rate of glucose transport in the cell. Glucose transport is mainly dependent on GLUT, and it plays an important role in regulating glucose transport and maintaining cardiac energy balance [54]. IGF1RS/IRS signaling regulates cellular energy metabolism through the PI3K/Akt signaling pathway, and its downstream genes have been found [55]. FOXO plays an important role in mediating the effects of insulin and growth factors on diverse physiological functions [56]. In the present study, we demonstrated that IGF1 knockdown significantly decreased the expression of GLUT3 and IGFBP, indicating that IGF1 suppression blocks energy metabolism. To further verify these results, we also detected the ATP content and oxygen consumption rate in two different groups; the results revealed that IGF1 knockdown can significantly impede the ability of using oxygen for cardiomyocytes, thereby reducing the production of ATP. Considering our results showing FOXO activation by IGF1 knockdown, we conclude that FOXO can inhibit myocardial development by interfering with myocardial energy metabolism.
Myogenic regulatory factors (MRFs) are a class of muscle-specific regulators that determine the function of cell-directed differentiation. MRFs are essential for muscle development in vertebrates [56]. MRFs include MYF5, MyoD, MyoG, and MRF4; the function of MRFs is transforming mesenchymal stem cells into myoblasts, which will further activate and maintain the differentiation state of myocytes. Each member of the MRF family plays different roles at different states. MyoD and MYF5 play an important role in the early proliferative phase of myocytes [57]. MYF5 is responsible for myoblast proliferation, and MyoD regulates the differentiation process for myoblasts. MyoG and MRF4 can drive terminal differentiation. The proliferation process of myoblasts is normal after knocking out MyoG, but the subsequent differentiation process is significantly inhibited [58]. Mesp1 and Mesp2 are key genes in regulating cardiac differentiation. Mesp1 can directly activate other genes in the cardiac core by binding to other gene promoters in cardiomyocytes. Mesp1 and Mesp2 reduction can impede heart development [59]. GATA4 and Nkx2.5 are transcription factors involved in heart development, and overexpression of GATA4 can accelerate myocardial differentiation [60]. Early embryo development requires the production and expenditure of large amounts of cellular energy for cell growth. Insulin can regulate skeletal muscle cell differentiation by mediating the activation of MAPK and PKB phosphorylation [61]. Montarras et al. demonstrated that insulin or IGF is necessary for cell differentiation [62]. Cellular energy metabolism that contains fatty acid is involved in fetal heart development [63]. In the present study, we found that IGF1 knockdown can significantly decrease the expression of MyoD, Mesp1, MYF5, MYF6, GATA4, GATA6, and Nkx2.5, indicating that IGF1 suppression can block myocardial development. We suggest that reducing energy metabolism inhibits the expression of myocardial development to factors through comprehensive results on energy metabolism.
In summary, we conclude that IGF1 knockdown hinders myocardial development through the energy metabolism dysfunction caused by ROS-dependent FOXO activation in the chicken heart. Our results indicate a novel hypothesis for IGF1 in the development of cardiomyocytes.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31872531), Earmarked Fund for China Agriculture Research System (No. CARS 35-04), Funding for State Key Laboratory of Animal Nutrition (2004DA125184F1725), Merit-based Funding for Returned Oversea Student of Heilongjiang Province (2018QD0005), Heilongjiang Postdoctoral Fund “Academic backbone” Project of Northeast Agricultural University (17XG11).
Abbreviations
- IGF1:
Insulin-like growth factor 1
- IGFs:
Insulin-like growth factors
- IGF1R:
Insulin-like growth factor 1 receptor
- IGF2:
Insulin-like growth factor 2
- GH:
Growth hormone
- BMP:
Bone morphogenic protein
- FGF4:
Fibroblast growth factor 4
- IR:
Insulin receptor
- IGFR:
Insulin-like growth factor receptor
- IRR:
Insulin receptor-related receptor
- TK:
Tyrosine kinase
- IRS:
Insulin receptor substrate
- PI3K:
Phosphatidylinositol 3-kinase
- VEGF:
Vascular endothelial growth factor
- ROS:
Reactive oxygen species
- H2O2:
Hydrogen peroxide
- GSH:
Glutathione
- GSH-Px:
Glutathione peroxidase
- CAT:
Catalase
- MDA:
Malondialdehyde
- iNOS:
Induced nitric oxide synthase
- SOD:
Superoxide dismutase
- T-AOC:
Total antioxidant capability
- IGFBP1:
Insulin-like growth factor-binding protein 1
- IGFBP2:
Insulin-like growth factor-binding protein 2
- IGFBP3:
Insulin-like growth factor-binding protein 3
- IGFBP4:
Insulin-like growth factor-binding protein 4
- IGFBP5:
Insulin-like growth factor-binding protein 5
- IGFBP7:
Insulin-like growth factor-binding protein 7
- GLUT1:
Glucose transporters 1
- GLUT3:
Glucose transporters 3
- GLUT8:
Glucose transporters 8
- Akt:
Threonine-protein kinase
- P-Akt:
Phosphorylation-threonine-protein kinase
- FOXO:
Forkhead box protein
- P-FOXO:
Phosphorylation-forkhead box protein
- JNK:
c-Jun N-terminal kinase
- P-JNK:
Phosphorylation-c-Jun N-terminal kinase
- GATA4:
GATA-binding protein 4
- GATA6:
GATA-binding protein 6
- Nkx2.5:
NK2 homeobox 5
- AMPK:
AMP-activated protein kinase
- MyoG:
Myogenin
- Mesp1:
Cardiac transcription factor mesoderm posterior 1
- MYF5:
Myogenic factor 5
- MYF6:
Myogenic factor 6
- O2−:
Superoxide anion
- GSSH:
Oxidized glutathione
- ATP:
Adenosine triphosphate
- IGFBPs:
Insulin-like growth factor-binding proteins
- MRFs:
Myogenic regulatory factors
- MAPK:
Mitogen-activated protein kinase
- Nrf2:
Nuclear factor erythroid 2-related factor 2
- PBS:
Phosphate-buffered solution
- GAPDH:
Glyceraldehyde-3-phosphate dehydrogenase.
Contributor Information
Honggui Liu, Email: liuhonggui1312@163.com.
Ziwei Zhang, Email: zhangziwei@neau.edu.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
All authors have read the manuscript and agreed to submit it in its current form for consideration for publication in the Journal.
Supplementary Materials
The graphical abstract of the entire manuscript.
References
- 1.Schmid C. Insulin-like growth factors. Cell Biology International. 1995;19(5):445–458. doi: 10.1006/cbir.1995.1088. [DOI] [PubMed] [Google Scholar]
- 2.Jehle P., Fussgaenger R., Blum W., et al. Differential autocrine regulation of intestine epithelial cell proliferation and differentiation by insulin-like growth factor (IGF) system components. Hormone and Metabolic Research. 1999;31(02/03):97–102. doi: 10.1055/s-2007-978705. [DOI] [PubMed] [Google Scholar]
- 3.Monzen K., Nagai R., Komuro I. A role for bone morphogenetic protein signaling in cardiomyocyte differentiation. Trends in Cardiovascular Medicine. 2002;12(6):263–269. doi: 10.1016/S1050-1738(02)00172-X. [DOI] [PubMed] [Google Scholar]
- 4.Schneider V. A., Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes & Development. 2001;15(3):304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musarò A., McCullagh K. J. A., Naya F. J., Olson E. N., Rosenthal N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature. 1999;400(6744):581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 6.Ueki K., Yamamoto-Honda R., Kaburagi Y., et al. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. The Journal of Biological Chemistry. 1998;273(9):5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- 7.Verspohl E. J., Roth R. A., Vigneri R., Goldfine I. D. Dual regulation of glycogen metabolism by insulin and insulin-like growth factors in human hepatoma cells (HEP-G2). Analysis with an anti-receptor monoclonal antibody. Journal of Clinical Investigation. 1984;74(4):1436–1443. doi: 10.1172/JCI111555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher R., Soos M. A., Schlessinger J., Brandenburg D., Siddle K., Ullrich A. Signaling-competent receptor chimeras allow mapping of major insulin receptor binding domain determinants. Journal of Biological Chemistry. 1993;268(2):1087–1094. [PubMed] [Google Scholar]
- 9.Engels M. C., Rajarajan K., Feistritzer R., et al. IGF promotes cardiac lineage induction by selective expansion of cardiogenic mesoderm in vitro. Circulation. 2013;128(22, article A17539) [Google Scholar]
- 10.Bisping E., Ikeda S., Sedej M., et al. Transcription factor GATA4 is activated but not required for insulin-like growth factor 1 (IGF1)-induced cardiac hypertrophy. Journal of Biological Chemistry. 2012;287(13):9827–9834. doi: 10.1074/jbc.M111.338749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliadis F., Kadoglou N., Didangelos T. Insulin and the heart. Diabetes Research and Clinical Practice. 2011;93(4942):S86–S91. doi: 10.1016/S0168-8227(11)70019-5. [DOI] [PubMed] [Google Scholar]
- 12.Laurino L., Wang X. X., de la Houssaye B. A., et al. PI3K activation by IGF-1 is essential for the regulation of membrane expansion at the nerve growth cone. Journal of Cell Science. 2005;118(16) Part 16:3653–3662. doi: 10.1242/jcs.02490. [DOI] [PubMed] [Google Scholar]
- 13.Zhu C., Qi X., Chen Y., Sun B., Dai Y., Gu Y. PI3K/Akt and MAPK/ERK1/2 signaling pathways are involved in IGF-1-induced VEGF-C upregulation in breast cancer. Journal of Cancer Research and Clinical Oncology. 2011;137(11):1587–1594. doi: 10.1007/s00432-011-1049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia G., Mitra A. K., Gangahar D. M., Agrawal D. K. Insulin-like growth factor-1 induces phosphorylation of PI3K-Akt/PKB to potentiate proliferation of smooth muscle cells in human saphenous vein. Experimental and Molecular Pathology. 2010;89(1):20–26. doi: 10.1016/j.yexmp.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Zhao H., Shao Y., et al. Copper (II) and/or arsenite-induced oxidative stress cascades apoptosis and autophagy in the skeletal muscles of chicken. Chemosphere. 2018;206:597–605. doi: 10.1016/j.chemosphere.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Zhao H., Guo M., et al. Arsenite renal apoptotic effects in chickens co-aggravated by oxidative stress and inflammatory response. Metallomics. 2018;10(12):1805–1813. doi: 10.1039/C8MT00234G. [DOI] [PubMed] [Google Scholar]
- 17.Papaconstantinou J. Insulin/IGF-1 and ROS signaling pathway cross-talk in aging and longevity determination. Molecular and Cellular Endocrinology. 2009;299(1):89–100. doi: 10.1016/j.mce.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papaiahgari S., Yerrapureddy A., Hassoun P. M., Garcia J. G. N., Birukov K. G., Reddy S. P. EGFR-activated signaling and actin remodeling regulate cyclic stretch–induced NRF2-ARE activation. American Journal of Respiratory Cell and Molecular Biology. 2007;36(3):304–312. doi: 10.1165/rcmb.2006-0131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jie Y., Hamid S., Cai J., Liu Q., Xu S., Zhang Z. Selenium deficiency-induced thioredoxin suppression and thioredoxin knockdown disbalanced insulin responsiveness in chicken cardiomyocytes through PI3K/Akt pathway inhibition. Cellular Signalling. 2017;38:192–200. doi: 10.1016/j.cellsig.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q., Yang J., Gong Y., Cai J., Zhang Z. Role of miR-731 and miR-2188-3p in mediating chlorpyrifos induced head kidney injury in common carp via targeting TLR and apoptosis pathways. Aquatic Toxicology. 2019;215:p. 105286. doi: 10.1016/j.aquatox.2019.105286. [DOI] [PubMed] [Google Scholar]
- 21.Yang J., Gong Y., Cai J., Liu Q., Zhang Z. lnc-3215 suppression leads to calcium overload in selenium deficiency-induced chicken heart lesion via the lnc-3215-miR-1594-TNN2 pathway. Molecular Therapy - Nucleic Acids. 2019;18:1–15. doi: 10.1016/j.omtn.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R., Jin C., Wang Z., Wang Z., Wang J., Wang L. Effects of manganese deficiency on the microstructure of proximal tibia and OPG/RANKL gene expression in chicks. Veterinary Research Communications. 2015;39(1):31–37. doi: 10.1007/s11259-015-9626-5. [DOI] [PubMed] [Google Scholar]
- 23.Fang N., Sun R., Fu B., et al. IGF-1 promotes the development and cytotoxic activity of human NK cells. Nature Communications. 2013;4(1):p. 1479. doi: 10.1038/ncomms2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhao H., Guo M., Fei D., Zhang L., Xing M. Targeting the miR-122/PKM2 autophagy axis relieves arsenic stress. Journal of Hazardous Materials. 2020;383:p. 121217. doi: 10.1016/j.jhazmat.2019.121217. [DOI] [PubMed] [Google Scholar]
- 25.Wang S., Li X., Wang W., Zhang H., Xu S. Application of transcriptome analysis: oxidative stress, inflammation and microtubule activity disorder caused by ammonia exposure may be the primary factors of intestinal microvilli deficiency in chicken. Science of the Total Environment. 2019;696, article 134035 doi: 10.1016/j.scitotenv.2019.134035. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H., Wang Y., Yang X., et al. Zinc alleviates arsenism in common carp: varied change profiles of cytokines and tight junction proteins among two intestinal segments. Fish & Shellfish Immunology. 2019;94:761–768. doi: 10.1016/j.fsi.2019.09.069. [DOI] [PubMed] [Google Scholar]
- 27.Yang J., Zhang Y., Hamid S., et al. Interplay between autophagy and apoptosis in selenium deficient cardiomyocytes in chicken. Journal of Inorganic Biochemistry. 2017;170:17–25. doi: 10.1016/j.jinorgbio.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y., Yang J., Cai J., Liu Q., Zhang J. ., Zhang Z. Effect of Gpx3 gene silencing by siRNA on apoptosis and autophagy in chicken cardiomyocytes. Journal of Cellular Physiology. 2019;234(6):7828–7838. doi: 10.1002/jcp.27842. [DOI] [PubMed] [Google Scholar]
- 29.Wu D., Yotnda P. Production and detection of reactive oxygen species (ROS) in cancers. Journal of Visualized Experiments. 2011;57, article e3357 doi: 10.3791/3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabha P. S., Das U. N., Koratkar R., Sagar P. S., Ramesh G. Free radical generation, lipid peroxidation and essential fatty acids in uncontrolled essential hypertension. Prostaglandins, Leukotrienes and Essential Fatty Acids. 1990;41(1):27–33. doi: 10.1016/0952-3278(90)90127-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhao X. J., Li Z. P., Wang J. H., et al. Effects of chelated Zn/Cu/Mn on redox status, immune responses and hoof health in lactating Holstein cows. Journal of Veterinary Science. 2015;16(4):439–446. doi: 10.4142/jvs.2015.16.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao X., Chen Z., Chen Y., et al. Effect on SOD, CAT and free radical in blood and liver of sepsis rats treated with edaravone. Clinical Medicine. 2008;28(12) [Google Scholar]
- 33.Wang S., Chi Q., Hu X., Cong Y., Li S. Hydrogen sulfide-induced oxidative stress leads to excessive mitochondrial fission to activate apoptosis in broiler myocardia. Ecotoxicology and Environmental Safety. 2019;183:p. 109578. doi: 10.1016/j.ecoenv.2019.109578. [DOI] [PubMed] [Google Scholar]
- 34.Li J., Xing T., Wang L., Tao J. P., Liu Z. P. Inhibitory effect of S-nitroso-glutathione on Eimeria tenella oocysts was mainly limited to the early stages of sporogony. Veterinary Parasitology. 2010;173(1-2):64–69. doi: 10.1016/j.vetpar.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Chen M., Li X., Shi Q., Zhang Z., Xu S. Hydrogen sulfide exposure triggers chicken trachea inflammatory injury through oxidative stress-mediated FOS/IL8 signaling. Journal of Hazardous Materials. 2019;368:243–254. doi: 10.1016/j.jhazmat.2019.01.054. [DOI] [PubMed] [Google Scholar]
- 36.Tumati S., Burger H., Martens S., van der Schouw Y. T., Aleman A. Association between cognition and serum insulin-like growth factor-1 in middle-aged & older men: an 8 year follow-up study. PLoS One. 2016;11(4, article e0154450) doi: 10.1371/journal.pone.0154450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huk D., Lincoln J. Oxidative stress in cardiac valve development. In: Rodriguez-Porcel M., Chade A., Miller J., editors. Studies on Atherosclerosis. Oxidative Stress in Applied Basic Research and Clinical Practice. Boston, MA: Humana Press; 2017. [DOI] [Google Scholar]
- 38.Grieve D. J., Byrne J. A., Cave A. C., Shah A. M. Role of oxidative stress in cardiac remodelling after myocardial infarction. Heart, Lung & Circulation. 2004;13(2):132–138. doi: 10.1016/j.hlc.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Puig O., Marr M. T., Ruhf M. L., Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & Development. 2003;17(16):2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pehmøller C., Treebak J. T., Birk J. B., et al. Genetic disruption of AMPK signaling abolishes both contraction- and insulin-stimulated TBC1D1 phosphorylation and 14-3-3 binding in mouse skeletal muscle. American Journal of Physiology-Endocrinology and Metabolism. 2009;297(3):E665–E675. doi: 10.1152/ajpendo.00115.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Yang J. H. Activation of the PI3K/Akt pathway by oxidative stress mediates high glucose-induced increase of adipogenic differentiation in primary rat osteoblasts. Journal of Cellular Biochemistry. 2013;114(11):2595–2602. doi: 10.1002/jcb.24607. [DOI] [PubMed] [Google Scholar]
- 42.Stitt T. N., Drujan D., Clarke B. A., et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Molecular Cell. 2004;14(3):395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 43.Xie Z., He C., Zou M. H. AMP-activated protein kinase modulates cardiac autophagy in diabetic cardiomyopathy. Autophagy. 2011;7(10):1254–1255. doi: 10.4161/auto.7.10.16740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwabuchi S., Kawahara K. Extracellular ATP-prinoceptor signaling and AMP-activated protein kinase regulate astrocytic glucose transporter 3 in an in vitro ischemia. Neurochemistry International. 2013;63(4):259–268. doi: 10.1016/j.neuint.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Peng K., Li Y., Long L., et al. Knockdown of _FoxO3a_ induces increased neuronal apoptosis during embryonic development in _zebrafish_. Neuroscience Letters. 2010;484(2):98–103. doi: 10.1016/j.neulet.2010.07.068. [DOI] [PubMed] [Google Scholar]
- 46.Yao H.-D., Wu Q., Zhang Z.-W., et al. Gene expression of endoplasmic reticulum resident selenoproteins correlates with apoptosis in various muscles of se-deficient chicks. Journal of Nutrition. 2013;143(5):613–619. doi: 10.3945/jn.112.172395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tosca L., Chabrolle C., Crochet S., Tesseraud S., Dupont J. IGF-1 receptor signaling pathways and effects of AMPK activation on IGF-1-induced progesterone secretion in hen granulosa cells. Domestic Animal Endocrinology. 2008;34(2):204–216. doi: 10.1016/j.domaniend.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Hinchy E. C., Gruszczyk A. V., Willows R., et al. Mitochondria-derived ROS activate AMP-activated protein kinase (AMPK) indirectly. Journal of Biological Chemistry. 2018;293(44):17208–17217. doi: 10.1074/jbc.RA118.002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sengupta A., Chakraborty S., Paik J., Yutzey K. E., Evans-Anderson H. J. FoxO1 is required in endothelial but not myocardial cell lineages during cardiovascular development. Developmental Dynamics. 2012;241(4):803–813. doi: 10.1002/dvdy.23759. [DOI] [PubMed] [Google Scholar]
- 50.Evans-Anderson H. J., Alfieri C. M., Yutzey K. E. Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circulation Research. 2008;102(6):686–694. doi: 10.1161/CIRCRESAHA.107.163428. [DOI] [PubMed] [Google Scholar]
- 51.Pawlikowska P., Gajkowska B., Hocquette J.-F., Orzechowski A. Not only insulin stimulates mitochondriogenesis in muscle cells, but mitochondria are also essential for insulin-mediated myogenesis. Cell Proliferation. 2006;39(2):127–145. doi: 10.1111/j.1365-2184.2006.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kricker J. A., Hyde C. E., van Lonkhuyzen D. R., et al. Mechanistic investigations into interactions between IGF-I and IGFBPs and their impact on facilitating cell migration on vitronectin. Growth Factors. 2010;28(5):359–369. doi: 10.3109/08977194.2010.494603. [DOI] [PubMed] [Google Scholar]
- 53.Collett-Solberg P. F., Cohen P. The role of the insulin-like growth factor binding proteins and the IGFBP proteases in modulating IGF action. Endocrinology and Metabolism Clinics of North America. 1996;25(3):591–614. doi: 10.1016/S0889-8529(05)70342-X. [DOI] [PubMed] [Google Scholar]
- 54.Jing H., Gao X., Xu L., Lin H., Zhang Z. H2S promotes a glycometabolism disorder by disturbing the Th1/Th2 balance during LPS-induced inflammation in the skeletal muscles of chickens. Chemosphere. 2019;222:124–131. doi: 10.1016/j.chemosphere.2019.01.136. [DOI] [PubMed] [Google Scholar]
- 55.Nadler S. T., Stoehr J. P., Rabaglia M. E., Schueler K. L., Birnbaum M. J., Attie A. D. Normal Akt/PKB with reduced PI3K activation in insulin-resistant mice. American Journal of Physiology. Endocrinology and Metabolism. 2001;281(6):1249–1254. doi: 10.1152/ajpendo.2001.281.6.e1249. [DOI] [PubMed] [Google Scholar]
- 56.Barthel A., Schmoll D., Unterman T. G. FoxO proteins in insulin action and metabolism. Trends in Endocrinology and Metabolism. 2005;16(4):183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Kablar B., Asakura A., Krastel K., et al. MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochemistry and Cell Biology. 1998;76(6):1079–1091. doi: 10.1139/o98-107. [DOI] [PubMed] [Google Scholar]
- 58.Valdez M. R., Richardson J. A., Klein W. H., Olson E. N. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Developmental Biology. 2000;219(2):287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 59.Kitajima S., Takagi A., Inoue T., Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127(15):3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 60.Hu D. L., Chen F. K., Liu Y. Q., et al. GATA-4 promotes the differentiation of P19 cells into cardiac myocytes. International Journal of Molecular Medicine. 2010;26(3):365–372. [PubMed] [Google Scholar]
- 61.Al-Khalili L., Kramer D., Wretenberg P., Krook A. Human skeletal muscle cell differentiation is associated with changes in myogenic markers and enhanced insulin-mediated MAPK and PKB phosphorylation. Acta Physiologica Scandinavica. 2004;180(4):395–403. doi: 10.1111/j.1365-201X.2004.01259.x. [DOI] [PubMed] [Google Scholar]
- 62.Montarras D., Pinset C., Pérez M. C., Ilan J., Gros F. Muscle differentiation: insulin-like growth factors as positive modulators of myogenic regulatory genes? Comptes Rendus de l'Académie des Sciences. Série III. 1993;316(9):1025–1031. [PubMed] [Google Scholar]
- 63.Warshaw J. B., Kimura R. E. Cellular energy metabolism during fetal development: V. Fatty acid synthesis by the developing heart. Developmental Biology. 1973;33(1):224–228. doi: 10.1016/0012-1606(73)90178-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The graphical abstract of the entire manuscript.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.