Abstract
A 4-oxalocrotonate tautomerase (4-OT) trimer has been isolated from Burkholderia lata and a kinetic, mechanistic, and structural analysis has been performed. The enzyme is the third described oligomer state for 4-OT along with a homo- and heterohexamer. The 4-OT trimer is part of a small subset of sequences (133 sequences) within the 4-OT subgroup of the tautomerase superfamily (TSF). The TSF has two distinct features: members are composed of a single β-α-β unit (homo- and heterohexamer) or two consecutively joined β-α-β units (trimer) and generally have a catalytic amino-terminal proline. The enzyme, designated fused 4-OT, functions as a 4-OT where the active site groups (Pro-1, Arg-39, Arg-76, Phe-115, Arg-127) mirror those in the canonical 4-OT from Pseudomonas putida mt-2. Inactivation by 2-oxo-3-pentynoate suggests that Pro-1 of fused 4-OT has a low pKa enabling the prolyl nitrogen to function as a general base. A remarkable feature of the fused 4-OT is the absence of P3 rotational symmetry in the structure (1.5 Å resolution). The asymmetric arrangement of the trimer is not due to the fusion of the two β-α-β building blocks because an engineered “unfused” variant that breaks the covalent bond between the two units (to generate a heterohexamer) assumes the same asymmetric oligomerization state. It remains unknown how the different active site configurations contribute to the observed overall activities and whether the asymmetry has a biological purpose or role in the evolution of TSF members.
Graphical Abstract
INTRODUCTION
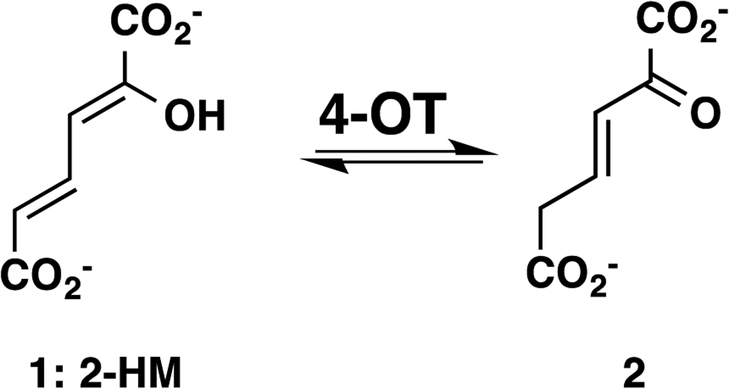
The canonical 4-oxalocrotonate tautomerase (4-OT) from Pseudomonas putida mt-2 converts 2-hydroxymuconate (1, 2-HM, Scheme 1) to 2-oxo-3-hexenedioate (2), which is one of the reactions in the bacterial meta-fission pathway for the degradation of aromatic hydrocarbons.1–3 The genes coding the enzymes in this pathway are grouped together on a plasmid (the TOL plasmid pWW0) and the presence of this plasmid allows the host bacteria to use simple alkyl-substituted aromatic hydrocarbons as sole sources of carbon and energy.2,3 A wealth of kinetic, mechanistic, and structural data has been collected for 4-OT, resulting in a reasonably good understanding of the catalytic mechanism as well as the roles of four key players in the mechanism (Pro-1, Arg-11, Arg-39, and Phe-50).4–8 4-OT is a homohexamer where each monomer is made up of 62 amino acids.9,10
Scheme 1.
4-OT-catalyzed Reaction
A second oligomeric arrangement was described for a heterohexamer 4-OT (hh4-OT) from the thermophilic organism, Chloroflexus aurantiacus J-10-fl.11 The hh4-OT consists of three α,β-dimers where both the α- and β-subunits have 72 amino acids. The genomic context suggests that the hh4-OT is part of a meta-fission pathway, and it has been shown to carry out the reaction shown in Scheme 1.11 The key players in the hh4-OT mechanism (βPro-1, αArg-12, αArg-40, αTrp-51) correspond to those in 4-OT.
The canonical 4-OT is the founding member of the tautomerase superfamily (TSF), which has two defining features: members are constructed using a β-α-β building block and generally have a catalytic N-terminal proline.12,13 The monomers in both the homo- and heterohexameric 4-OTs display the signature β-α-β motif as well as the N-terminal proline. Over the last few decades, the structural and catalytic properties of more than 30 TSF members have been determined. These studies defined the five major subgroups in the TSF, which are designated the 4-OT,12,13 5-(carboxymethyl)-2-hydroxymuconate isomerase,14 (CHMI), macrophage migration inhibitory factor (MIF),15 cis-3-chloroacrylic acid dehalogenase (cis-CaaD),16 and malonate semialdehyde decarboxylase (MSAD) subgroups.17 Members of the 4-OT subgroup are composed of a single β-α-β unit, whereas those of the other four subgroups are composed of two consecutively fused β-α-β units. The single β-α-β units generally make up homo- or heterohexamers and the fused β-α-β units result in trimers, each resulting in an overall highly similar quaternary arrangement of β-α-β units.
This long-standing observation about the TSF was examined in a global context across the superfamily in a sequence similarity network (SSN).18 The analysis confirmed that the pattern generally prevails. Using pairwise sequence identity as a metric in an all-by-all comparison, more than 11,000 protein sequences sorted into the five known subgroups, where the 4-OT subgroup is the largest one (4472 protein sequences).18 The predominance of this structural theme suggests that a gene fusion event took place early in the evolution of the TSF, followed by the diversification of function that is seen today.
In the course of an analysis of the 4-OT subgroup, a subset of sequences double the length of that of the canonical 4-OTs was identified.18 These “fused” 4-OTs form a separate subgroup that still connects to the short 4-OTs. These proposed trimeric 4-OTs represent a third oligomeric structure identified for 4-OT with features similar to possible progenitors for other subgroups in the TSF (the great majority of which are of the double length represented by the fused 4-OTs). In view of the potential significance of the members in this subset, a kinetic, mechanistic, and structural analysis was carried out on a representative of the fused 4-OTs from Burkholderia lata. This analysis shows that the enzyme functions as a 4-OT, the overall structure is trimeric, where each monomer is made up of two consecutively fused β-α-β units, and the active site is nearly superimposable on that of the canonical 4-OT. However, one striking property of the fused 4-OT, whose structure was determined at 1.5 Å resolution, is that it lacks a three-fold rotational symmetry, resulting in an asymmetric arrangement of the three active sites. There is no other known TSF member like this, and, to the best of our knowledge, such an observation has not been previously reported. In order to investigate whether “fusing” the two β-α-β units had resulted in the asymmetric arrangement, an engineered variant was made where the covalent bond between the two units was broken. This “unfused” 4-OT (1.8 Å resolution) also assumes an asymmetric arrangement. The biological and evolutionary implications of the asymmetry are unknown.
Experimental procedures
Materials.
All chemicals, biochemical, and enzyme substrates were obtained from sources described elsewhere.19,20 2-Oxo-3-pentynoate (2-OP) was synthesized as reported.21 Chromatographic resins (DEAE Sepharose fast flow and Bio-Gel P60 gel beads) and columns (PD-10 Sephadex G-25M, Econo-Column, and Spectra/Chrom LC) were acquired from sources described elsewhere.19 The Amicon stirred cell concentrators and the ultrafiltration membranes (10,000 Da, MW cutoff) were purchased from EMD Millipore Inc. (Billerica, MA). InstantBlue for staining SDS-PAGE gels was obtained from C.B.S. Scientific Company, Inc. (Del Mar, CA).
Bacterial Strains, Plasmids, and Growth Conditions.
Escherichia coli strain BL21-Gold(DE3) was obtained from Agilent Technologies (Santa Clara, CA). The gene encoding fused 4-OT from Burkholderia lata (ATCC 17760, UniProt accession: Q392K7) (designated “fused 4-OT”) was codon-optimized for expression in E. coli, synthesized, and cloned into the expression vectors pJ411 (pJ411-F4-OT) by DNA2.0, Inc., now ATUM (Newark, CA).19 Cells were grown overnight (~16 h) at 37 °C in Luria-Bertani (LB) media, supplemented with 25 mM Na2HPO4 and 25 mM KH2PO4, buffered to pH ~6.75, 5 mM Na2SO4, 2 mM MgSO4, and kanamycin (Kn, 30 μg/mL).
General Methods.
Steady-state kinetic assays were performed on an Agilent 8453 diode-array spectrophotometer at 22 °C. Nonlinear regression data analysis was performed using the program Grafit (Erithacus Software Ltd., Staines, U.K.). Protein concentrations were determined by the Waddell method.22 Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on denaturing gels containing 12% polyacrylamide.23 The PCR was carried out on a Labnet MultiGene OptiMax Thermal Cycler (Labnet International, Inc., Edison, NJ). Electrospray ionization mass spectrometry was performed on an LCQ electrospray ion-trap mass spectrometer (Thermo, San Jose, CA), housed in the Institute for Cellular and Molecular Biology (ICMB) Protein and Metabolite Analysis Facility at the University of Texas. 1H NMR spectroscopic experiments to examine possible dehalogenase activities were carried out as described elsewhere.24 Light scattering experiments were carried out as described.25
Construction of the SSN for the Level 2 4-OT Subgroup Containing the Canonical 4-OT.
The network was generated using an in-house version of the Pythoscape software,26 tailored for use with available hardware. All-by-all pairwise comparisons of 133 Level 2 4-OT subgroup sequences, as curated in the Structure-Function Linkage Database (SFLD) (Figure 1), were obtained using the BLAST algorithm. The nodes (each representing a sequence) are arranged using Prefuse force directed OpenCL layout provided by the Cytoscape software,27 weighted by BLAST bit score of the associated edges. Edges are drawn between two nodes only if the mean similarity between the two sequences is at least as significant as the bit score of 96 chosen to illustrate similarity relationships for this network.
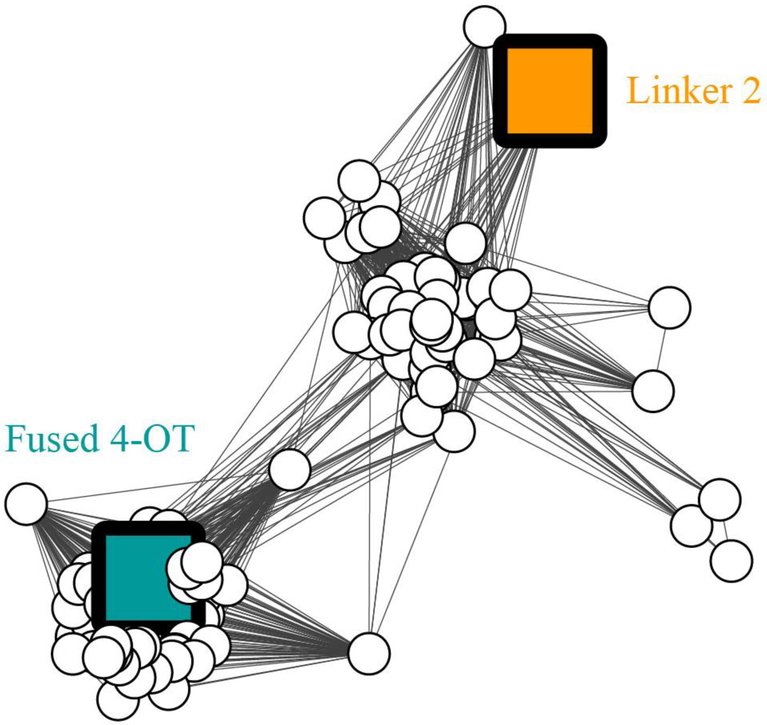
Figure 1. Sequence similarity network showing the fused 4-OTs in the 4-OT subgroup.
The SNN is part of the Level 2 4-OT subgroup containing the canonical 4-OT (Figures S1 and S2). The SSN represents 133 sequences with characteristics shown in Figure 2. Nodes representing the characterized sequences for “fused 4-OT” and “Linker 2” are colored and labeled. Edges are drawn between two nodes only if the mean similarity between the two sequences is at least as significant as the bit score of 96 chosen to illustrate similarity relationships for this network.
Construction of the P1A, R39A, R76A, R104A, and R127A Mutants of Fused 4-OT.
The fused 4-OT mutants were generated by the PCR using the pJ411-F4-OT plasmid as a template. The cycling parameters were 95 °C for 5 min followed by 35 cycles of 98 °C for 30 s, 45 °C for 20 s, and 72 °C for 20 s, with a final elongation step of 72 °C for 10 min. The P1A and R127A mutations were generated by direct amplification of the entire gene using the indicated primers. For the P1A mutant, the forward primer, which contains an NdeI restriction site (underlined) and the desired mutation (in bold), was 5′- ATAGCAGGTACATATGGCGACTCTTGAAGTC-3′, and the reverse primer was the universal T7 terminator primer. For the R127A mutant, the forward primer was the universal T7 promoter primer. The reverse primer, which contains a HindIII restriction site (underlined) and the desired mutation (in bold), was 5′- GTGATGTTATAAGCTTATCACGCGCCCAGTGCACG-3′. The R39A, R76A and R104A mutations were generated by overlap extension PCR28 using the same cycling parameters, and the following primers in combination with the universal T7 promoter and terminator primers. The desired mutations are in bold. For the R39A mutant: 5′- C GAG AGC GTC GCG GTG CTG CTG ACC-3′ (R39A-F) and 5′- GGT CAG CAG CAC CGC GAC GCT CTC G-3′ (R39A-R). For the R76A mutant: 5′-G ATC GCG GGT GCG ACC GAC GAG-3′ (R76A-F), and 5′- CTC GTC GGT CGC ACC CGC GAT C-3′ (R76A-R). For the R104A mutant: 5′- CAG GCG ACG GCG GTT ATG ATT-3′ (R104A-F), and 5′-AAT CAT AAC CGC CGT CGC CTG-3′ (R104A-R). Each PCR reaction mixture was made up in the Thermo Phusion High-fidelity Master Mix to which was added 100 ng of template DNA, and 50 ng of each primer. After the PCR, the products and pET-24(a) vector were treated with the NdeI and HindIII restriction enzymes. Subsequently, the vector was dephosphorylated with alkaline phosphatase. After purification, each PCR product and vector were ligated for 2 h at room temperature using T4 DNA ligase. An aliquot of the ligation mixture was transformed into E. coli BL21(Gold) DE3 cells. Transformants were selected at 37 °C on LB/Kn plates. Plasmid DNA was isolated from several colonies, and subsequently sequenced to verify that no mutations had been introduced during the amplification of the gene.
Construction of the Expression Vector for “Unfused” 4-OT.
The gene for the “unfused” 4-OT was generated using the PCR protocol described above where the pJ411-F4-OT plasmid was used as the template. The following primers were used to introduce a stop codon after the Leu-65 codon (bold, italics), a ribosomal binding site (bold), and start codon before the Pro-66 codon (bold, italics). The NdeI and HindIII restriction sites are underlined. The N-terminal β-α-β-subunit (Pro-1 to Leu-65) of unfused 4-OT was amplified using the primers 5’-A TAG CAG GTA CAT ATG CCA ACT CTT GAA GTC TTT CTG CC-3’ and 5’- CAT AAA TTT TAC CTC CTT TCA CAG GCT CGG CGG TGC-3’. The C-terminal β-α-β-subunit (Pro-66 to Arg-127) of unfused 4-OT was amplified using the primers 5’-G TGA AAG GAG GTA AAA TTT ATG CCG GTG ATC GTT GCG-3’ and 5’-G TGA TGT TAT AAG CTT TCA GCG GCC CAG TGC ACG GGC-3’. The resulting PCR products were gel-purified, and the full-length gene for unfused 4-OT was generated by the overlap extension PCR protocol (above) and the following primers as forward and reverse primers, respectively: 5’-A TAG CAG GTA CAT ATG CCA ACT CTT GAA GTC TTT CTG CC-3’ and 5’-G TGA TGT TAT AAG CTT TCA GCG GCC CAG TGC ACG GGC-3’. The resulting full-length gene for unfused 4-OT was cloned into the pJ411 expression vector as described above.
Expression and Purification of Wild-type Fused 4-OT, Unfused 4-OT, and Fused 4-OT Mutants.
The proteins were all purified based on the protocol described elsewhere for the cis-CaaD homologue encoded by PputUW4_01749 in Pseudomonas sp. UW4 (UniProt K9NIA5, designated Ps01740).19 After growth, induction, cell disruption, and centrifugation, the clear supernatant was applied to a DEAE-Sepharose column (~15 mL bed volume), which had previously been equilibrated in buffer A (10 mM Na2HPO4 buffer, pH 7.3). The column was washed with buffer A (3 × 15 mL), and retained proteins were subsequently eluted with a linear gradient of buffer A made 500 mM in NaCl. Typically, the target proteins eluted around 50 mM NaCl. The fractions from the DEAE-Sepharose column containing the target protein were pooled, and concentrated to ~3 mL using an Amicon concentrator. This sample was subsequently applied to a size-exclusion column (Bio-Gel P60 beads, 500 mL bed volume), which had previously been equilibrated in buffer A, and proteins were eluted with buffer A (0.5 mL per minute) at 22 °C. The protein-containing fractions (5 mL) were analyzed by SDS-PAGE, and those containing near-homogenous target protein were pooled, and concentrated to ~20 mg/mL using an Amicon concentrator. Aliquots were flash-frozen in liquid nitrogen and stored at −80 °C until further use.
Covalent modification of Fused 4-OT and Unfused 4-OT by 2-Oxo-3-pentynoate (2-OP) and Mass Spectral Analysis.
Stock solutions of 2-OP (60 mM) were prepared by dissolving the solid compound (4 mg) in 100 mM Na2HPO4 buffer, pH 9.2 (600 μL, final pH ~7). To 900 μL of a 1 mg/mL solution of protein in 50 mM Na2HPO4 buffer, pH 7.3, was added 100 μL of the stock solution of 2-OP. The reaction mixtures were incubated at 22 °C for 90 min (< 1% activity remaining). The 2-OP containing reaction mixture was made 50 mM in NaBH4 (from a 1M stock solution in ultrapure water), and incubated at 22 °C for 3 h. Subsequently, the protein in the reaction mixtures was buffer-exchanged into 10 mM Na2HPO4 buffer (pH 7.3) using a PD-10 column, flash-frozen in liquid nitrogen, and stored at −80 °C until further use. The sample was analysed by ESI-MS according to previously published procedures.29
Steady State Kinetics.
The assays were carried out at 22 °C in 10 mM Na2HPO4 buffer (pH 7.3) using 2-hydroxymuconate (2-HM), 5-(methyl)-2-hydroxymuconate (5-Me-2-HM), phenylenolpyruvate (PP), 2-hydroxy-2,4-pentadienoate (HPD), and 5-(carboxymethyl)-2-hydroxymuconate (CHM) as substrates.19,20 Stock solutions of 2-HM (25 mM), 5-Me-2-HM (10 and 25 mM), PP (100 mM), HPD (17 and 50 mM), and CHM (17 and 50 mM) were prepared by dissolving the appropriate amount of the crystalline free acid in absolute ethanol. Substrate concentrations ranged from 13–300 μM for 2-HM, 40–450 μM for 5-Me-2-HM, 26–1030 μM for PP, 170–900 μM for HPD, and 85–770 μM for CHM. Fused 4-OT was found to be unstable at the dilute concentrations used in the assay for >1 h. Therefore, the enzyme was diluted to the assay concentration (0.0033 μM) directly before use from a 0.016 mg/mL stock solution, at which concentration it is stable for at least 24 h. For 5-Me-2-HM, HPD, and CHM, the enzyme was diluted directly to the assay concentration (0.018 μM, 0.058 μM, and 1.4 μM, respectively) from a ~20 mg/mL stock solution. With the exception of the R104A fused 4-OT, which was assayed the same way as wild-type fused 4-OT, the mutant enzymes were directly diluted to their assay concentrations (0.14–1.0 μM) from a ~20 mg/mL stock solution, due to the low-level activity towards 2-HM. For unfused 4-OT, a 0.14 mg/mL or 0.7 mg/mL stock solution was prepared, respectively, to measure kinetics for 2-HM and PP (final enzyme assay concentrations were 0.011 μM and 0.194 μM, respectively).
The enol-keto tautomerization of substrates was monitored at wavelengths reported elsewhere using published extinction coefficients with the following exceptions.20 The enol-keto tautomerization of 2-HM was monitored by following the decay of the enol form at 300 nm (ε = 22.5 × 103 M−1 cm−1). The enol-keto tautomerization of PP was monitored by following the decay of the enol-form at 305 nm (ε = 2800 M−1 cm−1).20 Initial rates were determined, plotted against the substrate concentration and fitted to the Michaelis-Menten equation using Grafit.
Crystallization.
Initial crystallization conditions for fused 4-OT and unfused 4-OT were identified using sparse-matrix screening with a Phoenix crystallization robotic system (Art Robbins Instruments). After systematic optimization of the hits using the vapor-diffusion sitting drop method, reproducible crystals grew within a few days. Fused 4-OT crystallized in conditions consisting of 0.1 M HEPES buffer at pH 7.5 and 28–33% PEG 8000 at room temperature at a concentration of ~9 mg/mL. Unfused 4-OT crystallized in conditions consisting of 200 mM magnesium acetate and 28% PEG 3550 at room temperature at a concentration of ~26 mg/mL. Crystals were cryoprotected in mother liquor containing 25–30% glycerol.
Data collection, Processing, Structure Determination and Refinement.
X-ray data for fused 4-OT were collected at the Advanced Light Source beamline 5.0.3 (ALS, Berkeley, CA) and the diffraction data for the unfused 4-OT were collected at Advanced Photon Source (APS) beamline 23-ID-D. The data sets were indexed, integrated, and scaled using HKL-2000.30 The structures were determined by molecular replacement (MR) using Phaser-MR31 and Autobuild32 from the PHENIX suite of programs. The Cg10062 monomer crystal structure (19.4% sequence identity, PDB entry 3N4G) was used as a search model for the initial phases for the fused 4-OT structure factors. The monomeric alanine truncated version of the canonical 4-OT was used as a search model for the initial estimate of the unfused 4-OT structure factors. Subsequently, using high-resolution electron density maps, chain identity was assigned as the α-subunit (unfused 4-OT residues 1–65) and the β-subunit (unfused 4-OT 66–120). Structure refinement was performed using Phenix Refine.33 TLS parameter was included in the refinement of all structures.34 The final structures were evaluated during and after refinement using Molprobity.35 The refinement statistics for the structures are summarized in Table 1 (where F4-OT is fused 4-OT and uF4-OT is unfused 4-OT). All figures were prepared with PyMol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).36
Table 1:
Crystallographic Data Collection and Refinement Statistics
| PDB code | 6BLM | 6OGM |
|---|---|---|
| Data collection | F4-OT | uF4-OT |
| Space group | P212121 | P1211 |
| Cell dimensions | ||
| a, b, c (Å) | 47.8, 67.2, 94.8 | 39.6, 81.6, 96.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 95.6, 90.0 |
| Resolution (Å) | 42.69–1.49 (1.54–1.49)* | 47.88–1.86 (1.93–1.86)* |
| Wavelength (Å) | 1.0332 | 1.0332 |
| Rsym | 0.08751 (0.697)* | 0.117 (0.511)* |
| Rpim | 0.03551 (0.2877)* | 0.070 (0.313)* |
| CC1/2ϒ | 0.998 (0.836)ϒ | 0.991 (0.726)ϒ |
| I/ σ | 24.2 (2.6)* | 9.5 (2.0)* |
| Completeness (%) | 99.6 (98.7)* | 97.4 (93.3)* |
| Redundancy | 7.0 (6.8)* | 3.6 (3.5)* |
| Refinement | ||
| Resolution (Å) | 42.69–1.49 (1.543–1.49)* | 47.88–1.86(1.93–1.86)* |
| No. reflections | 356506(33851)* | 178400(17024)* |
| Rwork | 0.1968 (0.2581)* | 0.1831 (0.2440)* |
| Rfree± | 0.2153 (0.2906)* | 0.2237 (0.3058)* |
| No. atoms | 3075 | 5688 |
| Protein | 2751 | 5265 |
| Water | 324 | 423 |
| B-factors (Å2) | ||
| Protein | 25.91 | 24.99 |
| Water | 36.96 | 32.57 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.005 |
| Bond angles (°) | 1.10 | 0.82 |
| Ramachandran plot | ||
| Favored | 99.7 | 99.7 |
| Allowed | 0.3 | 0.3 |
| Outliers£ | 0.00 | 0.00 |
| Molprobity score^ | 1.27 / 93rd percentile¥ | 1.37 / 92nd percentile¥ |
Values for the corresponding parameters in the outermost shell in parenthesis.
CC1/2 is the Pearson correlation coefficient for a random half of the data, the two numbers represent the lowest and highest resolution shell respectively.
Rfree is the Rmodel calculated for 5% of the reflections randomly selected and omitted from refinement.
There is only one Ramachandran outlier which correspond to a proline residues (Pro 158) on one chain with strong electron density.
MolProbity score is calculated by combining clashscore with retainer and Ramachandran percentage and scaled based on X- ray resolution.
The percentage is calculated with 100th percentile as the best and 0th percentile as the worst among structures of comparable resolution.
Results and Discussion
Discovery of the Fused 4-OTs.
The structure-function relationships of more than 11,000 members in the TSF were visualized in a sequence similarity network (SSN).18 In the course of the analysis of a similarity path connecting the 4-OT and cis-CaaD subgroups and the characterization of two linking sequences, a group of fused 4-OTs was identified within the 4-OT subgroup, in that sequences in this subset are double that of the length of the short 4-OTs (~125 vs. 62 amino acids, respectively). Two representative sequences from this fused 4-OT subset were chosen and a preliminary characterization was reported.18 One sequence is from Burkholderia lata (strain ATCC 17760, UniProt accession: Q392K7) and the encoded protein is designated “fused 4-OT”. The other sequence is from Pusillimonas sp. (strain T7–7) (PT7_0534, UniProt accession F4GMX9) and the encoded protein is designated “Linker 2”.18 Both proteins are trimers, where each monomer is made up of two consecutively fused β-α-β units.18
The diversity of the 4-OT subgroup (4472 non-redundant sequences) in the previously reported SSN necessitated a further subgrouping of the main Level 1 subgroup to result in several Level 2 subgroups.18 An update of one of the Level 2 4-OT subgroups (the one that includes the canonical 4-OT) is reported herein (Figure S1) where an additional 776 sequences were added (to give a total of 1995 sequences). When the same network is color-coded by sequence length (less than 100 amino acids and more than 100 amino acids), three distinct clusters can be seen (Figure S2). A majority of sequences (133 sequences) falls into two of the clusters, identified by the “fused 4-OT” or “Linker 2” nodes (Figure 1). The third cluster (59 sequences) will be investigated at a later time. With a few exceptions (which might be due to a misannotated start codon), the remaining sequences are short ones (Figure S2).
Sequence Analysis.
The alignment of the fused 4-OT and canonical 4-OT sequences identified conserved and potential active site groups. As noted previously, the canonical 4-OT aligns best with the C-terminal half of fused 4-OT.18 Notably, fused 4-OT has Arg-76 (equivalent to Arg-11 in 4-OT), Arg-104 (equivalent to Arg-39 in 4-OT), Phe-115 (equivalent to Phe-50 in 4-OT), the GIGG region from 116–119 (equivalent to G51-G54 in 4-OT), and Arg-127 (equivalent to Arg-62 in 4-OT). In addition, fused 4-OT has the N-terminal proline (Pro-1) and Arg-39. The most significant observation is that Pro-66 (in the fused 4-OT) aligns with Pro-1 of the canonical 4-OT. Pro-66 is rarely conserved in TSF members and the positional conservation might reflect a gene fusion of two separate “short” 4-OTs and little divergence since the fusion.
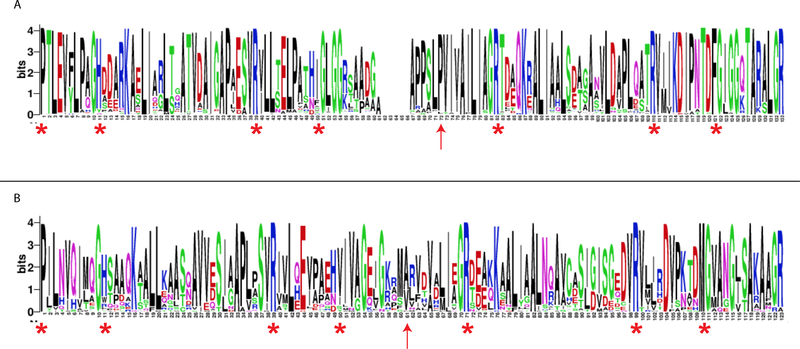
A multiple sequence alignment (MSA) of the 86 sequences in the fused 4-OT cluster and the 47 sequences in the Linker 2 cluster is visualized in two sequence logos (Figures 2A and 2B, respectively). The sequences in the fused 4-OT cluster (Figure 2A) are characterized by 4-OT-like sections joined in the middle by a linker-region, which results in a gap in the MSA. Although not identical in sequence, both 4-OT-like sections share similar conserved features (red asterisks). The N-terminal section shows the conserved N-terminal Pro characteristic of the TSF, a positively charged residue at positions 11 (His) and 39 (Arg), and a non-polar residue at position 50 followed by a GxGG-motif. This is similar to the core catalytic machinery, and the beta-hairpin motif important for the formation of the functional homohexamer structure, of the canonical 4-OT.9,13 The C-terminal section shows almost the same features, where two Arg-residues (logo-positions 82 and 110), and a nonpolar residue (position 121) followed by a GxGG-motif, are conserved at equivalent positions relative to the N-terminal section. Worth noting, the “fused” 4-OTs have a conserved Pro-residue, 10 positions before the first conserved Arg-residue (at logo-position 72, red arrow). This would exactly have been the position of an N-terminal Pro, had this C-terminal section not been fused to the N-terminal section.
Figure 2. Sequence logos for the fused 4-OT and Linker 2 clusters.
A) The sequence logo for the fused 4-OT cluster. The sequences of the “fused” 4-OTs (86 total) are characterized by two 4-OT-like sections joined in the middle by a linker-region, which results in a gap in the multiple sequence alignments (MSA). The red asterisks indicate the conserved features (described in the text) and the red arrow indicates a conserved proline residue (red arrow logo-position 72) that is 10 positions before the arginine at logo-position 82. This could represent the position of an N-terminal proline before fusion of two short 4-OTs in many of the sequences. B) The sequence logo for the Linker 2 cluster. The sequences show the conserved features (described in the text), but the positioning is not as striking. Moreover, there is not an obvious conservation of proline as the start of the C-terminal section (red arrow logo-position 61). The logos are graphical representations of multiple sequence alignments (MSAs) generated using WebLogo.37
The sequences in the Linker 2 cluster (Figure 2B) have many of the same conserved features (Pro-1, His-11, Arg-39, a hydrophobic residue, Ile, at position 50, Arg-71, Arg-99 and a hydrophobic residue, Met, at position 110) as indicated by the red asterisks. In both N-terminal and C-terminal section, the GxGG motif is less prominent. There is no gap in the MSA and no obvious proline that might represent the start of the second 4-OT-like sequence. Linker 2 has diverged from the fused 4-OT.18
Purification and Characterization of the Fused 4-OT.
The fused 4-OT was overproduced in E. coli BL21(DE3) and purified to near homogeneity. The typical yield was ~100 mg of fused 4-OT per liter of culture. ESI-MS analysis of the purified fused 4-OT showed a single major signal corresponding to a monoisotopic mass of 13,030 Da (calc. 13,161 Da) in the reconstructed mass spectrum. The mass difference of 131 Da between the theoretical and the observed mass indicates that the translation-initiating methionine was post-translationally removed, resulting in a mature protein (127 amino acids for the fused 4-OT) with an N-terminal proline.38 DLS analysis of the fused 4-OT showed a single peak (97.4% mass fraction) with a molar mass of 39,640 Da. This species corresponds best with the trimeric form of fused 4-OT (calculated mass of 39,090 Da).
Kinetic Properties of the Fused 4-OT.
The protein was screened for activity with known substrates for TSF members (Table 2). Of these substrates, the highest kcat/Km value is measured for 2-HM with lower values being measured for 5-methyl-2-HM, PP, and HPD (5.7-fold to 21.5-fold lower). The lowest value is measured for CHM (1217-fold lower). The enzyme could not be saturated with CHM. It appears that the bulkier substituent at C5 (and/or the charge) substantially affects binding and activity.
Table 2.
Steady-state Kinetic Parameters for Fused 4-OT with Indicated Substrates.a
| Substrate |
kcat (s−1) |
Km (μM) |
kcat/Kmb (M−1 s−1) |
|---|---|---|---|
| 2-HM | 580 ± 25 | 208 ± 16 | 2.8 × 106 |
| 5-Me-2-HM | 72 ± 2 | 147 ± 9 | 4.9 × 105 |
| PP | 136 ± 8 | 720 ± 75 | 1.9 × 105 |
| HPD | 170 ± 12 | 1330 ± 140 | 1.3 × 105 |
| CHM | - | - | 2.3 × 103 |
The kinetic parameters were measured using the assay described in the text.
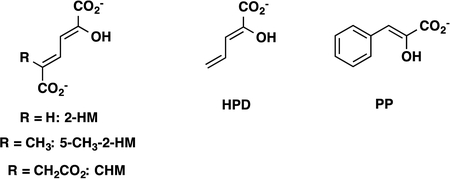
The enzyme was also tested for its ability to process the cis- and trans-isomers of 3-chloroacrylic acid.24 After 7 days, no product could be detected (by 1H NMR spectroscopy) when incubated with the cis-isomer. However, a trace amount of product (<5%) could be detected using the trans-isomer. The signals for products (malonate semialdehyde and acetaldehyde) are clearly present, but are too weak to be reliably integrated. For reference, 4-OT converts 76% of the trans-isomer to products (malonate semialdehyde, acetaldehyde and the hydrates) in the same time period.24 The lengthy incubation time results in the non-enzymatic decarboxylation of malonate semialdehyde.24
Irreversible Inhibition of Fused 4-OT by 2-OP.
The reaction of fused 4-OT with 2-OP was monitored at pH 7.0 for 90 min, and treated with NaBH4 subsequent to mass spectral analysis. For fused 4-OT, the reconstructed mass spectrum showed two major signals at 13,030 Da and 13,142 Da in a ratio of relative abundance of 1:3. These masses correspond to the native, unmodified protein (13,030 Da) and the protein modified by a single adduct resulting from incubation with 2-OP (13,142 Da where the mass difference is 112 Da). (For unknown reasons, treatment of the 2-OP-inactivated fused 4-OT with NaBH4 did not reduce the adduct.) These observations suggest that the prolyl nitrogen has a low pKa value such that it is largely unprotonated at pH 7.0 (behaving like 4-OT where the pKa of the prolyl nitrogen is 6.4).5,39 The introduction of NaBH4 raises the pH and likely causes removal of some label.
Mutagenic Analysis.
In order to identify the importance of five conserved residues (Pro-1, Arg-39, Arg-76, Arg-104, and Arg-127) to the mechanism, alanine mutants were constructed, expressed, and characterized. The kinetic properties were determined using 2-HM and are summarized in Table 3. Based on the kcat/Km values, Pro-1, Arg-39 and Arg-76 are the most important residues for catalysis (where changing Arg-76 to an alanine results in a 746-fold decrease in the kcat/Km value). Arg-104 is the least important (with little significant change in the kcat/Km value) followed by Arg-127 (98-fold decrease in the kcat/Km value).
Table 3.
Kinetic Parameters for the Fused 4-OT Mutants with 2-HM.a
| Enzyme | kcat (s−1) | rel. kcat | Km (μM) | rel. Km |
kcat/Km (M−1 s−1) |
rel. kcat/Km |
|---|---|---|---|---|---|---|
| WT | 580 ± 25 | 1.0 | 208 ± 16 | 1.0 | 2.79 × 106 | 1.0 |
| P1A | 3.4 ± 0.1 | 5.9 × 10−3 | 221 ± 11 | 1.06 | 1.54 × 104 | 5.5 × 10−3 |
| R39A | - | - | - | - | 8.34 × 103 | 3.0 × 10−3 |
| R76A | - | - | - | - | 3.74 × 103 | 1.3 × 10−3 |
| R104A | 313 ± 15 | 0.54 | 213 ± 18 | 1.02 | 1.47 × 106 | 0.53 |
| R127A | - | - | - | - | 2.84 × 104 | 1.0 × 10−2 |
The kinetic parameters were measured using the assay described in the text.
Construction of an Unfused 4-OT, Purification, Characterization, and Kinetic Properties.
Based on the observation that Pro-66 aligned with Pro-1 of the canonical 4-OT, an “unfused” version of the fused 4-OT was constructed. A stop codon was introduced after Leu-65 followed by a ribosome binding site and a start codon before Pro-66. Both “halves” were amplified by the PCR (where the α-subunit is defined as Pro-1 to Leu-65 and the β-subunit is defined as Pro-66 to Arg-127) using the pJ411-F4-OT plasmid. The two fragments were joined by overlap extension PCR, and the resulting proteins produced and purified using the same protocol described for the fused 4-OT.
The typical yield was ~10 mg of unfused 4-OT per liter of culture. ESI-MS analysis of the purified, unfused 4-OT showed three major signals corresponding to monoisotopic masses of 6,579, 6,599, and 6,627 Da (calc. 6,579 Da for the α-subunit and 6,468 Da for the β-subunit) in the reconstructed mass spectrum. The observed masses correspond to the demethionylated α-subunit (6,579 Da with an “unblocked” Pro-1), the β-subunit retaining the initiating methionine (6,599 Da), and the β-subunit retaining the initiating N-formyl methionine (6,627 Da).38 The signals corresponding to the α-subunit and the β-subunit with the initiating N-formyl methionine are comparable in intensity suggesting a 1:1 complex (for an α,β-dimer). The signal corresponding to the β-subunit retaining the initiating methionine is much less intense.
DLS analysis of the unfused 4-OT showed two peaks: the first peak (98.8% mass fraction) has a molar mass of 38,170 Da (calculated mass of 39,090 Da) and the second peak (1.2% mass fraction) has a molar mass of 13,090 Da. The first peak likely corresponds to the trimer made up of an α,β-dimer and the second peak could correspond to an α,β-dimer (or a contaminant).
When expressed separately, the α-subunit of unfused 4-OT expressed normally, but the β-subunit formed inclusion bodies suggesting instability. A stable complex was only obtained when the two subunits were cloned into the same vector and expressed simultaneously under the same promoter. On gel filtration, two main peaks eluted at apparent molecular weights of 39 kDa and 7 kDa. ESI-MS analysis showed that the first peak to elute contains both α and β subunits in a 1:1 ratio, while the second peak only contains the α subunit monomer (with no detectable tautomerase activity).
The kinetic properties of the unfused 4-OT were examined with 2-HM and PP (representing a di- and monocarboxylate substrate) and are summarized in Table 4. The enzyme was active with both substrates, but the catalytic efficiencies were somewhat less than those of the fused 4-OT. Using 2-HM, the kcat/Km is 4-fold less than that of fused 4-OT (due to the 5.5-fold decrease in the kcat). Using PP, the kcat/Km is 3.4-fold less than that of fused 4-OT (due to the 7-fold decrease in the kcat).
Table 4.
Kinetic Analysis of Unfused 4-OT with 2-HM and PP.a
| Substrate |
kcat (s−1) |
Km (μM) |
kcat/Km (M−1 s−1) |
|---|---|---|---|
| 2-HM | 105 ± 6 | 148 ± 18 | 7.1 × 105 |
| PP | 19.3 ± 0.7 | 346 ± 28 | 5.6 × 104 |
The kinetic parameters were measured using the assay described in the text.
Irreversible Inhibition of Unfused 4-OT by 2-OP.
The reaction of the unfused 4-OT with 2-OP was carried out under the same conditions as those indicated above for fused 4-OT. The reconstructed mass spectrum showed four major signals. Two signals (6579 and 6691 Da) are in a ratio of relative abundance of 1:2, and correspond to the native, unmodified α-subunit (6579 Da) and the α-subunit modified by a single adduct resulting from incubation with 2-OP (6691 Da where the mass difference is 112 Da). (Like the fused 4-OT, treatment of the 2-OP-inactivated unfused 4-OT with NaBH4 did not reduce the adduct.) The signals at 6,599 Da and 6,627 Da correspond to the β-subunit with the initiating methionine and the β-subunit with the initiating N-formyl methionine, respectively. These observations suggest that the prolyl nitrogen of the α-subunit has a low pKa value such that it is largely unprotonated at pH 7.0 (behaving like 4-OT where the pKa of the prolyl nitrogen is 6.4 and the fused 4-OT).5,39 The introduction of NaBH4 raises the pH and likely causes removal of some label. The “blocked” N-terminal proline of the β-subunit precludes modification by 2-OP.
Structural Analysis of the Fused 4-OT.
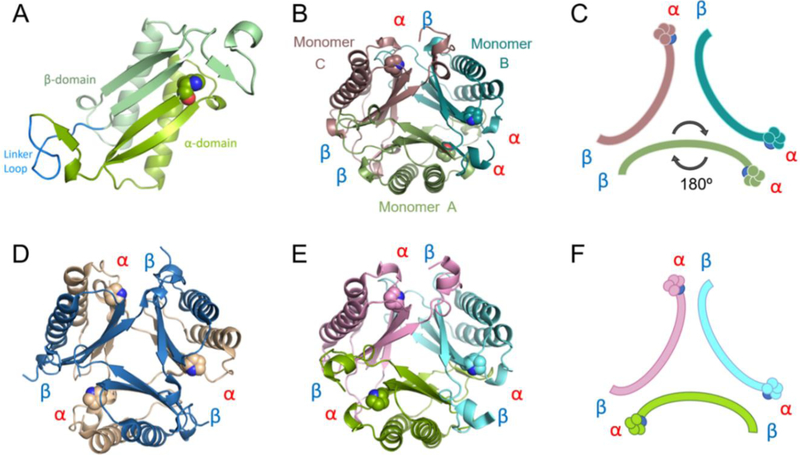
The crystal structure of fused 4-OT was determined by molecular replacement using a monomer from the trimeric cis-CaaD homologue Cg10062 (PDB code: 3N4G) as the search model. The final structure was refined to 1.49 Å resolution (Table 1). Consistent with the dynamic light scattering profile in solution, fused 4-OT exists as a trimer where each monomer consists of two connected β-α-β building blocks (Figure 3A). Throughout this paper, the first building block of fused 4-OT will be referred to as the α-domain (containing residues Pro-1 through Leu-65 in fused 4-OT) and the second building block will be referred to as the β-domain (containing Pro-66 through Arg-127 in fused 4-OT).
Figure 3.
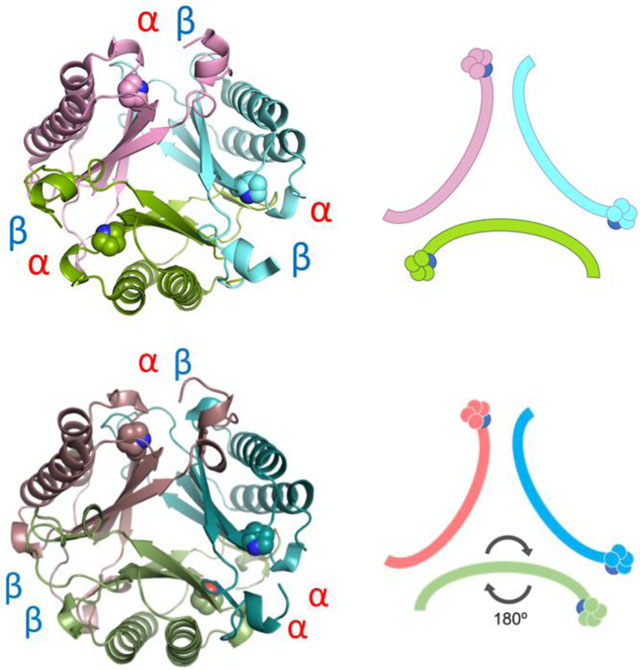
A) Ribbon diagram of one monomer of fused 4-OT. The α- and β-domains are labeled and colored in two shades of green. The linker loop is labeled and colored in bright blue. B) The fused 4-OT trimer, color coded and chains defined: monomer A in green (flipped monomer), monomer B in teal and monomer C in taupe. Interfaces defined between each monomer. C) Schematic of oligomerization arrangement of fused 4-OT. Color scheme is the same as in 3B and the position of Pro-1 is shown. D) Ribbon representation of the C. aurantiacus heterohexamer 4-OT with each interface defined.20 E) Ribbon representation of Linker 2 (a fused 4-OT) with each interface labeled.18 All interfaces are identical alpha-beta interfaces. F) In the same orientation as E, schematic of N-terminal β-strand containing Pro-1 of Linker 2, all monomers are symmetrical.
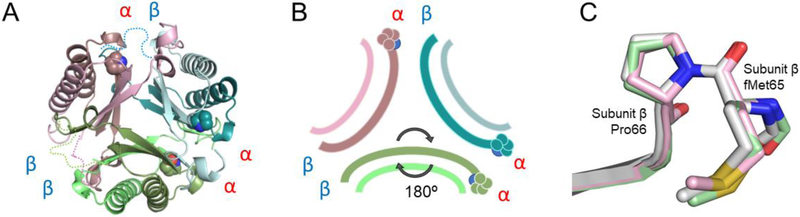
The overall fold of each monomer of fused 4-OT is highly similar to the canonical 4-OT homodimer from P. putida mt-2 with a root-mean-square deviation (RMSD) of 0.69 Å and the heterodimer of the hh4-OT from C. aurantiacus (Figure 3D) with a RMSD of 1.19 Å at the backbone atoms.9,20 The most surprising observation that sets the fused 4-OT apart from all TSF members is that the trimer lacks three-fold rotational symmetry observed in every other known trimeric structure in the TSF thus far.13,18 In this asymmetric trimer, one monomer (designated as monomer A) is “flipped” 180° relative to the other two monomers in the trimer, creating a mirror symmetry with monomer B (monomer A in Figures 3B and 3C). As a result, each protein-protein interface in the trimer is unique and composed of different residues (Figures 3B and 3C). One interface (termed the αβ interface) is formed between the α and β domains from neighboring monomers and has one N-terminal proline residue, as is found in other TSF trimers. The other two interfaces are dramatically different from other TSF trimers due to the asymmetric trimerization. An interface, designated as the αα interface, has two N-terminal proline residues. The other unique interface, the ββ interface, does not have an N-terminal proline residue.
This is the first time such a mode of oligomerization has ever been observed in the TSF. In the 4-OT subgroup, two identical monomers, each made up of a single β-α-β building block, can form a homodimer and these dimers then form a trimer resulting in a homohexameric organization.9,13 Some 4-OT subgroup members form heterohexamers, such as the C. aurianticus 4-OT, which consists of a trimer of heterodimers formed by α and β subunits that vary in sequence, but conserve the TSF building block (Figure 3D).20 The structure of Linker 2 has been reported and it has similar P3 symmetry forming the trimer (Figures 3E and 3F).18 In the other four subgroups, the members are trimers that consist of two consecutively fused β-α-β building blocks.13,18 However, in all cases, the structures have P3 symmetry. The asymmetric trimerization state of fused 4-OT prompted us to explore the factors that might be responsible for such a unique oligomerization state.
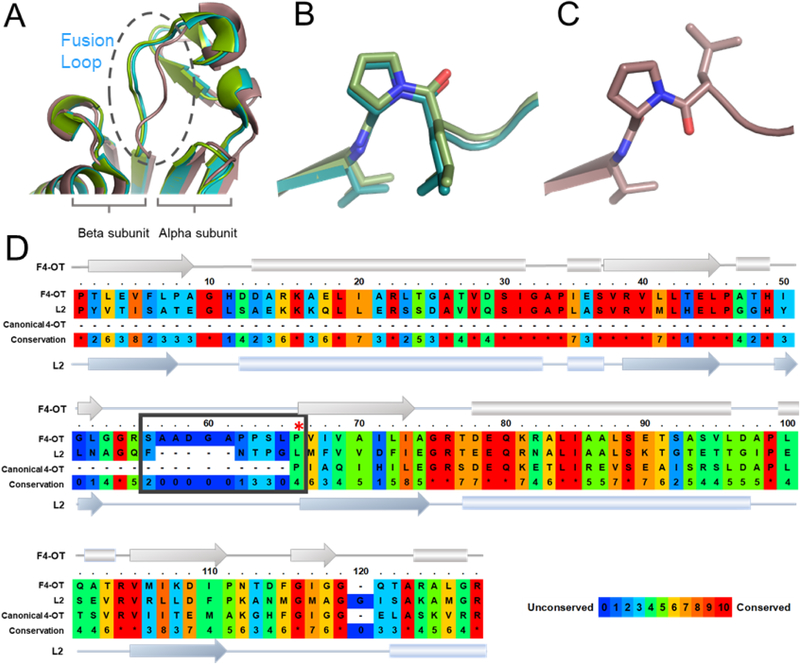
Superimposition of the three monomers of fused 4-OT revealed that the overall folding of the monomers is highly similar except for a 10-amino acid linker loop connecting the α- and β- domains of each monomer (Figure 4A). The linker loops of monomers A (flipped) and B overlay nicely, but monomer C shows a conformational difference at this linker loop (Figure 4A). The structural deviation is greatest near the beginning of the β-domain where the Leu-65/Pro-66 peptide bond is in the cis configuration for both monomer A and monomer B (Figure 4B), but is in the trans configuration for monomer C (Figure 4C). In canonical 4-OT, the monomers of the homohexamer are not covalently restricted. Hence, these observations prompted us to explore whether the linker region, in particular, the cis-prolyl peptide, might specify the oligomerization state of the asymmetric arrangement.
Figure 4.
A) Superimposition of fused 4-OT monomers, close-up to the linker loop region. B) Stick view of L65/P66 motif overlay of monomers A and B showing cis-proline configuration. C) Stick view of L65/P66 motif monomer C showing Pro-66 in the trans configuration. D) Sequence alignment of fused 4-OT, Linker 2, and the canonical 4-OT (labeled as F4-OT, L2, and canonical 4-OT). The secondary structure representation is labeled in the same way. Linker loop is enclosed in a box. A red asterisk shows Pro-1 of canonical 4-OT aligned to Pro-66 of fused 4-OT. The PRALINE sequence alignment tool was used to make this figure.40
The linker region connecting the α and β domains shows a significant difference between the sequences in the fused 4-OT and Linker 2 clusters (Figures 2 and 4D). The sequences for the fused 4-OT cluster (86 total) show a loop of ~10 amino acids between the α- and β-domains. This loop is followed by the β-domain that shows high sequence identity with the canonical 4-OT (Figure 4D). The proline residue whose prolyl nitrogen functions as a nucleophile for canonical homohexamer 4-OT catalysis is also preserved in the β domain of fused 4-OT, although this proline residue (Pro-66) no longer has a free prolyl nitrogen and cannot act as nucleophile. In contrast, the sequences for the members of the Linker 2 cluster show a much shorter linker loop of only 4 amino acids and the β domain does not begin with a proline residue (Figures 2 and 4D). As noted above, the structure of Linker 2 shows P3 rotational symmetry (Figure 3E).18 The differences in the linker loop that connects α and β domains of the fused 4-OTs (length and the existence of cis-prolyl peptide) raised the question of whether this linker governs the oligomerization arrangement of members of fused 4-OT.
To evaluate the effect of this linker loop on the oligomeric state of fused 4-OT, an “unfused” 4-OT was engineered by removing the covalent bond between the α and β domains. This unfused 4-OT decouples the linker loop so that the β subunit (α/β subunits corresponding to the α/β domains of fused 4-OT) now has the N-terminal proline, as is found in both the canonical 4-OT and the hh4-OT. The structure of the unfused 4-OT heterohexamer was determined by x-ray crystallography at a resolution of 1.8 Å. Molecular replacement was carried out using a canonical 4-OT homohexamer with each residue changed to an alanine in the search model. Two hexamers were identified in each asymmetric unit with a Matthews coefficient of 36.7%.41 With the backbone conformation determined, the electron density was calculated to determine the identity of the side chains after being initially modeled as alanines. Since the α- and β-subunits are nearly identical in primary sequence with only 6 residues out of 62 being significantly different, we focused on these residues to establish the identity of the subunit independently using the high resolution diffraction data. The final model of unfused 4-OT is a heterohexamer containing three α-subunits and three β-subunits. The structure reveals the same asymmetric arrangement as was observed in the wild type fused 4-OT with one α-β heterodimer flipped relative to the other two heterodimers (Figures 5A and 5B). The linker loop (residues Ser-56 to Leu-65) is highly flexible so that the electron density is not visible. However, other parts of the structure are almost identical (RMSD of the flipped A dimer to the B dimer is 0.34 Å and C dimer to flipped A dimer is 0.30 Å in the backbone atoms). Interestingly, the initiating N-formyl methionine for the β subunit was not removed and the electron density for this methionine residue is clearly visible in all three β subunits (Figure S3). Furthermore, the prolyl peptide bond connecting the initiating methionine and Pro are all in the cis-configuration (Figure 5C), unlike the trans-proline observed in monomer C of fused 4-OT (Figure 4B). Thus, neither the linker loop nor the proline isomerization state at the β-subunit is responsible for the asymmetric oligomerization.
Figure 5.
A) Ribbon diagram of unfused 4-OT. Heterodimer A colored two shades of green, heterodimer B is colored two shades of teal and heterodimer C colored taupe/pink. The arrangement is an asymmetric trimer of dimers. The interfaces remain αα, αβ, and ββ, as labeled. The missing linker loop for each dimer is shown in dotted lines in corresponding colors. B) Schematic diagram of unfused 4-OT showing the flipped dimer and the corresponding interfaces. C) Overlay of three β-subunits of unfused 4-OT showing stick mode of the first three amino acids. The Leu-Pro sequence in the fused 4-OT is replaced by the initiating methionine followed by proline (Pro-66 in the fused 4-OT), in all β-subunits of the unfused 4-OT. The Met-Pro is in the trans configuration.
Three questions remain. First, what factors are responsible for the asymmetry in the structure of fused 4-OT? One possibility is the specific interactions at the three interfaces. Identification of these interactions can be used to identify other asymmetric trimers in this subgroup of fused 4-OTs, if they exist. This possibility is being explored. A second question is whether the asymmetric trimer has any biological relevance or if it is simply a biological oddity. The actual biological function of the fused 4-OT is not known, but it appears to be part of a degradative pathway for aromatic hydrocarbons based on the similar genomic context to that of the canonical 4-OT. It also functions as a 4-OT using 2-HM (23-fold decrease in kcat/Km vs. that for 4-OT)18 and PP (2-fold decrease in kcat/Km vs. that for 4-OT),18 so that the different active site configurations don’t affect the overall activity. Although the activities for the individual active sites will (almost certainly) be different, it’s not immediately obvious how to measure them or what biological relevance this could have. The final question concerns the evolutionary implications of the fused 4-OTs. The fused 4-OT is a clear representative of what a progenitor for the members of the other four subgroups (CHMI, MIF, cis-CaaD, and MSAD) in the TSF might have looked like. It’s the obvious “jumping-off point”. It would be curious to find out what role the asymmetry played in the process, if any.
Supplementary Material
ACKNOWLEDGEMENTS
The protein mass spectrometry analysis was conducted in the Institute for Cellular and Molecular Biology Protein and Metabolite Analysis Facility at the University of Texas at Austin. Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at the University of Texas at Austin. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We are indebted to Steve D. Sorey (Department of Chemistry, University of Texas at Austin) for his expert assistance in the acquisition of the 1H NMR spectra and to Dr. Tamer S. Kaoud for his assistance with the light scattering experiments.
Funding
This research was supported by the National Institutes of Health Grant (GM-129331 to CPW, YJZ, and PCB, GM-41239 to CPW, GM-104896 to YJZ, and GM-60595 to PCB) and the Robert A. Welch Foundation (F-1334 to CPW and F-1778 to YJZ).
ABBREVIATIONS
- CHM
5-(carboxymethyl)-2-hydroxymuconate
- CHMI
5-(carboxymethyl)-2-hydroxymuconate isomerase
- cis-CaaD
cis-3-chloroacrylic acid dehalogenase
- ESI-MS
electrospray ionization mass spectrometry
- hh4-OT
heterohexamer 4-oxalocrotonate tautomerase
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid
- 2-HM
2-hydroxymuconate
- HPD
2-hydroxy-2,4-pentadienoate
- Kn
kanamycin
- LB
Luria-Bertani
- MIF
macrophage migration inhibitory factor
- MSAD
malonate semialdehyde decarboxylase
- 5-Me-2-HM
5-(methyl)-2-hydroxymuconate
- MR
molecular replacement
- MAD
multi-wavelength anomalous dispersion
- MSA
multiple sequence alignment
- 2-OP
2-oxo-3-pentynoate
- 4-OT
4-oxalocrotonate tautomerase
- PP
phenylenolpyruvate
- PEG
polyethylene glycol
- PCR
polymerase chain reaction
- RMSD
root-mean-square deviation
- SSN
sequence similarity network
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SFLD
Structure-Function Linkage Database
- TSF
tautomerase superfamily
Footnotes
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank: PDB entry 6BLM for fused 4-OT and 6OGM for unfused 4-OT.
SUPPLEMENTARY INFORMATION
Supporting Information Available: This material is available free of charge via the Internet. Sequence similarity networks for the Level 2 4-OT subgroup containing the canonical 4-OT and representative omit map of unfused 4OT β-domain.
The authors declare no competing financial interest.
REFERENCES
- (1).Whitman CP, Aird BA, Gillespie WR, and Stolowich NJ (1991) Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J. Am. Chem. Soc 113, 3154–3162. [Google Scholar]
- (2).Dagley S (1978) Pathways for the utilization of organic growth substrates, in The Bacteria: A Treatise on Structure and Function (Ornston LN and Sokatch JR, Eds), pp. 305–388, Academic Press, New York. [Google Scholar]
- (3).Harayama S, Rekik M, Ngai K-L, and Ornston LN (1989) Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida, J. Bacteriol 171, 6251–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Stivers JT, Abeygunawardana C, Mildvan AS, Hajipour G, Whitman CP, and Chen LH (1996) Catalytic role of the amino-terminal proline in 4-oxalocrotonate tautomerase: affinity labeling and heteronuclear NMR studies. Biochemistry 35, 803–813. [DOI] [PubMed] [Google Scholar]
- (5).Stivers JT, Abeygunawardana, Mildvan AS, Hajipour G, and Whitman CP (1996) 4-Oxalocrotonate tautomerase: pH dependences of catalysis and pKa values of active site residues. Biochemistry 35, 814–823. [DOI] [PubMed] [Google Scholar]
- (6).Harris TK, Czerwinski RM, Johnson WH Jr., Legler PM, Abeygunawardana C, Massiah MA, Stivers JT, Whitman CP, and Mildvan AS (1999) Kinetic, stereochemical, and structural effects of mutations of the active site arginine residues in 4-oxalocrotonate tautomerase. Biochemistry 38, 12343–12357. [DOI] [PubMed] [Google Scholar]
- (7).Czerwinski RM, Harris TK, Johnson WH Jr., Legler PM, Stivers JT, Mildvan AS, and Whitman CP (1999) Effects of mutations of the active site arginine residues in 4-oxalocrotonate tautomerase on the pKa values of active site residues and on the pH dependence of catalysis. Biochemistry 38, 12358–12366 [DOI] [PubMed] [Google Scholar]
- (8).Czerwinski RM, Harris TK, Massiah MA, Mildvan AS, and Whitman CP (2001) The structural basis for the perturbed pKa of the catalytic base in 4-oxalocrotonate tautomerase: kinetic and structural effects of mutations of Phe-50. Biochemistry 40, 1984–1995. [DOI] [PubMed] [Google Scholar]
- (9).Taylor AB, Czerwinski RM, Johnson WH Jr., Whitman CP, and Hackert ML (1998) Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 Å resolution: analysis and implications for the mechanism of inactivation and catalysis. Biochemistry 37, 14692–14700. [DOI] [PubMed] [Google Scholar]
- (10).Chen LH, Kenyon GL, Curtin F, Harayama S, Bembenek ME, Hajipour G, and Whitman CP (1992) 4-Oxalocrotonate tautomerase, an enzyme composed of 62 amino acid residues per monomer, J. Biol. Chem 267, 17716–17721. [PubMed] [Google Scholar]
- (11).Burks EA, Yan W, Johnson WH Jr., Li W, Schroeder, Min C, Gerratana B, Zhang Y, and Whitman CP (2011) Kinetic, crystallographic, and mechanistic characterization of TomN: elucidation of a function for a 4-oxalocrotonate tautomerase homologue in the tomaymycin biosynthetic pathway. Biochemistry 35, 7600–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Murzin AG (1996) Structural classification of proteins: new superfamilies. Curr. Opin. Struct. Biol 6, 386–394. [DOI] [PubMed] [Google Scholar]
- (13).Poelarends GJ, Veetil VP, and Whitman CP (2008) The chemical versatility of the β-α-β fold: Catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cell. Mol. Life Sci 65, 3606–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Subramanya HS, Roper DI, Dauter Z, Dodson EJ, Davies GJ, Wilson KS, and Wigley DB (1996) Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry 35, 792–802. [DOI] [PubMed] [Google Scholar]
- (15).Bloom J, Sun S, and Al-Abed Y (2016) MIF, a controversial cytokine: a review of structural features, challenges, and opportunities for drug development. Expert Opin. Ther. Targets 20, 1463–1475. [DOI] [PubMed] [Google Scholar]
- (16).Poelarends GJ, Serrano H, Person MD, Johnson WH Jr., Murzin AG, and Whitman CP (2004) Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the mechanistic, structural, and evolutionary relationship between isomer-specific 3-chloroacrylic acid dehalogenases. Biochemistry 43, 759–772. [DOI] [PubMed] [Google Scholar]
- (17).Poelarends GJ, Johnson WH Jr., Murzin AG, and Whitman CP (2003) Mechanistic characterization of a bacterial malonate semialdehyde decarboxylase: identification of a new activity on the tautomerase superfamily. J. Biol. Chem 278, 48674–48683. [DOI] [PubMed] [Google Scholar]
- (18).Davidson R, Baas B-J, Akiva E, Holliday G, Polacco BJ, LeVieux JA, Pullara CR, Zhang YJ, Whitman CP, and Babbitt PC (2018) A global view of structure-function relationships in the tautomerase superfamily, J. Biol. Chem 293, 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).LeVieux JA, Baas B-J, Kaoud TS, Davidson R, Babbitt PC, Zhang YJ, Whitman CP, Kinetic and structural characterization of a cis-3-chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily, Arch Biochem Biophys, 636, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Burks EA, Yan W, Johnson WH Jr., Li W, Schroeder GK, Min C, Gerratana B, Zhang Y, and Whitman CP (2011) Kinetic, crystallographic, and mechanistic characterization of TomN: elucidation of a function for a 4-oxalocrotonate tautomerase homologue in the tomaymycin biosynthetic pathway, Biochemistry 35, 7600–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Johnson WH Jr., Czerwinski RM, Fitzgerald MC, and Whitman CP (1997) Inactivation of 4-oxalocrotonate tautomerase by 2-oxo-3-pentynoate. Biochemistry 36, 15724–15732. [DOI] [PubMed] [Google Scholar]
- (22).Waddell WJ (1956) A simple ultraviolet spectrophotometric method for the determination of protein. J. Lab. Clin. Med 48, 311–31 [PubMed] [Google Scholar]
- (23).Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- (24).Wang SC, Johnson WH Jr., and Whitman CP (2003) The 4-oxalocrotonate tautomerase- and YwhB-catalyzed hydration of 3E-haloacrylates: implications for the evolution of new enzymatic activities. J. Am. Chem. Soc 125, 14282–14283. [DOI] [PubMed] [Google Scholar]
- (25).Kaoud TS, Devkota AK, Harris R, Rana MS, Abramczyk O, Warthaka M, Lee S, Girvin ME, Riggs AF, and Dalby KN (2011) Activated ERK2 is a monomer in vitro with or without divalent cations and when complexed to the cytoplasmic scaffold PEA-15. Biochemistry 50, 4568–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Barber AE 2nd, and Babbitt PC (2012) Pythoscape: a framework for generation of large protein similarity networks. Bioinformatics 28, 2845–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ho SN, Hunt HD, Horton RM, Pullen JK, and Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- (29).Wang SC, Person MD, Johnson WH Jr., and Whitman CP (2003) Reactions of trans-3-chloroacrylic acid dehalogenase with acetylene substrates: consequences of and evidence for a hydration reaction, Biochemistry 42, 8762–8773. [DOI] [PubMed] [Google Scholar]
- (30).Otwinowski Z, and Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- (31).McCoy A (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr., Sect D: Biol. Crystallogr 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung L-W, Read RJ, and Adams PD (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr., Sect D: Biol. Crystallogr 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr., Sect D: Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr., Sect D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect D: Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).DeLano WL (2002) The PyMol molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- (37).Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004) WebLogo: A sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hirel PH, Schmitter MJ, Dessen P, Fayat G, and Blanquet S (1989) Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A 86, 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Baas B-J, Zandvoort E, Wasiel AA, Quax WJ, and Poelarends GJ (2011) Characterization of a newly identified mycobacterial tautomerase with promiscuous dehalogenase and hydratase activities reveals a functional link to a recently diverged cis-3-chloroacrylic acids dehalogenase. Biochemistry 50, 2889–2899. [DOI] [PubMed] [Google Scholar]
- (40).Dijkstra M Bawono P, Abeln S, Feenstra KA, Fokkink W, and Herings J (2018) Motif-aware PRALINE: Improving the alignment of motif regions. PLoS Comput Biol 14(11):e1006547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Matthews BW (1968) Solvent content of protein crystals. J. Mol. Biol 33, 491–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.