Supplemental Digital Content is available in the text
Keywords: enterocolitis, extremely low birth weight, gastrointestinal microbiome, neonatal intensive care units, premature infant
ABSTRACT
Objectives:
Microbial communities influencing health and disease are being increasingly studied in preterm neonates. There exists little data, however, detailing longitudinal microbial acquisition, especially in the most extremely preterm (<26 weeks’ gestation). This study aims to characterize the development of the microbiota in this previously under-represented cohort.
Methods:
Seven extremely preterm infant-mother dyads (mean gestation 23.6 weeks) were recruited from a single neonatal intensive care unit. Oral and endotracheal secretions, stool, and breast milk (n = 157 total), were collected over the first 60 days of life. Targeted 16S rRNA gene sequencing identified bacterial communities present.
Results:
Microbiota of all body sites were most similar immediately following birth and diverged longitudinally. Throughout the sampling period Escherichia, Enterococcus, Staphylococcus, and an Enterobacteriaceae were dominant and well dispersed across all sites. Temporal divergence of the stool from other microbiota was driven by decreasing diversity and significantly greater proportional abundance of Bifidobacteriaceae compared to other sites.
Conclusions:
Four taxa dominated all anatomical sampling sites. Rare taxa promoted dissimilarity. Cross-seeding between upstream communities and the stool was demonstrated, possibly relating to buccal colostrum/breast milk exposure and indwelling tubes. Given the importance of dysbiosis in health and disease of extremely preterm infants, better understanding of microbial acquisition within this context may be of clinical benefit.
What Is Known
Microbial colonization in early life can affect later health status with preterm infants at greater risk of microbially impacted disease states such as necrotizing enterocolitis and late-onset sepsis.
Preterm infants within the neonatal intensive care unit do not face typical colonization exposures.
Previous comparative studies show conflicting findings with either individual patient in closer-to-term infants, or sampling site in more preterm infants explaining greatest difference between microbial communities.
What Is New
This study represents the first to characterize microbial acquisition and development exclusively in a cohort of extremely preterm infants born <24 weeks gestational age.
Development of microbial communities in intubated, extremely preterm infants occurs holistically, across all body-sites with longitudinal specialization toward divergent communities.
Breast milk and oral and endotracheal microbiota shows decreasing associations with stool communities over time, with oral communities exhibiting greatest impact and substantial environmental influence.
The 24 months after birth represent a period of turbulent microbiota development as neonates transition to a more stable state (1,2). This turbulence is even more apparent in preterm infants at risk of disease states such as late-onset infection and necrotizing enterocolitis (NEC).
Neonatal exposures essential to preterm infant care impact microbiota development. Exposures include increased time to full milk feeds, parenteral nutrition, administration of antibiotics or antifungals (3–6), and use of indwelling medical devices (7). Extremely preterm infants also have a naïve immune system and intestinal function (8). All such factors contribute to a proinflammatory state (9), increasing likelihood of NEC development. In addition, indwelling medical devices increase the likelihood of biofilm formation which may provide a source of persistent, pathogenic, antibiotic resistant bacteria (10).
Previous studies in more mature infants have identified individual-specific signatures, irrespective of body-site sampling (11,12). This is in agreement with the findings of the human microbiome project, which included exclusively term infants (13). Few studies have attempted to identify environmental sources of microbial colonization in preterm infants (14,15), although results highlight similarities between ward surfaces and infants’ microbiota.
Typically, studies have used stool as a proxy for the gut lumen to compare microbiota between delivery modes (16), and in cases of NEC (17), and late-onset sepsis (LOS) (18), increasing our understanding of the gut microbiota in health and disease states. This study explores further the acquisition of the gut microbiota by supplementing stool microbial profiling with identification of exclusively viable bacterial communities in several other sampling sites. These include oral (ORS), and endotracheal (ETS), samples, as well as maternally expressed breast milk (EBM), samples taken from the feeding tubes to generate a more holistic view of microbiota development and track succession of microbiota acquisition throughout the gastrointestinal tract. Our sample salvaging method facilitated use of suctioned samples from these areas, performed as part of routine clinical care. We focused on the most immature infants since they are currently least well represented in the literature and most vulnerable to the preterm associated diseases NEC and LOS. This cohort includes both infants who developed these diseases and those who did not.
METHODS
Sample Collection
Longitudinal samples from 4 distinct sites (oral and endotracheal secretions, maternal breast milk from feeding tubes and stool) were collected from a cohort of extremely preterm (<26 weeks gestational age (GA), at birth), infant-mother dyads (n = 7), meeting the inclusion criteria (Methods, SDC1, Supplemental Digital Content), including 1 twin pair, over the first 60 days of life (figure, SDC2, Supplemental Digital Content). All were cared for simultaneously in the neonatal intensive care unit (NICU), at the Royal Victoria Infirmary, Newcastle upon Tyne, England, ventilated from birth and received breast milk feeds. The NICU employs standardized approaches to nutrition including administration of maximal amounts of maternal EBM progressed at standard rates as far as infant tolerance permits and standardized management of sepsis. Infants routinely received probiotic supplementation consisting of a liquid preparation of Lactobacillus acidophilus, Bifidobacterium bifidum, and B infantis (Labinic, Biofloratech, UK), as soon as minimal enteral nutrition was tolerated (Fig. SDC3, Supplemental Digital Content). Samples were collected under ethical permission granted by NRES Committee North East—Newcastle & North Tyneside 2 (10/H0908/39), and analysed with written parental consent.
Maternal EBM collected from either the nasogastric feeding tube or from residual volume in bottles and syringes was collected into sterile cryovials. Oral and endotracheal secretions were collected in a sterile catheter via routine suction when clinically indicated. Stool samples were collected from the nappy in a sterile glass pot. Sample collections were opportunistic rather than predefined. All samples were stored frozen at −20°C until transfer to −80°C, usually within 7 days, until processing.
Relevant clinical details were collected from each infant's medical notes and are displayed in Table 1. NEC and sepsis were diagnosed and categorized by agreement of senior research clinicians by reviewing clinical, laboratory, x-ray, and operative findings.
TABLE 1.
Patient Demographics
| Gestational age | Birth weight | Mode of delivery | DoL full feeds | DoL probiotic start | Breast milk fortifier start (DoL) | Key disease | DoL NEC onset | DoL +ve BC | Antibiotics (DoL administered) | Total days Abx administered | Survival to discharge | ||
| Patient | |||||||||||||
| 1 | 24+3 | 600 | Vaginal | 11 | 4 | 56 | Medical NEC | 41 | - | 0,9,38,40,41 | 23 | Yes | |
| 2 | 25+2 | 700 | Vaginal | 14 | 7 | 21 | - | - | - | 0,4,14,26 | 14 | Yes | |
| 3 | 23+3 | 520 | Vaginal | 13 | 3 | 17 | - | - | - | 0,3,18,41,51 | 23 | Yes | |
| 4 | 23+2 | 580 | Vaginal | 10 | 4 | 44 | Sepsis | - | 40 | 0,40 | 16 | Yes | |
| 5 | 23+1 | 520 | Vaginal | 50 | 3 | NR | Surgical NEC, sepsis | 14 (op18) | 23 | 0,7,14,15,18,25,37,40 | 38 | No | |
| 6 | 23+1 | 590 | Vaginal | 21 | 7 | 40 | - | - | - | 0,25,37 | 41 | Yes | |
| 7 | 23+4 | 500 | Caesarean | 15 | 6 | NR | Sepsis | - | 7 | 0,7,15 | 9 | Yes | |
| Mean | 23.4 | 573 | |||||||||||
Patient demographic data for all 7 patients sampled longitudinally in this study. Averages for continuous variables are expressed as the mean. Abx = antibiotics, BC = Blood culture, DoL = Day of life, +ve = Positive, NEC = Necrotising Enterocolitis, NR = not received, op = operated, dashes used to represent no diagnosed diseases.
Propidium Monoazide Treatment and DNA Extraction
Samples were processed within randomized batches to reduce confounding effects of contaminant DNA. All samples were treated with propidium monoazide to exclude nonviable bacterial cell DNA before extraction of microbial DNA, as previously described (19). Microbial DNA was extracted from breast milk, oral and endotracheal suction samples using QIAGEN PowerFood DNA Microbial Isolation Kits (Hilden, DE), and from stool samples using QIAGEN PowerLyzer PowerSoil DNA Microbial Isolation Kits. All DNA extractions were done as per manufacturers’ instructions but supplemented with a prolonged bead beating step, to ensure lysis of Gram-positive bacteria.
Library Preparation and Sequencing
Because of high variability in extracted bacterial DNA yield (figure, SDC4, Supplemental Digital Content), presequencing normalization of sample DNA content was performed by quantitative PCR (Methods, SDC5, Supplemental Digital Content). Following normalization, nested, full-length 16S rRNA gene PCR was performed to maximize nonviable DNA exclusion (19).
Sequencing libraries were prepared for targeted sequencing of the V4 region of the 16S rRNA gene from PCR amplicons as per the Schloss standard operating procedure (SOP), (20), using primers 515F and 806R (21), (table, SDC6, Supplemental Digital Content). Libraries were sequenced on the Illumina MiSeq platform (CA), using V2 (2 × 250), chemistry.
Paired end reads were trimmed, merged, and processed by alignment to the SILVA database, followed by de novo clustering in to operational taxonomic units, and taxonomic assignment in Mothur, following the MiSeq SOP (20).
Extraction kit and sequencing negative controls were prepared and sequenced simultaneously with all samples (Methods, SDC7, Supplemental Digital Content).
Statistical Analysis
To compare microbial community compositions alpha and beta diversity indices (Shannon diversity, weighted Bray-Curtis dissimilarity), were calculated using the vegan community ecology package in R (22). Kruskal-Wallis rank-sum test and pairwise Mann-Whitney-Wilcoxon test were employed to compare means of continuous data. Adonis PERMANOVA was used to identify metadata variables impacting microbiota composition. Multivariate comparisons were performed by pairwise PERMANOVA. To stratify dominant and rare taxa within sampling site communities, distribution abundance relationships were calculated (23) (methods explained in figure, SDC8, Supplemental Digital Content). Differentially abundant bacteria between anatomical sites were identified by LEfSe. Influence of the microbiota of upstream anatomical sites on that of the stool was performed using SourceTracker(24), and linear regressions of bacterial relative abundances. Bonferroni correction was applied during pairwise comparisons across all sites.
RESULTS
Patient Demographics
EBM (n = 40), oral secretions (ORS: n = 40), endotracheal secretions (ETS: n = 35), and stool (n = 38), samples were analysed. Both mean birth weight (584 g, standard deviation [SD] = 66 g), and mean gestational age (23.6 weeks, SD = 0.8 weeks), were lower than in previous preterm infant microbiota studies (11,15,17,18,25). Two patients developed NEC, 3 patients developed LOS, and 3 patients developed neither. Patient demographics are described in Table 1.
Sequence Data Analysis
A total of 7.74 × 106 sequence reads from 3109 operational taxonomic units were observed in 153 PMA-treated samples (mean reads per sample = 4.90 × 104, SD = 5.91 × 104). Filtering of any taxa not classified to at least the class level and 86 potential contaminant taxa yielded 6.72 × 106 reads (mean per sample = 4.39 × 104, SD = 4.72 × 104) (table, SDC9, Supplemental Digital Content). Remaining reads were normalized by expression of total counts as relative abundances per sample. Comparisons of results obtained from PMA-treated samples with non-PMA-treated samples are available as supplementary materials (table, SDC10; Figure, SDC11, Supplemental Digital Content).
Stool Microbiota Diverges From Upstream Microbiota Over Time
Longitudinal progression of microbiota composition was observed, with multiple shared taxa observed between different patients (Fig. 1).
FIGURE 1.
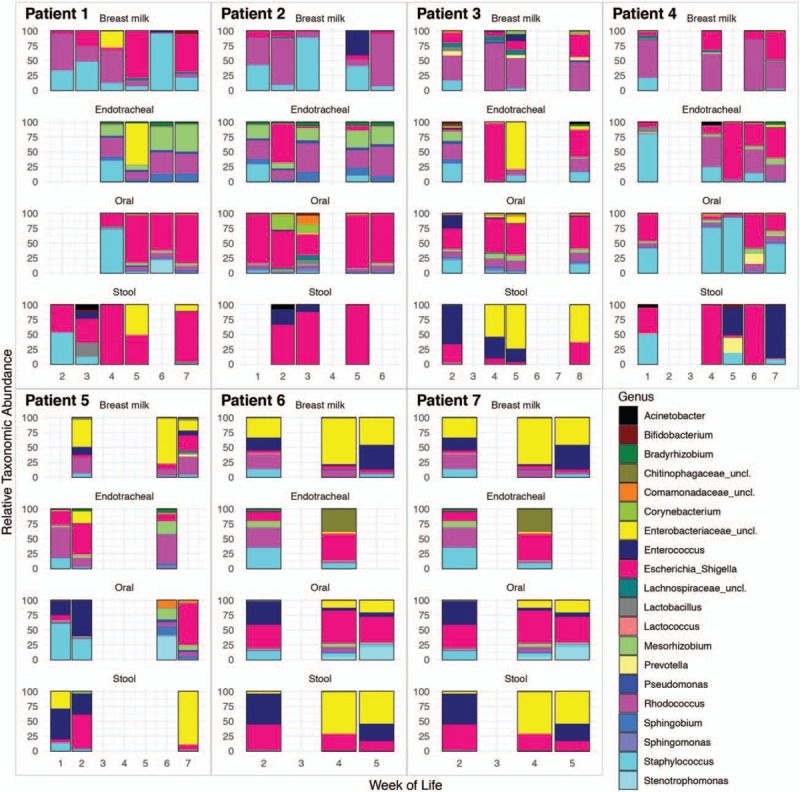
Longitudinal microbiota development of the top 20 most abundant bacterial taxa across all patients and body sites on a weekly scale. Where multiple samples were collected in the same week relative abundances were summed before rescaling.
Over the first 60 days of life a general increasing trend in alpha diversity was observed in all upstream sites, but stool diversity decreased over this time (Fig. 2A and B). During the first 6 weeks of life all body site microbiota showed similar diversity, but by week of life (WoL) 7 and 8 significant differences were observed in diversity between the breast milk, oral (P = 0.05), and stool (P = 0.001) communities (table, SDC12, Supplemental Digital Content). Stool microbiota showed significantly lower bacterial alpha diversity than both breast milk and endotracheal communities when all time points were considered (P < 0.05).
FIGURE 2.
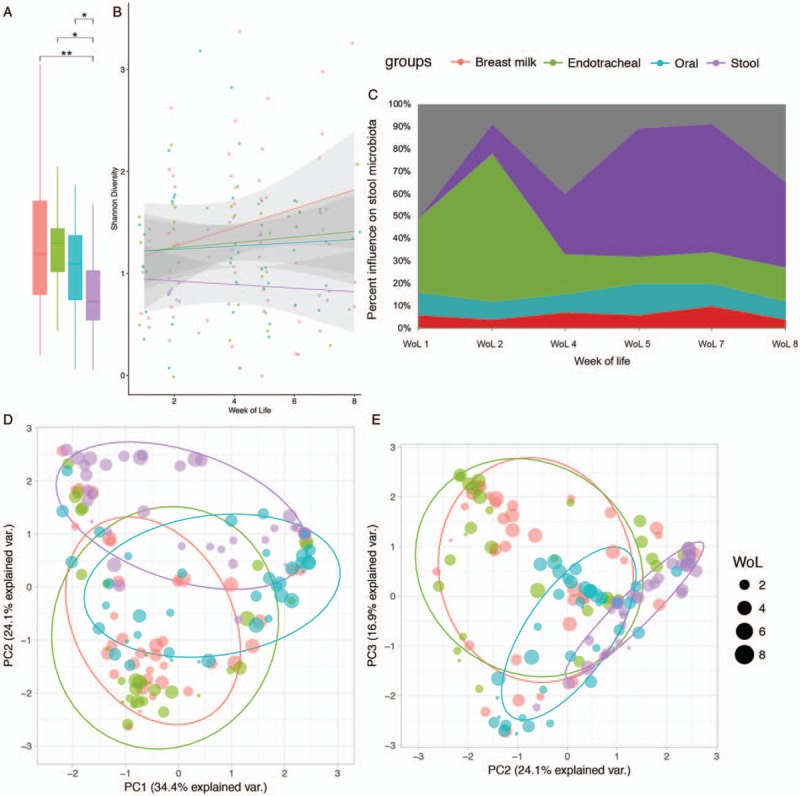
Longitudinal community differences between breast milk (red), oral (blue), endotracheal (green), and stool (purple), microbiota. Stool was significantly less diverse (Shannon diversity), than breast milk (MWW: P.adj = 0.009), endotracheal (MWW: P.adj = 0.014), and oral (MWW: P.adj = 0.024) communities (A), with longitudinally decreasing diversity (B). Potential sources of the stool microbiota, including unknown sources (grey), as calculated by SourceTracker are illustrated longitudinally (C). Beta diversity (Bray-Curtis dissimilarity) between microbiota is illustrated in ordinations (D, E), and explains 74.5% of sample variance. Point size in ordinations depicts week of life. Results of the respective pairwise PERMANOVA are available online (table, SDC13, Supplemental Digital Content). PC = principle component; WoL = week of life.
At earlier time points the microbiota of upstream sampling sites and stool show much greater similarity (combined similarity with stool: 49% at WoL 1 and 2), than at later time points (combined similarity with stool: 27% at WoL 8) (Fig. 2C). Over time stool microbiota showed increasing similarity to previous stools (13% at WoL 2; 57% at WoL 5 and 7). The oral microbiota was the most similar to the stool microbiota, with an average similarity of 26% compared to breast milk (6%), and endotracheal samples (10%). Unknown, likely environmental, sources, also play a substantial role in shaping the stool microbiota, comprising as much as 51% at WoL 1. Established stool microbiota had greater average similarity (32%), than any upstream or unknown sources. The greater similarity of upstream and unknown sources in the immediate neonatal period suggests community plasticity diminishes with time and may have important implications for timing of interventions designed to influence the microbiota.
Substantial overlap of community composition was observed between separate sampling sites (Fig. 2D/E). Greatest overlap between site microbiota was observed between breast milk/feeding tube and endotracheal communities, suggesting seeding of the respiratory microbiome by breast milk (Figure SDC14, Supplemental Digital Content).
Adonis PERMANOVA based on Bray-Curtis dissimilarity, identified both sampling site (R2 = 14%, P = 0.001), and day of life (R2 = 18%, P = 0.02), as the main individual factors impacting microbiota dissimilarity (table, SDC10, Supplemental Digital Content). When combined these 2 factors demonstrated the greatest impact on community composition (R2 = 30%, P = 0.04) (table, SDC10, Supplemental Digital Content). Despite small sample sizes, NEC and LOS had an insignificant impact on community composition when stratified by sampling site (R2 = 1%, P = 0.04), suggesting no difference was observed within site microbiota between diseased babies and controls.
Few Dominant Taxa Are Well Dispersed Across All Sampling Sites
Four dominant taxa were shared across all sampling sites: Escherichia, Enterococcus, Staphylococcus, and an unclassified Enterobacteriaceae. These 4 conserved dominant taxa were the only dominant taxa found in the stool microbiota. Significant positive relationships identified between distribution and abundance (P < 0.001), of bacterial taxa enabled classification of all taxa in the upper quantile of occupancy as dominant and all other taxa as rare community members (figure, SDC8, Supplemental Digital Content).
Multiple other taxa were shared between sampling sites (Fig. 1), with a Rhodococcus spp. identified by both culture and sequencing as dominant in all upstream mucosal sites despite not having been previously associated with such environments.
Homogenizing Effects of Dominant Taxa Masks Differential Influence of Rare Community Members
The prevalence and abundance of the 4 dominant taxa within all anatomical sites obscured the divergent impact of the rare taxa within the microbiota. Despite the apparent overlap in microbiota observed by ordination, significant differences between all anatomical site communities were identified by pairwise PERMANOVA (P.adj = 0.006 [table, SDC13, Supplemental Digital Content]).
Stratification of the microbiota into common and rare communities enabled identification of rare features of the microbiota responsible for the significant dissimilarity observed between anatomical sites. LEfSe analysis (Fig. 3), identified proportionally greater Actinobacteria (P < 0.001), specifically the Nocardiaceae (P < 0.001) in breast milk. The Rhodococcus spp., already identified as being ubiquitous throughout upstream mucosal samples in this study belongs to the family Nocardiaceae. The absence of Rhodococcus from stool microbiota indicates that it is unable to survive passage through either the (acidic) stomach or the hypoxic gut.
FIGURE 3.
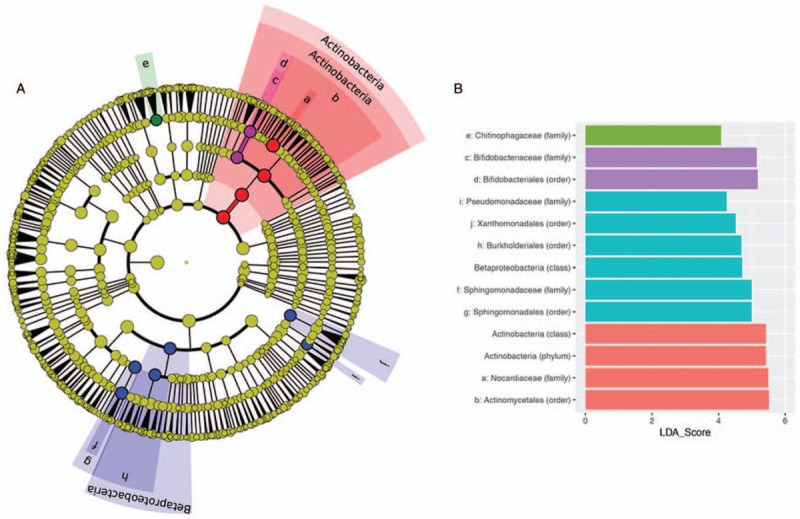
Results of LEfSe analysis, identifying the discriminative rare taxonomic features between the 4 sampling sites investigated (A), and describing the effect size of each discriminative feature (B). The cladogram is rooted at the kingdom level and discriminative features identified following removal of 4 dominant common taxa are coloured by site as described in Figure 2. Where space was not available to plot the taxon name on the cladogram (A) a letter was assigned. These correspond to the taxa, as detailed in (B).
The obligate anaerobic family of Bifidobacteriaceae were observed in significantly greater proportional abundance (P < 0.001) in the anaerobic gut environment than the aerobic environments of the upper sampling sites or breast milk (Fig. 3). The greater proportional abundance of Bifidobacteriaceae in the gut may be due to the administration of probiotics to the patients studied here and specialization of the genus for colonization of the human milk oligosaccharide rich, hypoxic environment of the infant gut.
DISCUSSION
The majority of studies in extremely preterm infants explore the relationship between gut microbiota and disease states (17,18), nosocomial surfaces (14,15,26), or maternal breast milk (27,28). None have previously focused exclusively on such an extremely premature infant cohort (11,15,17,18,25,29). To our knowledge this is the first study to characterize microbial acquisition in a cohort of infants with a mean GA <24 weeks. This study also represents the first to utilize viability determination to explore holistic gastrointestinal microbiota assembly. We deliberately included both clinically well infants and those with LOS and NEC since up to 1 in 3 of extremely preterm infants will experience these complications of prematurity.
Despite considerable overlap between the gut microbiota and upstream sites, divergence is already apparent within the first 60 days of life. We highlight strong homogenizing influences of 4 dominant taxa, which persist from initial sampling, throughout this study. These results reflect the findings of previous studies (11,12), which describe ubiquitous communities across preterm anatomical sites, and the Human Microbiome Project, which observed patient-specific microbial signatures in healthy, term infants (13). In contrast, but in agreement with Costello et al (30), we observed sitewise and longitudinal differences to be the major determinant of variation between extremely preterm infant microbiota.
The longitudinal sampling performed in this study highlights site microbiota are more similar during early life and diverge over time, thus suggesting bacterial seeding at birth is “body wide,” not site specific. Individual site microbial communities then evolve toward adult-like profiles over time. We build upon the work of previous studies by demonstrating the potential of rare members of the microbiota to drive processes of environmental selection, expanding divergence between patient and site microbiota.
This study shows anatomical sites in closer proximity share more similar bacterial communities than those anatomically disparate from one another. Indeed, breast milk and endotracheal microbiota shared the greatest similarity in abundance of individual taxa, whereas the least similarity was observed between the microbiota of breast milk and stool. Transit through the acidic stomach environment may be a factor in the selectivity and limited dispersal from the upstream sites to the lower gut microbiota. This could also explain the greater observed similarity between the sites at earlier time points (31).
The 4 taxa conserved between all body sites (Enterococcus, Enterobacteriaceae, Staphylococcus, and Escherichia), were the only dominant taxa identified in the stool by distribution abundance relationship and largely responsible for observed homogenisation between sample microbiota. This may suggest a bi-directional translocation of bacteria between upstream and downstream gastrointestinal environments, potentially contributed to through NICU practices and handling by parents and staff (26). Moossavi et al (32) recently demonstrated that indirect breast-feeding may be linked with greater abundance of environmental microbes such as Enterobacteriaceae and Enterococcaceae. Given NICU feeds are all delivered indirectly this provides an explanation for the dominance of such bacteria observed in this study.
The large impact of unknown sources on stool microbiota composition identified here suggests the sampling strategy employed failed to capture all possible influences of the infant gut microbiota. Likely unsampled sources of bacteria are well summarized in a review by Hartz et al (26), including care givers and their equipment, maternal and paternal skin, and nosocomial surfaces.
Stratification of communities in to common and rare members highlighted several discriminative taxa between sampling site microbiota. More specifically, the Nocardiaceae family exhibited differentially greater relative abundance in breast milk samples. This is of interest because of the prevalence of Rhodococcus within the upstream microbiota of all infants despite no previous identification of this.
To our knowledge, Rhodococcus has not been previously reported in breast milk, nor is it archived within the Human Oral Microbiome Database. Rhodococcus is usually associated with the environment (33,34), although member species have been identified as potential human pathogens (35–37), especially in immunocompromised individuals, with whom immunologically naïve extremely preterm infants may share similar features.
A 737 bp sequence of DNA extracted from isolated colonies cultured from breast milk showed greatest similarity to a type strain of Rhodococcus quingshengii via Nucleotide BLAST (Methods, SDC7, Supplemental Digital Content). Rhodococcus quingshengii has previously been shown to have azole degrading capacity (38). Fluconazole is a triazole antifungal agent routinely orally delivered in this NICU for prophylactic prevention of candidiasis (39). Failure to isolate Rhodococcus from the probiotics suggests it is not a contaminant of the probiotic supplements. Instead, isolation from breast milk fed to these infants through in situ feeding tubes highlights the need for further studies to explore links between Rhodococcus, antifungal prophylaxis, and use of in-situ feeding tubes in preterm infants.
As with any observational study there exist several limitations to these findings. Firstly, patient numbers are relatively small. This is due to the high rates of morbidity and mortality of such an extremely premature infant cohort (mean GA = 23.4 weeks; mean BW = 576 g), which restricted the number of patients able to provide sufficient samples to fulfil the inclusion criteria. In addition, the opportunistic sampling employed restricts the power of comparisons regarding longitudinal microbiota development. Systematic sampling may facilitate further insights in future studies.
Importantly, this study represents the first to exclusively characterize such an extremely preterm population. Although nuances of care and individual circumstances dictate there may not be a “typical” infant born <24 weeks GA, we demonstrate within individual hospital wards there may be a typical pattern of colonization, characterized initially by few dominant bacterial genera and temporally developing dissimilarity driven by rare community members.
Larger cohort studies would help validate and further explore these findings. Inclusion of neonates from multiple NICUs, exposed to different clinical approaches, such as different approaches to probiotic and nutritional care may substantially impact the results and should be explored further. Greater sequencing resolution may also elucidate directionality of microbial transfer. Access to resected gut lumen tissue would enable further validation of the “multiple-site” hypothesis. All these form part of our ongoing programme of work.
The gestational age and longitudinal sampling of infants in this study are unique in the current literature, despite extremely preterm infants being the most sick and complex. This study demonstrates several associations between the microbiota of stool and multiple upstream mucosal and other environmental communities within the first 60 days of life. Given the importance of dysbiosis in health and disease of extremely preterm infants better understanding of microbial acquisition and development arising from standard NICU exposures and practices may be of clinical benefit.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the work of all clinical staff on the NICU at the Royal Victoria Infirmary, Newcastle upon Tyne for their work in recruiting and collecting samples, specifically Julie Groombridge and Gayle Gills. Dr. Andrew Nelson of NuOmics and Dr Christopher Stewart at Newcastle University were constant sources of help and guidance.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal's Web site (www.jpgn.org).
Ethics approval and consent to participate: This study was carried out in accordance with the recommendations of “NRES Committee North East—Newcastle & North Tyneside 2” with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the “NRES Committee North East—Newcastle & North Tyneside 2.”
The datasets generated and/or analysed during the current study are available in the EBI Metagenomic portal (MGnify) repository (https://www.ebi.ac.uk/metagenomics/) under the study accession number PRJEB27807.
This work was supported by Northumbria University (grant number 10031605/2 awarded to G.R.Y.).
The authors declare that they have no competing interests.
REFERENCES
- 1.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci 2011; 108:4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018; 562:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bose C, Carlo W, Conde-Agudelo A, et al. WHO recommendaion on interventions to improve preterm birth outcomes 2015; 12–51. [Google Scholar]
- 4.Shah D, Sinn JK. Antibiotic regimens for the empirical treatment of newborn infants with necrotising enterocolitis. Cochrane Database Syst Rev 2012; CD007448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev 2014; 4:CD005496. [DOI] [PubMed] [Google Scholar]
- 6.Pammi M, Abrams SA. Oral lactoferrin for the prevention of sepsis and necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev 2011; CD007137. [DOI] [PubMed] [Google Scholar]
- 7.Finer NN, Carlo WA, Walsh MC, et al. SUPPORT Study Group of the Eunice Kennedy Shriver NICHD Neonatal Research Network. Early CPAP versus surfactant in extremely preterm infants. N Engl J Med 2010; 362:1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melville JM, Moss TJM. The immune consequences of preterm birth. Front Neurosci 2013; 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol 2007; 7:379–390. [DOI] [PubMed] [Google Scholar]
- 10.Petersen SM, Greisen G, Krogfelt KA. Nasogastric feeding tubes from a neonatal department yield high concentrations of potentially pathogenic bacteria— even 1 d after insertion. Pediatr Res 2016; 80:395. [DOI] [PubMed] [Google Scholar]
- 11.Olm MR, Brown CT, Brooks B, et al. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res 2017; 27:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu DM, Ma J, Prince AL, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 2017; 23:314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks B, Olm MR, Firek BA, et al. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat Commun 2017; 8:1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks B, Firek BA, Miller CS, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2014; 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart CJ, Embleton ND, Clements E, et al. Cesarean or vaginal birth does not impact the longitudinal development of the gut microbiome in a cohort of exclusively preterm infants. Front Microbiol 2017; 8:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warner BB, Deych E, Zhou Y, et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 2016; 387:1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart CJ, Marrs ECL, Nelson A, et al. Development of the preterm gut microbiome in twins at risk of necrotising enterocolitis and sepsis. PLoS One 2013; 8:e73465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young GR, Smith DL, Embleton ND, et al. Reducing viability bias in analysis of gut microbiota in preterm infants at risk of NEC and sepsis. Front Cell Infect Microbiol 2017; 7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 2011; 108:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oksanen J, Guillaume Blanchet F, Kindt R, et al. Vegan: Community Ecology Package. R package version 2.5-6. 2019. http://cran.r-project.org/package=vegan. Accessed 1 September 2019. [Google Scholar]
- 23.Van der Gast CJ, Ager D, Lilley AK. Temporal scaling of bacterial taxa is influenced by both stochastic and deterministic ecological factors. Environ Microbiol 2008; 10:1411–1418. [DOI] [PubMed] [Google Scholar]
- 24.Knights D, Kuczynski J, Charlson ES, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 2011; 8:761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costello EK, Carlisle EM, Bik EM, et al. Microbiome assembly across multiple body sites in low-birthweight infants. MBio 2013; 4:e00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartz LE, Bradshaw W, Brandon DH. Potential NICU environmental influences on the neonate's microbiome. Adv Neonatal Care 2015; 15:324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory KE, Samuel BS, Houghteling P, et al. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 2016; 4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannaraj PS, Li F, Cerini C, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017; 171:647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mai V, Young CM, Ukhanova M, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One 2011; 6:e20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costello EK, Carlisle EM, Bik EM, et al. Microbiome assembly across multiple body sites in low-birthweight infants. MBio 2013; 4:e00782–e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sondheimer JM, Clark DA, Gervaise EP. Continuous gastric pH measurement in young and older healthy preterm infants receiving formula and clear liquid feedings. J Pediatr Gastroenterol Nutr 1985; 4:352–355. [DOI] [PubMed] [Google Scholar]
- 32.Moossavi S, Sepehri S, Robertson B, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019; 25:324.e4–335.e4. [DOI] [PubMed] [Google Scholar]
- 33.Cornelis K, Ritsema T, Nijsse J, et al. The plant pathogen Rhodococcus fascians colonizes the exterior and interior of the aerial parts of plants. Mol Plant Microbe Interact 2001; 14:599–608. [DOI] [PubMed] [Google Scholar]
- 34.Muscatello G, Lowe JM, Flash ML, et al. Review of the epidemiology and ecology of Rhodococcus equi. Rev Epidemiol Ecol Rhodococcus equi 2007; 53:214–217. [Google Scholar]
- 35.Guerrero R, Bhargava A, Nahleh Z. Rhodococcus equi venous catheter infection: a case report and review of the literature. J Med Case Rep 2011; 5:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones AL, Brown JM, Mishra V, et al. Rhodococcus gordoniae sp. nov., an actinomycete isolated from clinical material and phenol-contaminated soil. Int J Syst Evol Microbiol 2004; 54:407–411. [DOI] [PubMed] [Google Scholar]
- 37.Weinstock DM, Brown AE. Rhodococcus equi: an emerging pathogen. Clin Infect Dis 2002; 34:1379–1385. [DOI] [PubMed] [Google Scholar]
- 38.Xu J-L, He J, Wang Z-C, et al. Rhodococcus qingshengii sp. nov., a carbendazim-degrading bacterium. Int J Syst Evol Microbiol 2007; 57:2754–2757. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman D, Boyle R, Hazen KC, et al. Fluconazole prophylaxis against fungal colonization and infection in preterm infants. N Engl J Med 2001; 345:1660–1666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


