Abstract
p300 and CBP are highly related histone acetyltransferase (HAT) enzymes that regulate gene expression, and their dysregulation has been linked to cancer and other diseases. p300/CBP is composed of a number of domains including a HAT domain which is inhibited by the small molecule A-485 and an acetyl-lysine binding bromodomain which was recently found to be selectively antagonized by the small molecule I-CBP112. Here we show that the combination of I-CBP112 and A-485 can synergize to inhibit prostate cancer cell proliferation. We find that the combination confers a dramatic reduction in p300 chromatin occupancy compared to the individual effects of blocking either domain alone. Accompanying this loss of p300 on chromatin, combination treatment leads to the reduction of specific mRNAs including androgen-dependent and pro-oncogenic prostate genes such as KLK3 (PSA) and c-Myc. Consistent with p300 chromatin binding directly affecting gene expression, mRNAs that are significantly reduced by combination treatment also exhibit a strong reduction in p300 chromatin occupancy at their gene promoters. The relatively few mRNAs that are up-regulated upon combination treatment show no correlation with p300 occupancy. These studies provide support for the pharmacologic advantage of concurrent targeting of two domains within one key epigenetic modification enzyme.
Graphical Abstract:

Antagonists of p300 HAT and bromodomain synergize to induce p300 dissociation from chromatin globally and lead to decreased expression of key prostate cancer and DNA replication genes. TF=transcription factor. I-CBP112 is a bromodomain inhibitor and A-485 is a p300 HAT inhibitor. Ac-Nuc=acetylated nucleosomes. Yellow circles indicate acetyl-Lys.
Introduction
The influence of epigenetic regulation on cell growth and gene regulation in normal and disease states is now intensively studied in biomedical research.1–6 Among the post-translational modifications that mark histones on key lysine (Lys) residues, acetylation has emerged as pivotal in determining chromatin states and impacting gene expression.7–10
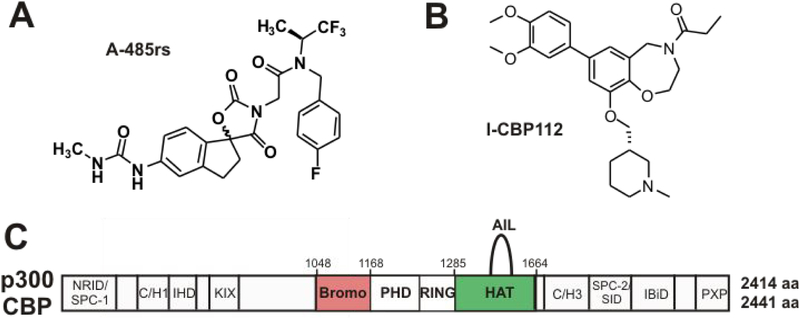
Lysine acetyltransferase enzymes utilize acetyl-CoA to catalyze Lys acetylation and include several small families and among these, p300 and CBP have been of high interest to the epigenetics community due to their important roles in chromatin-mediated gene regulation.9 These closely related human paralogs, p300 and CBP (often written as p300/CBP), are large multi-domain enzymes that contain a centrally located histone acetyltransferase (HAT) domain flanked by several protein-protein interaction domains including a bromodomain (Brd) on its N-terminal side (Figure 1)..9,11,12 Bromodomains are approximately 100 amino acid autonomously folding units located in several dozen human proteins and have been found to bind one or more acetyl-Lys residues.13–16 The HAT domain is a “writer” domain since it deposits acetyl marks while the bromodomain is a “reader” domain which binds acetyl marks.6,13 p300 and CBP are well-established as transcriptional coactivators that can acetylate more than 1000 cellular Lys sites and have been shown to be principally responsible for acetylation of histone H3K18 on chromatin.17–19
Figure 1:
p300/CBP small molecule modulators used here and the domain architecture of p300. (A) Structure of A-458rs which is a 1:1 diastereomeric mixture of spirocycycles, the histone acetyltransferase (HAT) domain inhibitor, and (B) I-CBP112, the bromodomain (Brd) antagonist. (C) Annotated domains of p300 and CBP. The bromodomain is an acetyl-Lys “reader” domain. The HAT active site “writes” Lys acetylation. The autoinhibitory loop (AIL) regulates the activity of the HAT domain depending upon its autoacetylation state (Ref. 9).
Dysregulation of p300/CBP has been linked to pro-oncogenic properties in a variety of cancers such as acute leukemias, prostate cancer, and other malignancies.9,20–23 In addition, loss of function mutations of p300/CBP are found in non-Hodgkins lymphoma and Rubenstein-Taybi syndrome.9 Efforts to develop potent and selective pharmacologically useful inhibitors of p300/CBP and HATs in general have lagged behind other epigenetic modifying processes, such as histone deacetylase inhibitors, due to pharmacological challenges of the HAT structure. Thus, the potential of p300/CBP inhibitors as anti-cancer therapeutics has been largely unexplored. However, recent advances in the development of p300/CBP modulators include the discoveries of the potent and selective p300/CBP HAT inhibitor A-485 and the bromodomain antagonist I-CBP112.24–26 A-485 is a spiro-oxazolidinedione derivative that has been crystallized with p300 and blocks-acetyl-CoA binding in a stereospecific manner.24,25 I-CBP112 shows selectivity for the p300/CBP bromodomain and competes with acetyl-Lys binding.26 Both of these p300/CBP modulators have shown anti-proliferative activity against human prostate cancer cell lines.25,27,28 A-485 leads to widespread reduction in cellular acetylation at approximately 1000 different acetyl-Lys sites; similar to a p300/CBP genetic knockdown.17 In contrast, I-CBP112 shows only a small impact on cellular acetylation and notably can enhance p300-mediated acetylation of nucleosomes.17,28,29
Given their distinct mechanisms of action on p300/CBP modulation, we hypothesized that these ligands might show synergistic actions in cellular pharmacology. Indeed, recent studies have suggested beneficial pharmacological effects by targeting two epigenetic enzymes in one complex.30 In this study, we explore this possibility in the context of prostate cancer. We investigate the effects of I-CBP112 and A-485rs (a 1:1 diastereomeric mixture of A-485) alone and in combination on prostate cancer cell growth, p300 chromatin binding, and gene expression. The results point to independent effects of bromodomain and HAT domain antagonism on reducing chromatin occupancy of p300, and synergistic effects of these compounds on the transcription of key genes. These studies highlight the therapeutic potential of concomitantly targeting these two p300/CBP domains in cancer.
Materials and Methods
Materials
Recombinant Xenopus histone H3 was expressed in and purified from E. coli as described previously.31 Reconstituted nucleosomes contained semisynethtic histone H3 containing H3K9/K14/K18 triacetylation which were prepared using F40 sortase as described previously.31 Histone octamer was composed of Xenopus histone H2a, H2b, H3, and H4 and was mixed with 146 bp 601 Widom DNA and purified as described.31 Recombinant full-length human p300 (His-p300-Strep2-Flag) was expressed and purified from insect cells as described and visualized previously.32 Cell lines were kindly provided by the Yegnasubramanian lab (LNCaP-FGC and PC3) or the Luo lab (CWR22RV1) at Johns Hopkins. I-CBP112 was purchased from Cayman Chemical. A-485rs was synthesized as described in Lasko et al25 and Michaelides et al24 except that the step involving chiral column separation of the spirocycle diasteromers was omitted resulting in a 1:1 mixture of the two stereoisomers. Antibodies were from commercial sources with the vendors noted below. Other reagents were obtained as reagent grade and were used without further purification.
Immunoblot Histone Acetyltransferase Assays
Acetylation of histone H3K18 (H3K18ac) was assessed in vitro by acetyltransferase assays in which histone acetylation was quantified by immunoblot. Reactions were performed in buffer composed of 50 mM HEPES pH 7.9, 50 mM NaCl, 1 mM TCEP, and 25 μg/mL BSA at 30 °C and initiated by the addition of acetyl-CoA. All reactions were performed under initial rate conditions (<30% conversion of the limiting substrate). Acetylation reactions contained 10 μM histone H3, 2 nM p300, and 0–10 μM A-485rs with or without 20 μM I-CBP112. Reactions were pre-incubated on ice for 30 min, initiated by the addition of 200 nM acetyl-CoA, and allowed to proceed for 5 min. All ligands were dissolved in 10% DMSO and the final reaction concentration maintained at 1.25% DMSO, a concentration previously determined to not affect enzyme function.28 Reactions were quenched with 4× SDS loading dye [200 mM Tris-HCl (pH 6.8), 40% glycerol, 8% SDS, 50 mM EDTA, 4% β-mercaptoethanol, and 0.08% bromophenol blue] and mixtures heated at 95 °C for 5 min before separation on 15% tris-glycine gels. Proteins were transferred by iBLOT to nitrocellulose, and probed overnight at 4°C with 1:1000 H3K18ac (Millipore 07–354) in 2.5% BSA-TBST followed by anti-rabbit HRP at 1:5000 for 1 h at room temperature in 2.5% BSA-TBST. All assays were performed at least twice (n = 2) and the average and standard error of replicates reported.
Cell Proliferation Assays
All cells were maintained in RPMI 1640 with 10% FBS and 1% penicillin-streptomycin at 37°C in 5% CO2. Cell plating titrations were performed to determine the linear range of cell plating number for 72 hr assays. Based on these results, 5000 LNCaP-FGC cells, 3000 CWR22RV1, and 700 PC3 cells were plated in 96-well flat-bottom plates 24 h before treatment. After 24 h, cells were treated with the respective small molecule inhibitor compounds (I-CBP112, A-485rs, OTX015) dissolved in DMSO to a final concentration of 0.18%, which was shown to have negligible cell growth effects. After 66 h, 0.476% [3H]thymidine was added to the cells per well and the cells were allowed to proliferate for an additional 6 h. Cells were then harvested, and the counts of 3H in each well performed on a 1450 Microbeta scintillation counter were taken relative to those treated with vehicle alone to quantify the effect of the ligand on proliferation. Experiments were conducted in triplicate or greater, and repeated on two or more occasions. Each titration was normalized to zero titrated ligand. IC50 values were calculated using a standard dose-response
| equation: |
where x represents the ligand concentration, IC50 the concentration of agonist at half response, y the % counts relative to the average DMSO control, and h the Hill coefficient.
Synergy Analysis of Two Compound Treatments
Synergy effects between two compound treatments was determined by using the method of Chou-Talalay. Values were normalized to that of the average for no ligand exposure =1 and analyzed by the CompuSyn program. Combination index values below 0.8 were considered synergistic and below 0.3 considered strongly synergistic.33
Cellular H3K18ac
LNCaP cells were plated at 2–4 × 105 cells/well in 6-well plates, allowed to adhere, and treated at ~70% confluence with increasing amounts of A-485rs with or without 20 μM I-CBP112 at a constant 0.07% DMSO for 24 h. After washing with PBS, cells were harvested by lysis in 2x SDS loading buffer [100 mM Tris-HCl (pH6.8), 20% glycerol, 4% SDS, 50 mM EDTA, 2% β-mercaptoethanol, and 0.04% bromophenol blue]. Lysate was heated to 95°C for 10 min and equal amounts separated on 15% tris-glycine gels. Equal loading was confirmed by total H3 histone blotting. Total H3 blots were probed with 1:5000 anti-total histone H3 (Abcam 1791) in 5% BSA-TBST for 1 h followed by anti-rabbit HRP at 1:5000 for 1 h at room temperature in 2.5% BSA-TBST. H3K18ac blots were probed overnight at 4°C with 1:1000 H3K18ac (Millipore 07–354) in 2.5% BSA-TBST followed by anti-rabbit HRP as above. The average relative acetylation of quadruplicate biological replicates was determined and a representative Western blot is shown.
ChIP-qRT-PCR/ ChIP-Sequencing
LNCaP cells were plated in 2–10 cm plates per condition, allowed to adhere overnight, and grown to ~65% confluency. Cells were treated with the respective dosage of ligand in a constant final 0.0624% DMSO concentration. After 24 h, formaldehyde was added to a final concentration of 1% and allowed to crosslink for 10 min at room temperature with shaking. Crosslinking was quenched with a final concentration of 125 mM glycine for 5 min following which cells were washed twice in cold 1x PBS. Cells were scraped from the plate, excess PBS removed, and the cell pellet resuspended in cell lysis buffer [10 mM Tris-HCl pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.4% NP-40, 10 mM sodium butyrate]. After cell lysis by pipetting, the nuclear pellet was resuspended in 1 volume SDS cell lysis buffer and incubated on ice for 10 min after which 3 volumes of non-SDS cell lysis buffer was added. The chromatin was sheared with a Diagenode Bioruptor® on high for 20 cycles (30 sec on, 30 sec off). The supernatant was precleared with 2.5 μg of mouse IgG (Santa Cruz) and Protein G Dyanbeads (Invitrogen) for 2 h at 4°C. The supernatants were diluted in ChIP dilution buffer [20mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.01% SDS] to equal concentrations for all treatments, 1% of input was removed, and the remainder immunoprecipitated with mouse IgG (as above) or p3oo (Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa; ENCITp300–1 was deposited to the DSHB by Wold, B. / Vielmetter, J. (DSHB Hybridoma Product ENCITp3 0 0–1)).34 Immunoprecipitations were rotated overnight at 4°C. Subsequently Protein G Dynabeads™ were added and the slurry incubated an additional 3 h at 4°C. The bound chromatin was separated on a magnet and the pellet washed with low salt wash buffer [20 mM Tris-HCl pH 8, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 0.1% SDS], high salt wash buffer [20 mM Tris-HCl pH 8, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA, 0.1% SDS], LiCl wash buffer [0.25 M LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0] and TE wash buffer [10 mM Tris-HCl pH 8.0, 1 mM EDTA] sequentially. The specifically bound chromatin was eluted with elution buffer [10% SDS, 100 nM sodium bicarbonate] at 50°C with agitation for 30 min and repeated. The eluates were combined and crosslinks reversed with 200 mM NaCl overnight. RNA was digested with 40 μg/ml RNAse A and proteins with 3.2 U of Proteinase K for 2 h at 50°C. DNA was isolated by phenol-chloroform extraction and ethanol precipitation and resuspended in 50 μl diH2O. qPCR was performed with Power SYBR™ Green PCR Master Mix (Thermo Fisher Scientific) on a BioRad CFX C1000 thermocyler and analyzed with BioRad CFX Manager 3.1. Primers are listed in Supplementary Table S1. For ChIP-Seq, libraries were made by SWIFT library prep, and single end sequencing was performed by the Center for Functional Cancer Epigenetics of the Dana Farber Cancer Institute, normalized to sequencing depth, and analyzed by the ChiLin pipeline.35 Concentrations were chosen for ChIP-Seq were chosen based on the IC50’s for KLK3 mRNA in the qRT-PCR assays and greater than the IC50’s for the 72 h cell proliferation assays.
RT-qPCR and RNA-Sequencing
Cells were treated exactly as above in the ChIP-Seq experiments for 24 h and harvested in TRIzol™ Reagent (Invitrogen). RNA was isolated by phenol-chloroform extraction and ethanol precipitation. qRT-PCR was performed with qScript™ One-Step SYBR® Green qRT-PCR Kit (QuantaBio) on a BioRad CFX C1000 thermocyler and analyzed with BioRad CFX Manager 3.1 utilizing the primers listed in Supplemental Table S2. All fold changes reported were normalized to GAPDH. For RNA-Seq, samples were enriched by polyA selection. Library prep and sequencing were performed by the Center for Functional Cancer Epigenetics of the Dana Farber Cancer Institute. Data were processed through the VIPER pipeline. Briefly, raw reads were aligned to hg19 with BowTie, assembled into transcripts and normalized with Cufflinks, batch corrected with ComBat, and differential expression determined by DESeq2. Quality checks were performed and samples clustered based on expression levels.36
Nucleosome Pulldown Assays
Anti-FLAG® M2 Magnetic Beads (Sigma) were washed 3x with 1x TBS and blocked overnight in 5% BSA-TBS at 4°C. Full length FLAG-tagged p300 of 142 nM final concentration was incubated with 2.3 μM triacetylated nucleosome (H3K9ac,K14ac,K18ac) in 50 mM HEPES pH 7.9, 100 mM NaCl, 0.1 mM TCEP, 50 μg/ul BSA, 2 mM sodium butyrate, 0.42% DMSO overnight at 4°C with or without 80 μM 1-CBP112. Subsequently, the beads were added to the binding reaction and allowed to incubate together for an additional 3 h at 4°C. Non-specifically bound proteins were removed from the beads by three washes with TBS. Remaining specifically bound proteins were eluted with 4x SDS loading dye, separated on a 15% tris-glycine gel, and probed for total histone H3 as above.
Results
A-485rs and I-CBP112 combination effects on p300 and prostate cancer growth
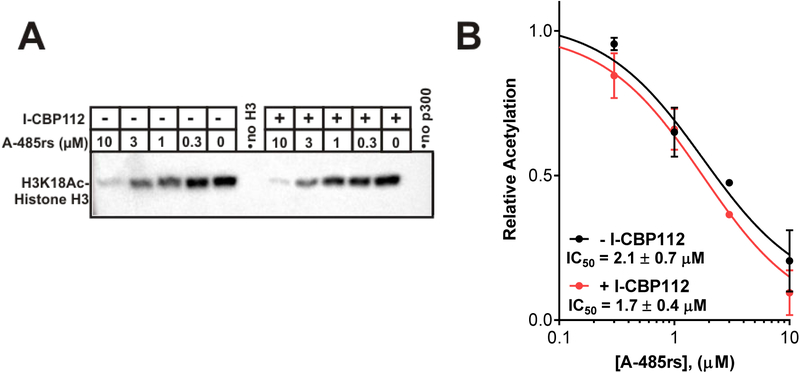
We used A-485 as a mixture of two diastereomers at the spiro-oxaolidinedione quaternary carbon (here called A-485rs) which is synthetically simpler to prepare than the enantiomerically pure material as it has been shown that the R-isomer is about 10-fold more active than the S-isomer.24,25 We used full-length p300 protein and purified histone H3 to assess p300 inhibitory properties of A-485rs in the presence or absence of I-CBP112, monitoring H3K18ac by western blot. These experiments showed that A-485rs has an IC50 of ~2 μM in blocking p300 acetyltransferase activity toward free histone H3 (Figure 2). This A-485rs potency was not significantly affected by the presence of I-CBP112, although the bromodomain antagonist boosted the baseline acetyltransferase activity on nucleosomes as described previously.28
Figure 2:
A-485rs inhibitory effects on p300 acetyltransferase activity in the presence or absence of I-CBP112 in a purified system. (A) Dose-dependent inhibition of full-length p300 catalyzed acetylation of 10 μM histone H3 by 0–10 μM A-485rs in the presence or absence of 20 μM I-CBP112 monitored by Western blot (anti-H3K18Ac). (B) Quantification of replicate results (n = 2) in (A) were fit to a standard dose response curve to determine the respective IC50’s. Error bars represent standard error of replicates; IC50 comparison p = 0.54.
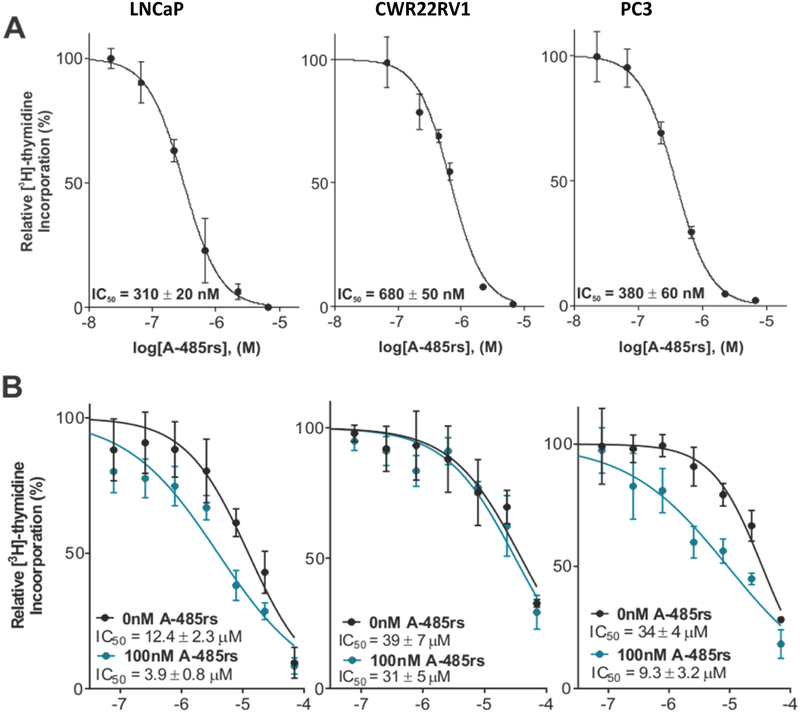
We next explored A-485rs effects on prostate cancer cells using LNCaP (androgen receptor positive), PC3 (androgen insensitive), and CWR22RV1 (splice variant, constitutively active androgen receptor) prostate cancer cell lines37–41 by measuring 3H-thymidine incorporation after 72 h. A-485rs showed potent inhibition of the growth of each of these cell lines with IC50’s in the range of 400–800 nM (Figure 3A). These values are in fairly good agreement with previously reported studies with A-485 for the LNCaP and splice variant prostate cancer cells25 but A-485rs was more potent in PC3 cells than expected25 perhaps because of the distinct methods for determining cell proliferation. It should also be noted that the fast PC3 doubling time may accentuate the differences of 3H-thymidine incorporation used here versus a metabolic assay employed in the prior study.25,42 To determine whether there might be synergistic effects of A-485rs and I-CBP112 on cell growth, we employed a relatively low concentration (100 nM) of A-485rs and measured the dose-response curve to I-CBP112 for all three prostate cancer cell lines. Interestingly, both LNCaP and PC3 cells showed that A-485rs treatment enhanced the anti-proliferative potencies of I-CBP112 about 3-fold. With I-CBP112 the IC50 reduces from 12.4 μM without A-485rs to 3.9 μM with A-485rs in LNCaP cells and from 34 μM to 9.3 μM in PC3 cells. In contrast, the anti-proliferative I-CBP112 IC50’s in CWR22RV1 prostate cancer cells were not significantly affected by 100 nM A-485rs treatments (39 μM without A-485rs vs. 31 μM plus A-485rs) (Figure 3B). (Biological replicates are shown in Supplementary Figure S1.) Using Chou-Talalay analysis,33 we determined that co-administration of A-485rs and I-CBP112 showed a mean combination index (CI) of 0.50 and 0.58 in LNCaP and PC3 cells respectively, indicating moderate to strong synergy. Meanwhile CWR22RV1 cells had a mean CI = 1.04, consistent with merely additive rather than synergistic effects.
Figure 3:
A-485rs and I-CBP112 effects on prostate cancer cell proliferation. (A) Cell proliferation assays (3H-thymidine incorporation) for LNCaP, CWR22RV1, and PC3 cells after 72 h of A-485rs treatment; n = 3. (B) Treatment of the aforementioned cells with varying I-CBP112 concentrations in the presence or absence of 100 nM A-485rs for 72 h; n = 3. Mean CI values = 0.50, 1.04, 0.58 respectively.
Given the apparent synergy with A-485rs and I-CBP112 in LNCaP cells, we further investigated the pharmacology of LNCaPs with dual treatment of A-485rs and the BET bromodomain antagonist OTX0 1 5.43–45 These studies revealed little difference between the A-485rs treated cells and those not receiving the HAT inhibitor regarding OTX015 IC50 values (Supplementary Figure S2A), suggesting that the synergy between I-CBP112 and A-485rs is due to a p300/CBP bromodomain specific effect of I-CBP112. In addition, the combination of the general transcription initiation (TFIIH) inhibitor triptolide46 with I-CBP112 was investigated in LNCaP cells (Supplementary Figure S2B). These experiments showed no impact of triptolide on the I-CBP112 IC50 values, indicating a lack of synergy and an anti-proliferative mechanism beyond general transcription inhibition for A-485rs.
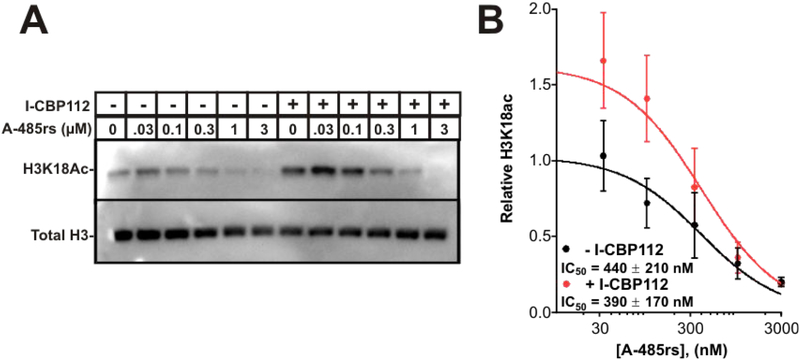
We considered that the A-485rs/I-CBP112 combination might exert synergy through more powerful inhibition of global histone acetylation. An A-485rs dose-response analysis of histone H3K18ac acetylation levels in LNCaPs was performed with or without I-CBP112 addition. As reported previously, I-CBP112 induced a modest increase in H3K18ac levels by western blot.28 However, the potency of A-485rs was similar whether I-CBP112 (20 μM) was present or not, with IC50 of ~400 nM in both cases (p= 0.85) (Figure 4). These cellular acetylation inhibition results are consistent with the in vitro enzymology findings showing no apparent synergy of A-485rs and I-CBP112 on p300 acetyltransferase action.
Figure 4:
Influence of I-CBP112 and A-485rs on histone H3K18ac in LNCaP cells. (A) LNCaP cells were treated with increasing concentrations of A-485rs with or without 20 μM I-CBP112. Equal amounts of cell lysates were separated on 15% SDS-PAGE gels and H3K18ac visualized by Western blot. (B) Quantification of (A). Error bars represent the SEM, n = 4; IC50 comparison p = 0.85.
Gene expression and chromatin analysis of A-485rs and I-CBP112
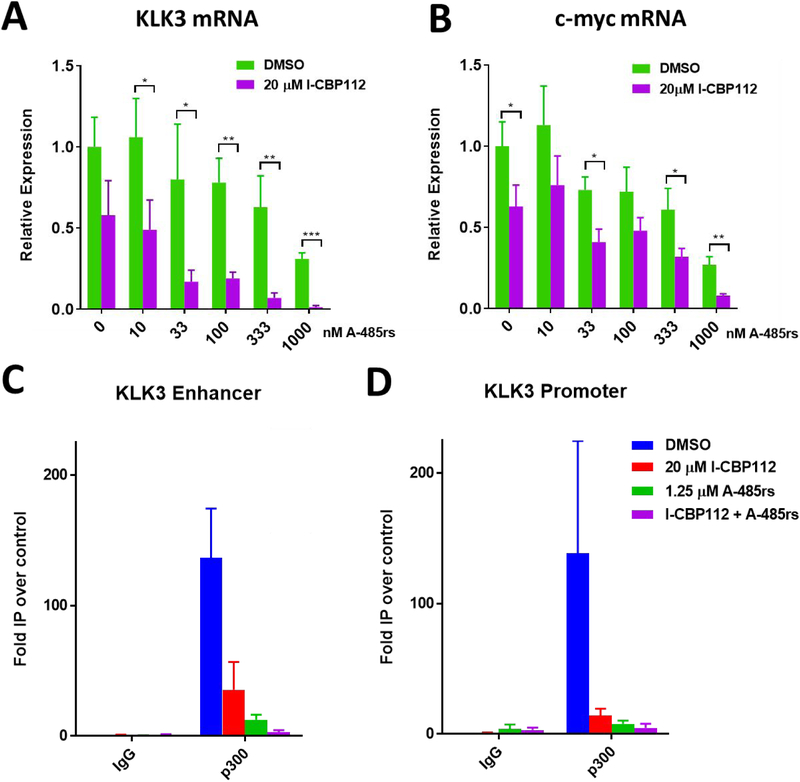
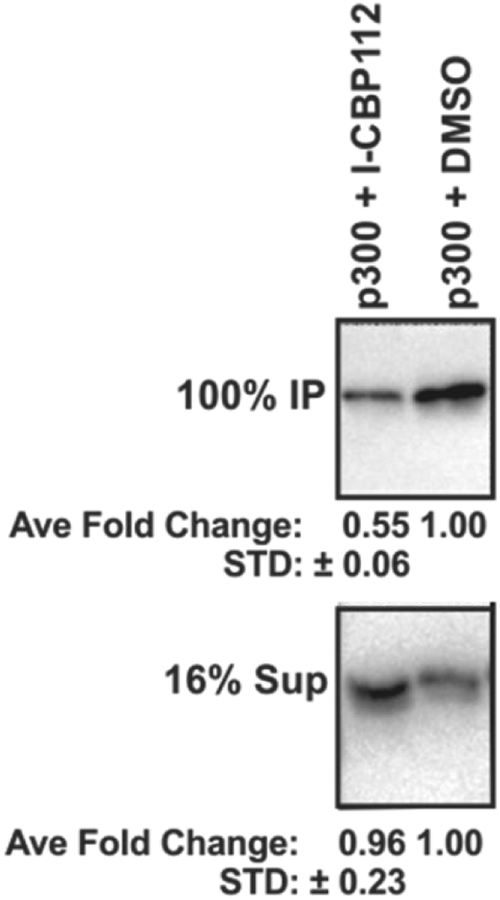
To gain insight into the effects of the combination of A-485rs and I-CBP112 on gene expression changes, we examined two genes previously shown in prostate cancer to be suppressed by p300/CBP HAT inhibition, KLK3 (aka PSA) and c-myc.22,47–49 These experiments revealed that co-treatment of LNCaP cells with I-CBP112 and A-485rs led to a more powerful and synergistic gene expression silencing impact across a range of A-485rs concentrations, most notably on KLK3 (median CI = 0.12), but also on c-myc (median CI = 0.49) (Figure 5A,B). No synergy was found on expression of other A-485rs regulated genes (Supplementary Figure S3 A,B).25 We hypothesized that the I-CBP112 effect may have been related to its binding the p300 bromodomain and antagonizing p300 association with acetylated chromatin. To test this directly, we carried out a protein pulldown experiment and found that I-CBP112 treatment reduced the efficiency of histone H3 tri-acetylated nucleosomes to bind full length p300 in a purified, reconstituted system (Figure 6). We next investigated p300 chromatin occupancy across the KLK3 promoter and enhancer regions in LNCaPs by ChlP-qPCR. Treatment of cells with A-485rs and I-CBP112 individually led to reduced occupancy of p300 at the KLK3 promoter and enhancer, whereas the combination treatment nearly abrogated p300 association with these chromatin regions (Figure 5C,D).
Figure 5:
Effects of A-485rs and I-CBP112 on mRNA levels and p300 occupancy of KLK3 and c-myc promoter and enhancer regions in LNCaP cells. mRNA levels of KLK3 (A) and c-myc (B) were measured by qRT-PCR after 24 h of treatment with increasing concentrations of A-485rs with or without 20 μM I-CBP112. LNCaP cells were treated with the indicated ligands for 24 h followed by ChlP-PCR of p300-bound KLK3 enhancer (C) and promoter (D). Representative plot of biological duplicates each done with n = 3. * p < 0.05, ** p < 0.005, *** p < 0.0005
Figure 6:
Triacetylated nucleosomes were pulled down by p300 less efficiently in the presence I-CBP112. Full length p300 (142 nM) bound by anti-FLAG magnetic beads was used to pull down triacetylated (K9ac,K14ac,K18ac) nucleosomes (2.3 μM) in the absence or presence of 80 μM I-CBP112. p300-bound nucleosomes were quantified by anti-H3 total Western blotting and compared to the total H3 remaining in the supernatant. Average and SEM with n = 3.
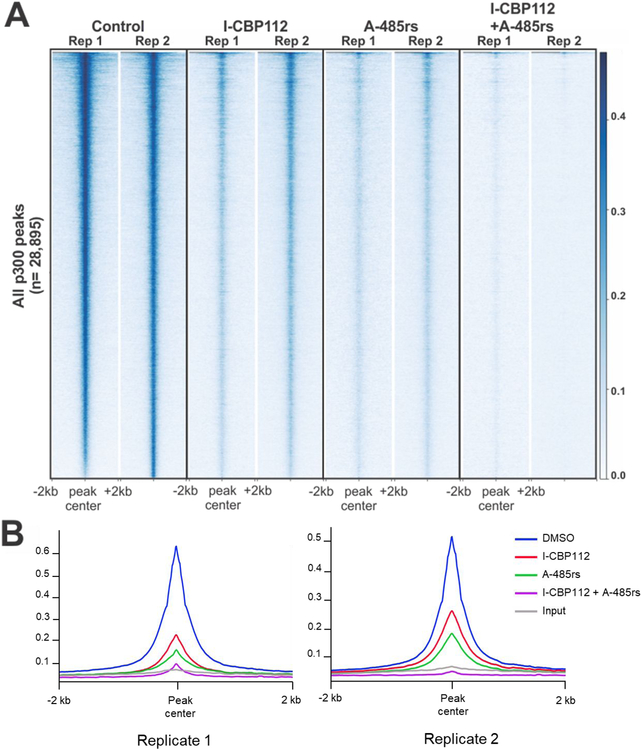
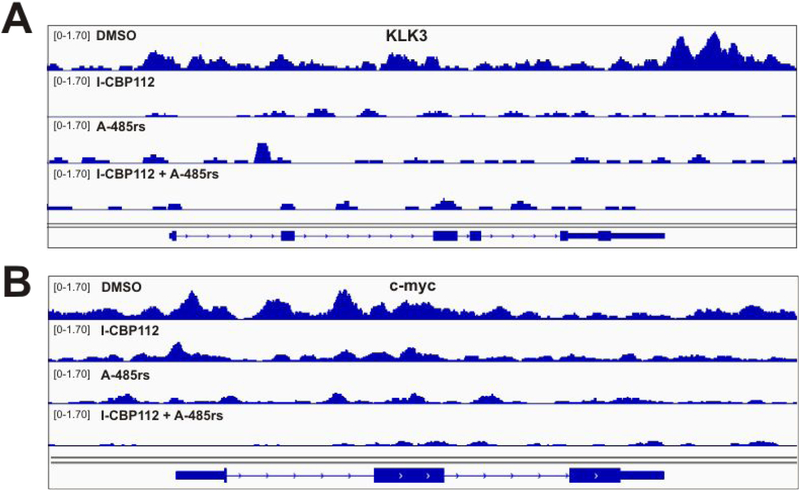
To more broadly assess the effects of A-48.Srs and T-CRP112 across the genome, we performed ChIP-Seq and RNA-Seq analysis in LNCaPs treated with these agents. ChIP-Seq revealed that individual treatment with A-485rs or I-CBP112 induced a broad reduction in p300 across its binding sites (Figures 7 and 8). A similar broad reduction is seen when plotting p300 binding across all TSSs (Supplementary Figure S4A). As with KLK3 and c-myc, the combination treatment with A-485rs and I-CBP112 conferred an even more dramatic and widespread reduction in p300 occupancy to near background across the genome (Figure 7). The combination treatment with A-485rs and I-CBP112 led to the a much greater degree of p300 dissociation at transcription start sites (TSS) than the more modest changes induced by either compound alone (Supplementary Figure S4B). Further, genes with the highest p300 occupancy in the control exhibited the greatest p300 loss in each treatment (Supplementary Figure S4C).
Figure 7:
A-485rs and I-CBP112 effects on p300 occupancy in chromatin. (A) ChIP-Seq was performed on LNCaP cells treated for 24 h. Heatmap shows p300 ChIP signal across peaks (n = 28,895) called for p300 under any condition. Plots are aligned by peak center and +/− 2kb is shown. Rep 1 and Rep 2 are results from two biological replicates. (B) Metagene plot of the average p300 signal across all peaks (n = 28,895) in each treatment.
Figure 8:
Effects of A-485rs and I-CBP112 on p300 occupancy of KLK3 and c-myc gene bodies determined by ChIP-Seq. Genome browser view of KLK3 (A) and c-myc (B) tracks after 24 h ligand treatment, y-axis scale shows normalized enrichment.
RNA-Seq analysis was performed under the same conditions, and biological replicates showed strong correlation within replicates for all treatments (Supplementary Figure S5). These data showed that I-CBP112 treatment of LNCaP cells had only a very slight effect on gene expression, with only 3 mRNA transcripts showing a greater than 2-fold reduction. In contrast, A-485rs treatment alone led to significant reduction of 360 mRNAs including the 3 mRNAs affected by I-CBP112 alone (Figure 9A,B). Combination of I-CBP112 and A-485rs led to a much larger set (811) of significantly reduced mRNAs, several of which showed a more dramatic degree of downregulation when compared to either agent alone. The set of mRNAs reduced by the combination of A-485rs and I-CBP112 showed substantial overlap with those reduced by A-485rs alone (Figure 9B). In addition, a relatively small number of mRNAs transcripts showed increased expression with A-485rs and the A-485rs/I-CBP112 combination (Figure 9C).
Figure 9:
Impact of A-485rs and I-CBP112 on gene expression determined using RNA-Seq. (A) mRNAs in LNCaP cells after 24 h treatment with compounds displayed using a volcano plot with the log2 fold change relative to DMSO shown on the x-axis and the p-value shown on the y-axis. Genes significantly downregulated (adjusted p-value < 0.05 and greater than 2-fold change) are highlighted in blue and genes significantly upregulated (adjusted p-value < 0.05 and greater than 2-fold change) are highlighted in red. Select mRNAs with the highest p values are labelled for each treatment. Venn diagram of peaks more than 2-fold downregulated (B) or upregulated (C). n = 2.
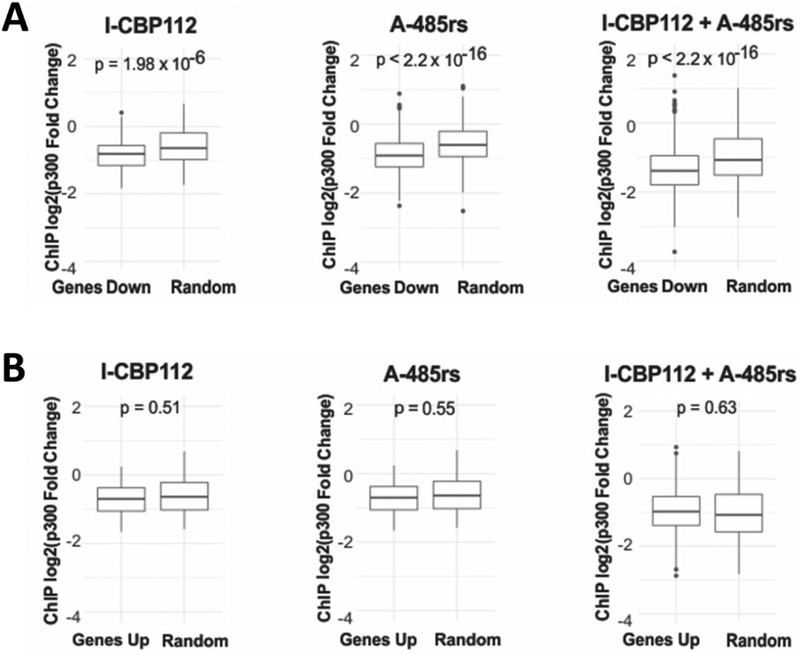
To understand a potential link between p300 chromatin binding and gene expression, we compared changes in p300 binding across the promoters of genes that were altered at the mRNA level. This showed that in each treatment group, genes downregulated at the mRNA level were more likely to have dissociated p300 than a randomly chosen set of genes (Figure 10A). This result is consistent with p300’s classical coactivator role. In contrast, genes upregulated by p300 inhibition showed no correlation with changes in p300 chromatin occupancy (Figure 10B), suggesting that increases in gene expression induced by A-485rs and I-CBP112 are regulated in a more complex fashion.
Figure 10:
Correlation between ChIP-Seq and RNA-Seq data. (A) Box and whisker plots of the log2 fold change in p300 binding for each treatment across downregulated genes (non-adjusted p-value < 0.01) or a random set of genes of equal number. Midline equals the median, top of box equals 75th percentile, and bottom of box equals 25th percentile. Upper and lower whisker equal 95th and 5th percentile, respectively. (B) As in (A) but with up-regulated genes.
Gene ontology analysis
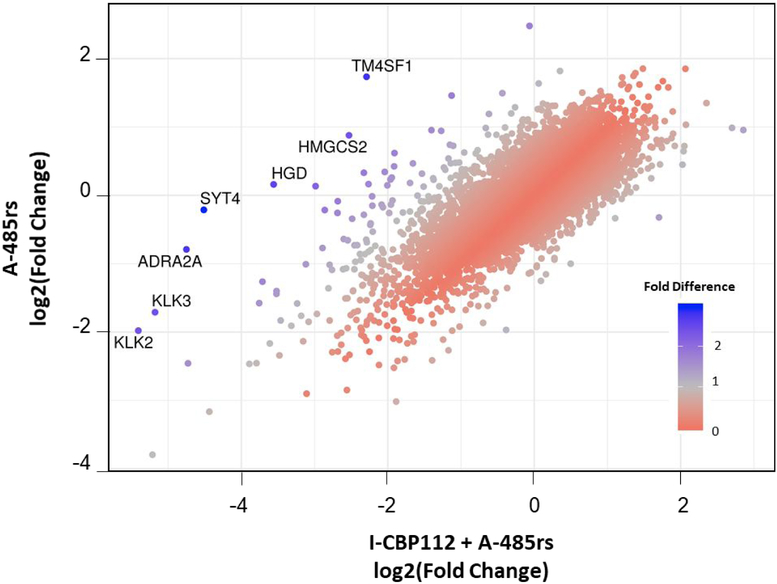
As the I-CBP112 + A-485rs treatment group showed a number of specific genes that were substantially more downregulated than in A-485rs treatment alone group (Figure 11), we performed gene ontology (GO) analysis using GOstats50 to gain possible insights into the functional differences. GO analysis revealed that the combination treatment specifically and strongly affected DNA replication pathways which were not significantly affected by A-485rs alone (Figure 12). These downregulated DNA replication genes or gene sets may help explain the more pronounced anti-proliferative effects of combining A-485rs and I-CBP112 in LNCaP cells.
Figure 11:
Scatterplot of the fold change of each gene upon A-485rs + I-CBP112 treatment vs. A-485rs treatment alone. Dots denoting genes are colored by the absolute value of fold change in expression between the two treatment groups. Those with the greatest difference in their expression upon the addition of I-CBP112 are named.
Figure 12:
Patterns of gene expression changes after I-CBP112 and A-485rs treatment of LNCaP using gene ontology (GO) analysis. Bubble plot of the top 5 GO pathways in each treatment group scored based on their p value with the % of genes in the category effected indicated by bubble size.
Discussion
Prior work has suggested that that the p300/CBP bromodomain may influence HAT activity, sometimes in a substrate-specific manner.28,51–54 Our new findings broaden our understanding of the functional interdependence between the p300/CBP bromo- and HAT domains, showing that bromodomain antagonism synergizes with HAT activity inhibition to alter expression levels of key mRNAs and abrogate p300 chromatin association. We find that there are distinct mechanisms by which p300’s bromo- and HAT mechanisms regulate gene expression: a few solely by p300 “reader” function, a larger number by the p300 “writer” function, and many more by both functions operating together.
We demonstrate that antagonists of the p300/CBP HAT and bromo-domains show synergistic action on slowing prostate cancer cell growth in two out of three cancer cell lines examined. In our studies, a relatively low concentration of the p300/CBP HAT inhibitor was able to sensitize LNCaP and PC3 cells, but not the androgen receptor splice variant CWR22RV1 prostate cancer cells, to inhibit growth by the bromodomain ligand. These results indicate that the presence of the androgen receptor per se is not necessary for the dual treatment anti-proliferative synergy since PC3 cells lack androgen receptor expression. It is unclear why the non-androgen dependent CWR22RV1 prostate cancer cells do not show such synergy but it may relate to the constitutive activation of the splice variant androgen receptor.
Among the mechanisms that could account for synergistic activity between the “reader” and “writer” domains, a direct enhancement of affinity model is unlikely since the potency of A-485rs inhibition of p300/CBP HAT activity is not significantly affected by the presence of I-CBP112. Nevertheless, as I-CBP112 can modestly stimulate p300/CBP catalyzed nucleosome acetylation, the presence of A-485rs negates this I-CBP112 pharmacologic impact. However, in LNCaP cells, our data suggest that a principal mechanism of the synergy during dual treatment involves a sharp reduction in p300 occupancy of chromatin brought on by the compound combination.
I-CBP112 inhibition of p300 association with chromatin broadly across the genome resembles the behavior of several other bromodomain antagonists that have led to the dissociation of their cognate proteins from chromatin.55–57 It is noteworthy that A-485rs acting on its own targeting the HAT “writer domain” also shows this behavior. One explanation for this is that A-485rs blocks histone acetylation and this in turn reduces the affinity of p300 for histones, via bromodomain interactions. The fact that p300/CBP bromodomain prefers binding multiply acetylated proteins11,53 may help explain the impact of A-485rs on p300’s chromatin occupancy. Another potential factor is that p300 mediated acetylation of transcription factors9,17,58 and itself59,60 may augment p300/CBP’s association with chromatin. Given the widespread nature of p300/CBP catalyzed acetylation of the proteome, it may be difficult to distinguish among these possible mechanisms.
The correlation of decreased p300 chromatin occupancy and reduced RNA levels is consistent with a cause and effect relationship involving transcriptional regulation. It is noteworthy that despite I-CBP112’s major impact on p300 occupancy, it showed only a very small impact on gene expression, consistent with the results of others.26 This leads to the interesting concept that p300 occupancy on chromatin via the bromodomain does not linearly correlate with transcriptional activity.61–64 Rather, recruitment of transcriptional machinery and RNA synthesis appears to proceed relatively normally without full p300 chromatin loading, suggesting there may be some redundancy under normal circumstances or only a modest quantity of p300 required for normal transcription. Alternatively, transient and reversible association of p300/CBP with chromatin65 may be sufficient for histone acetylation and gene transcription. In contrast, inhibition of the “writer” domain by A-485rs had a much more profound effect on gene expression. This can be rationalized, as A-485rs blocks both p300/CBP catalytic activity and its chromatin occupancy. It is noteworthy that the dual blockade of both the “writer” and “reader” domains of p300/CBP shows an even more dramatic impact on gene expression than the HAT inhibitor alone, especially on a specific subset of genes. Several possible mechanisms could explain this behavior. In one model, a subset of genes primarily requires p300’s catalytic function. The expression of another set of genes requires both p300/CBP bromodomain association with chromatin and the acetyltransferase enzymatic function for robust gene transcription. In this model, both the “reader” function of p300/CBP and the acetylation “writer” function join forces to drive the transcription of a subset of genes. Such multi-faceted coactivation models for p300/CBP have been proposed previously.64,66 In a second distinct mechanism, important genes, including those involved in DNA replication,67 are more resilient to minor cellular perturbations and are therefore downregulated only if p300 occupancy falls below a threshold level.
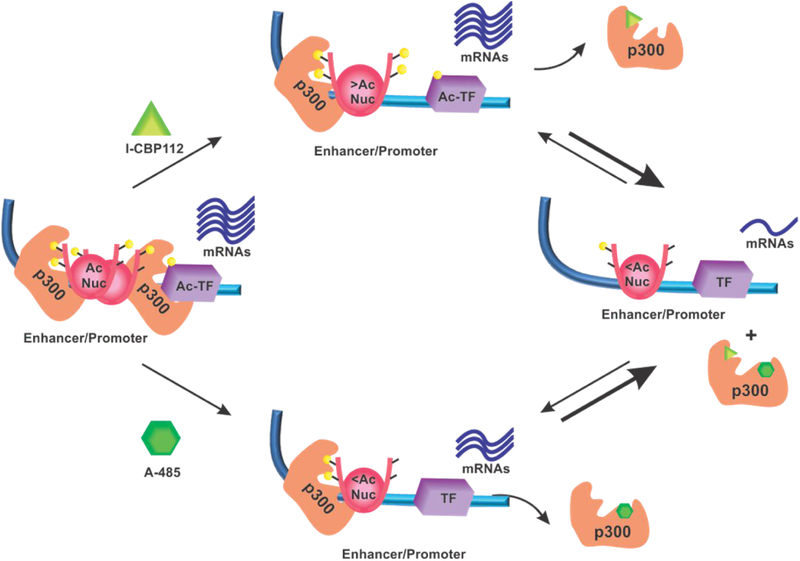
In summary, concomitant blockade of both the HAT and bromodomains in p300 leads to strong p300 dissociation and decreased gene expression (Figure 13). Whether this coordination between the HAT and bromodomains generalizes to other proteins like GCN5 and PCAF7 will require improved chemical matter to interrogate the roles of these specific domain contributions to their functions. In future studies, it will be exciting to examine if combining a p300/CBP bromodomain antagonist and a p300/CBP selective HAT inhibitor may show an enhanced therapeutic window in the setting of cancer animal models and human clinical studies.
Figure 13:
Proposed model of p300 ligand function. I-CBP112 binding of p300 to leads to increased acetylation of histones, decreased p300-chromatin association, and no change in mRNA production. A-485 decreases nucleosome and transcription factor acetylation, p300-chromatin association, and mRNA production. Together the ligands abrogate p300-chromatin association and downregulate many mRNAs. Yellow circles indicate acetyl-Lys. All other shapes as labeled.
Supplementary Material
Acknowledgments
We would like to thank the members of the Epigenetics Center at Johns Hopkins School of Medicine, members of the Bulyk and Cichowski labs of BWH, and members of the Kuroda and Perrimon labs of HMS for helpful discussions and instrumental time. We are grateful to the Jun Luo and Vasan Yegnasubramanian labs of Johns Hopkins for prostate cancer cells and input. The Dana Farber Center for Functional Cancer Epigenetics kindly performed ChIP-Seq and RNA-Seq and the basic analysis thereof. Dr. Jay Kalin and Dr. Shri Bhat of the Cole lab also assisted with helpful advice.
Funding
This work was supported by the NIH (GM62437 and DK118266 to P.A.C., 5K99GM124357 to B.E.Z., and GM126944 to M.I.K.), Leukemia and Lymphoma Society, Jane Coffin Childs Memorial Fund, and FAMRI Foundation for financial support.
Footnotes
Data Availability
The RNA-Seq and ChIP-Seq have been deposited in GEO with superseries record GSE124644
Conflict of Interest
P.A.C. is a cofounder of Acylin Therapeutics and has been a consultant to Abbvie which have had research programs in p300 HAT inhibitors.
REFERENCES
- (1).Dawson MA; Kouzarides T Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150 (1), 12–27. 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- (2).Prachayasittikul V; Prathipati P; Pratiwi R; Phanus-Umporn C; Malik AA; Schaduangrat N; Seenprachawong K; Wongchitrat P; Supokawej A; Prachayasittikul V; et al. Exploring the Epigenetic Drug Discovery Landscape. Expert Opin Drug Discov 2017, 12 (4), 345–362. 10.1080/17460441.2017.1295954. [DOI] [PubMed] [Google Scholar]
- (3).Kanwal R; Gupta K; Gupta S Cancer Epigenetics: An Introduction In Cancer Epigenetics: Risk Assessment, Diagnosis, Treatment, and Prognosis; Verma M, Ed.; Methods in Molecular Biology; Springer New York: New York, NY, 2015; pp 3–25. [Google Scholar]
- (4).Arrowsmith CH; Bountra C; Fish PV; Lee K; Schapira M Epigenetic Protein Families: A New Frontier for Drug Discovery. Nat Rev Drug Discov 2012, 11 (5), 384–400. 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- (5).Zucconi BE; Cole PA Allosteric Regulation of Epigenetic Modifying Enzymes. Curr Opin Chem Biol 2017, 39, 109–115. 10.1016/jxbpa.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ganesan A Multitarget Drugs: An Epigenetic Epiphany. ChemMedChem 2016, 11 (12), 1227–1241. 10.1002/cmdc.201500394. [DOI] [PubMed] [Google Scholar]
- (7).Voss AK; Thomas T Histone Lysine and Genomic Targets of Histone Acetyltransferases in Mammals. Bioessays 2018, 40 (10), e1800078 10.1002/bies.201800078. [DOI] [PubMed] [Google Scholar]
- (8).Wellen KE; Thompson CB A Two-Way Street: Reciprocal Regulation of Metabolism and Signalling. Nature Reviews Molecular Cell Biology 2012, 13 (4), 270–276. 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- (9).Dancy BM; Cole PA Protein Lysine Acetylation by P300/CBP. Chem. Rev 2015, 115 (6), 2419–2452. 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim SC; Sprung R; Chen Y; Xu Y; Ball H; Pei J; Cheng T; Kho Y; Xiao H; Xiao L; et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Molecular Cell 2006, 23 (4), 607–618. 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- (11).Delvecchio M; Gaucher J; Aguilar-Gurrieri C; Ortega E; Panne D Structure of the P300 Catalytic Core and Implications for Chromatin Targeting and HAT Regulation. Nat Struct Mol Biol 2013, 20 (9), 1040–1046. 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- (12).Manning ET; Ikehara T; Ito T; Kadonaga JT; Kraus WL P300 Forms a Stable, Template-Committed Complex with Chromatin: Role for the Bromodomain. Mol. Cell. Biol 2001, 21 (12), 3876–3887. 10.1128/MCB.21.12.3876-3887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Taverna SD; Li H; Ruthenburg AJ; Allis CD; Patel DJ How Chromatin-Binding Modules Interpret Histone Modifications: Lessons from Professional Pocket Pickers. Nat Struct Mol Biol 2007, 14 (11), 1025–1040. 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Smith SG; Zhou M-M The Bromodomain: A New Target in Emerging Epigenetic Medicine. ACS Chem. Biol 2016, 11 (3), 598–608. 10.1021/acschembio.5b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Moustakim M; Clark PGK; Hay DA; Dixon DJ; Brennan PE Chemical Probes and Inhibitors of Bromodomains Outside the BET Family †The Authors Declare No Competing Interests. Medchemcomm 2016, 7 (12), 2246–2264. 10.1039/c6md00373g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Vidler LR; Brown N; Knapp S; Hoelder S Druggability Analysis and Structural Classification of Bromodomain Acetyl-Lysine Binding Sites. J. Med. Chem 2012, 55 (17), 7346–7359. 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Weinert BT; Narita T; Satpathy S; Srinivasan B; Hansen BK; Schölz C; Hamilton WB; Zucconi BE; Wang WW; Liu WR; et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/P300 Acetylome. Cell 2018, 174 (1), 231–244.e12. 10.1016/j.cell.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Horwitz GA; Zhang K; McBrian MA; Grunstein M; Kurdistani SK; Berk AJ Adenovirus Small E1a Alters Global Patterns of Histone Modification. Science 2008, 321 (5892), 1084–1085. 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jin Q; Yu L-R; Wang L; Zhang Z; Kasper LH; Lee J-E; Wang C; Brindle PK; Dent SYR; Ge K Distinct Roles of GCN5/PCAF-Mediated H3K9ac and CBP/P300-Mediated H3K18/27ac in Nuclear Receptor Transactivation. EMBOJ 2011, 30 (2), 249–262. 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Culig Z; Santer FR Androgen Receptor Signaling in Prostate Cancer. Cancer Metastasis Rev 2014, 33 (2–3), 413–427. 10.1007/s10555-013-9474-0. [DOI] [PubMed] [Google Scholar]
- (21).Isharwal S; Miller MC; Marlow C; Makarov DV; Partin AW; Veltri RW P300 (Histone Acetyltransferase) Biomarker Predicts Prostate Cancer Biochemical Recurrence and Correlates with Changes in Epithelia Nuclear Size and Shape. Prostate 2008, 68 (10), 1097–1104. 10.1002/pros.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ianculescu I; Wu D-Y; Siegmund KD; Stallcup MR Selective Roles for CBP and P300 as Coregulators for Androgen-Regulated Gene Expression in Advanced Prostate Cancer Cells. J. Biol. Chem 2012, 287 (6), 4000–4013. 10.1074/jbc.M111.300194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Heemers HV; Sebo TJ; Debes JD; Regan KM; Raclaw KA; Murphy LM; Hobisch A; Culig Z; Tindall DJ Androgen Deprivation Increases P300 Expression in Prostate Cancer Cells. Cancer Res. 2007, 67 (7), 3422–3430. 10.1158/0008-5472.CAN-06-2836. [DOI] [PubMed] [Google Scholar]
- (24).Michaelides MR; Kluge A; Patane M; Van Drie JH; Wang C; Hansen TM; Risi RM; Mantei R; Hertel C; Karukurichi K; et al. Discovery of Spiro Oxazolidinediones as Selective, Orally Bioavailable Inhibitors of P300/CBP Histone Acetyltransferases. ACS Med. Chem. Lett 2018, 9 (1), 28–33. 10.1021/acsmedchemlett.7b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lasko LM; Jakob CG; Edalji RP; Qiu W; Montgomery D; Digiammarino EL; Hansen TM; Risi RM; Frey R; Manaves V; et al. Discovery of a Selective Catalytic P300/CBP Inhibitor That Targets Lineage-Specific Tumours. Nature 2017, 550 (7674), 128–132. 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Picaud S; Fedorov O; Thanasopoulou A; Leonards K; Jones K; Meier J; Olzscha H; Monteiro O; Martin S; Philpott M; et al. Generation of a Selective Small Molecule Inhibitor of the CBP/P300 Bromodomain for Leukemia Therapy. Cancer Res 2015, 75 (23), 5106–5119. 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Conery AR; Centore RC; Neiss A; Keller PJ; Joshi S; Spillane KL; Sandy P; Hatton C; Pardo E; Zawadzke L; et al. Bromodomain Inhibition of the Transcriptional Coactivators CBP/EP300 as a Therapeutic Strategy to Target the IRF4 Network in Multiple Myeloma. eLife 2016, 5, e10483 10.7554/eLife.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zucconi BE; Luef B; Xu W; Henry RA; Nodelman IM; Bowman GD; Andrews AJ; Cole PA Modulation of P300/CBP Acetylation of Nucleosomes by Bromodomain Ligand I-CBP112. Biochemistry 2016, 55 (27), 3727–3734. 10.1021/acs.biochem.6b00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Park S; Stanfield RL; Martinez-Yamout MA; Dyson HJ; Wilson IA; Wright PE Role of the CBP Catalytic Core in Intramolecular SUMOylation and Control of Histone H3 Acetylation. Proc. Natl. Acad. Sci. U.S.A. 2017, 114 (27), E5335–E5342. 10.1073/pnas.1703105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kalin JH; Wu M; Gomez AV; Song Y; Das J; Hayward D; Adejola N; Wu M; Panova I; Chung HJ; et al. Targeting the CoREST Complex with Dual Histone Deacetylase and Demethylase Inhibitors. Nature Communications 2018, 9 (1), 53 10.1038/s41467-017-02242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wu M; Hayward D; Kalin JH; Song Y; Schwabe JW; Cole PA Lysine-14 Acetylation of Histone H3 in Chromatin Confers Resistance to the Deacetylase and Demethylase Activities of an Epigenetic Silencing Complex. eLife 7 10.7554/eLife.37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Henry RA; Kuo Y-M; Andrews AJ Differences in Specificity and Selectivity between CBP and P300 Acetylation of Histone H3 and H3/H4. Biochemistry 2013, 52 (34), 5746–5759. 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chou T-C; Talalay P Quantitative Analysis of Dose-Effect Relationships: The Combined Effects of Multiple Drugs or Enzyme Inhibitors. Advances in Enzyme Regulation 1984, 22, 27–55. 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- (34).Gasper WC; Marinov GK; Pauli-Behn F; Scott MT; Newberry K; DeSalvo G; Ou S; Myers RM; Vielmetter J; Wold BJ Fully Automated High-Throughput Chromatin Immunoprecipitation for ChIP-Seq: Identifying ChIP-Quality P300 Monoclonal Antibodies. Sci Rep 2014, 4, 5152 10.1038/srep05152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Qin Q; Mei S; Wu Q; Sun H; Li L; Taing L; Chen S; Li F; Liu T; Zang C; et al. ChiLin: A Comprehensive ChIP-Seq and DNase-Seq Quality Control and Analysis Pipeline. BMC Bioinformatics 2016, 17 10.1186/s12859-016-1274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cornwell M; Vangala M; Taing L; Herbert Z; Köster J; Li B; Sun H; Li T; Zhang J; Qiu X; et al. VIPER: Visualization Pipeline for RNA-Seq, a Snakemake Workflow for Efficient and Complete RNA-Seq Analysis. BMC Bioinformatics 2018, 19 (1), 135 10.1186/s12859-018-2139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bonaccorsi L; Muratori M; Carloni V; Zecchi S; Formigli L; Forti G; Baldi E Androgen Receptor and Prostate Cancer Invasion1. International Journal ofAndrology 2003, 26 (1), 21–25. 10.1046/j.1365-2605.2003.00375.x. [DOI] [PubMed] [Google Scholar]
- (38).Antonarakis ES; Lu C; Wang H; Luber B; Nakazawa M; Roeser JC; Chen Y; Mohammad TA; Chen Y; Fedor HL; et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N. Engl. J. Med 2014, 371 (11), 1028–1038. 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sprenger CCT; Plymate SR The Link between Androgen Receptor Splice Variants and Castration Resistant Prostate Cancer. Horm Cancer 2014, 5 (4), 207–217. 10.1007/s12672-014-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hu R; Lu C; Mostaghel EA; Yegnasubramanian S; Gurel M; Tannahill C; Edwards J; Isaacs WB; Nelson PS; Bluemn E; et al. Distinct Transcriptional Programs Mediated by the Ligand-Dependent Full-Length Androgen Receptor and Its Splice Variants in Castration-Resistant Prostate Cancer. Cancer Res 2012, 72 (14), 3457–3462. 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nakazawa M; Antonarakis ES; Luo J Androgen Receptor Splice Variants in the Era of Enzalutamide and Abiraterone. Horm Cancer 2014, 5 (5), 265–273. 10.1007/s12672-014-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Adan A; Kiraz Y; Baran Yusuf. Cell Proliferation and Cytotoxicity Assays. Current Pharmaceutical Biotechnology 2016, 17 (14), 1213–1221. [DOI] [PubMed] [Google Scholar]
- (43).Brand M; Measures AM; Wilson BG; Cortopassi WA; Alexander R; Höss M; Hewings DS; Rooney TPC; Paton RS; Conway SJ Small Molecule Inhibitors of Bromodomain-Acetyl-Lysine Interactions. ACS Chem. Biol 2015, 10 (1), 22–39. 10.1021/cb500996u. [DOI] [PubMed] [Google Scholar]
- (44).Garnier J-M; Sharp PP; Burns CJ BET Bromodomain Inhibitors: A Patent Review. Expert Opinion on Therapeutic Patents 2014, 24 (2), 185–199. 10.1517/13543776.2014.859244. [DOI] [PubMed] [Google Scholar]
- (45).Noel JK; Iwata K; Ooike S; Sugahara K; Nakamura H; Daibata M Abstract C244: Development of the BET Bromodomain Inhibitor OTX015. Mol Cancer Ther 2013, 12 (11 Supplement), C244–C244. 10.1158/1535-7163.TARG-13-C244. [DOI] [Google Scholar]
- (46).Titov DV; Gilman B; He Q-L; Bhat S; Low W-K; Dang Y; Smeaton M; Demain AL; Miller PS; Kugel JF; et al. XPB, a Subunit of TFIIH, Is a Target of the Natural Product Triptolide. Nat. Chem. Biol 2011, 7 (3), 182–188. 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Santer FR; Höschele PPS; Oh SJ; Erb HHH; Bouchal J; Cavarretta IT; Parson W; Meyers DJ; Cole PA; Culig Z Inhibition of the Acetyltransferases P300 and CBP Reveals a Targetable Function for P300 in the Survival and Invasion Pathways of Prostate Cancer Cell Lines. Mol Cancer Ther 2011, 10 (9), 1644–1655. 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- (48).Debes JD; Sebo TJ; Lohse CM; Murphy LM; Haugen DAL; Tindall DJ P300 in Prostate Cancer Proliferation and Progression. Cancer Res 2003, 63 (22), 7638–7640. [PubMed] [Google Scholar]
- (49).Faiola F; Liu X; Lo S; Pan S; Zhang K; Lymar E; Farina A; Martinez E Dual Regulation of C-Myc by P300 via Acetylation-Dependent Control of Myc Protein Turnover and Coactivation of Myc-Induced Transcription. Mol. Cell. Biol 2005, 25 (23), 10220–10234. 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Falcon S; Gentleman R Using GOstats to Test Gene Lists for GO Term Association. Bioinformatics 2007, 23 (2), 257–258. 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- (51).Rack JGM; Lutter T; Kjæreng Bjerga GE; Guder C; Ehrhardt C; Varv S; Ziegler M; Aasland R The PHD Finger of P300 Influences Its Ability to Acetylate Histone and NonHistone Targets. Journal of Molecular Biology 2014, 426 (24), 3960–3972. 10.1016/j.jmb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- (52).Raisner R; Kharbanda S; Jin L; Jeng E; Chan E; Merchant M; Haverty PM; Bainer R; Cheung T; Arnott D; et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep 2018, 24 (7), 1722–1729. 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- (53).Nguyen UTT; Bittova L; Müller MM; Fierz B; David Y; Houck-Loomis B; Feng V; Dann GP; Muir TW Accelerated Chromatin Biochemistry Using DNA-Barcoded Nucleosome Libraries. Nat Meth 2014, 11 (8), 834–840. 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zhang Y; Xue Y; Shi J; Ahn J; Mi W; Ali M; Wang X; Klein BJ; Wen H; Li W; et al. The ZZ Domain of P300 Mediates Specificity of the Adjacent HAT Domain for Histone H3. Nature Structural & Molecular Biology 2018, 25 (9), 841 10.1038/s41594-018-0114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Asangani IA; Dommeti VL; Wang X; Malik R; Cieslik M; Yang R; Escara-Wilke J; Wilder-Romans K; Dhanireddy S; Engelke C; et al. Therapeutic Targeting of BET Bromodomain Proteins in Castration-Resistant Prostate Cancer. Nature 2014, 510 (7504), 278–282. 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Chapuy B; McKeown MR; Lin CY; Monti S; Roemer MGM; Qi J; Rahl PB; Sun HH; Yeda KT; Doench JG; et al. Discovery and Characterization of Super-Enhancer Associated Dependencies in Diffuse Large B-Cell Lymphoma. Cancer Cell 2013, 24 (6), 777–790. 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Garcia-Carpizo V; Ruiz-Llorente S; Sarmentero J; Graña-Castro O; Pisano DG; Barrero MJ CREBBP/EP300 Bromodomains Are Critical to Sustain the GATA1/MYC Regulatory Axis in Proliferation. Epigenetics & Chromatin 2018, 11 (1), 30 10.1186/s13072-018-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Faus H; Haendler B Androgen Receptor Acetylation Sites Differentially Regulate Gene Control. J. Cell. Biochem 2008, 104 (2), 511–524. 10.1002/jcb.21640. [DOI] [PubMed] [Google Scholar]
- (59).Thompson PR; Wang D; Wang L; Fulco M; Pediconi N; Zhang D; An W; Ge Q; Roeder RG; Wong J; et al. Regulation of the P300 HAT Domain via a Novel Activation Loop. Nat Struct Mol Biol 2004, 11 (4), 308–315. 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- (60).Ortega E; Rengachari S; Ibrahim Z; Hoghoughi N; Gaucher J; Holehouse AS; Khochbin S; Panne D Transcription Factor Dimerization Activates the P300 Acetyltransferase. Nature 2018, 562 (7728), 538–544. 10.1038/s41586-018-0621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Soutoglou E; Viollet B; Vaxillaire M; Yaniv M; Pontoglio M; Talianidis I Transcription Factor-Dependent Regulation of CBP and P/CAF Histone Acetyltransferase Activity. EMBO J. 2001, 20 (8), 1984–1992. 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Bedford DC; Brindle PK Is Histone Acetylation the Most Important Physiological Function for CBP and P300? Aging (Albany NY) 2012, 4 (4), 247–255. 10.18632/aging.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Holmqvist P-H; Mannervik M Genomic Occupancy of the Transcriptional Co-Activators P300 and CBP. Transcription 2013, 4 (1), 18–23. 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Kasper LH; Qu C; Obenauer JC; McGoldrick DJ; Brindle PK Genome-Wide and SingleCell Analyses Reveal a Context Dependent Relationship between CBP Recruitment and Gene Expression. Nucleic Acids Res. 2014, 42 (18), 11363–11382. 10.1093/nar/gku827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Liu X; Wang L; Zhao K; Thompson PR; Hwang Y; Marmorstein R; Cole PA The Structural Basis of Protein Acetylation by the P300/CBP Transcriptional Coactivator. Nature 2008, 451 (7180), 846–850. 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- (66).Kraus WL; Manning ET; Kadonaga JT Biochemical Analysis of Distinct Activation Functions in P300 That Enhance Transcription Initiation with Chromatin Templates. Mol. Cell. Biol 1999, 19 (12), 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Ramos YFM; Hestand MS; Verlaan M; Krabbendam E; Ariyurek Y; van Galen M; van Dam H; van Ommen G-JB; den Dunnen JT; Zantema A; et al. Genome-Wide Assessment of Differential Roles for P300 and CBP in Transcription Regulation. Nucleic Acids Res. 2010, 38 (16), 5396–5408. 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.