Abstract
While gene transfer using recombinant adeno-associated viral (rAAV) vectors has shown success in some clinical trials, there remain many tissues that are not well transduced. Because of the recent success in reprogramming islet-derived cells into functional β cells in animal models, we constructed 2 highly complex barcoded replication competent capsid shuffled libraries and selected for high-transducing variants on primary human islets. We describe the generation of a chimeric AAV capsid (AAV-KP1) that facilitates transduction of primary human islet cells and human embryonic stem cell–derived β cells with up to 10-fold higher efficiency compared with previously studied best-in-class AAV vectors. Remarkably, this chimeric capsid also enabled transduction of both mouse and human hepatocytes at very high levels in a humanized chimeric mouse model, thus providing a versatile vector that has the potential to be used in both preclinical testing and human clinical trials for liver-based diseases and diabetes.
Keywords: Therapeutics
Keywords: Diabetes, Embryonic stem cells, Gene therapy
Chimeric AAV vectors that show enhanced transduction in human islets result in high levels of transduction of hepatocytes In a murine chimeric liver model.
Introduction
An estimated 30.3 million US Americans are affected by either type 1 or type 2 diabetes mellitus (1). Various strategies to cure diabetes have been evaluated over the years with limited success. For example, transplantation of cadaveric human islets into the hepatic duct has been used to replace β cells in type 1 diabetic patients, so far with low efficiencies (reviewed in ref. 2). Islets are highly vascularized and require large amounts of oxygen to survive. Because revascularization of the transplanted islets takes several weeks, those transplants suffer from a large number of cell death due to oxygen deprivation. In order to improve graft survival and function, different approaches of ex vivo gene therapy, either by supplying or repressing certain transcription factors using different viral or nonviral delivery systems (for example adeno-associated virus (AAV), adenovirus, lentivirus, various lipids, or nonlipid polymers), may be used (reviewed in ref. 3). Recombinant AAV–mediated (rAAV-mediated) overexpression of Follistatin, a protein with important roles in the proper functioning of the reproductive, endocrine, and muscoskeletal systems, has been shown to promote β cell proliferation and maintain pancreatic islet mass in a diabetic mouse model (4). In addition to the approaches described above, it is of the utmost importance to prevent loss of the transplanted islets due to recurrent autoimmune destruction. Recently, a study described the use of rAAV to overexpress Igf1 in diabetic mice (5). Igf1 is a prosurvival factor and β cell mitogen that has important roles in β cell maturation and function and is also involved in the interplay between the endocrine and the immune system. Over a period of 30 weeks, this gene therapy strategy was successfull in counteracting progression to autoimmune diabetes. One strategy for diabetes treatment is the conversion of α cells or other endocrine or exocrine pancreatic cell types into β cells (reviewed in ref. 6). Cell conversion has been achieved by overexpression or repression of certain transcription factors, such as Pdx1, Ngn3, MafA, Pax4, and Arx (7–15). Most of those in vivo studies involved transgenic mouse models or used adenoviral vectors to deliver expression cassettes. However, rAAV vectors packaged with AAV8 capsid were also employed (11), as this serotype had previously been found to transduce murine β cells with high efficiency (16, 17). Recently, i.p. delivery of an AAV8-based vector expressing IL-2 under control of the β cell type–specific insulin promoter has been described to achieve highly specific transduction of β cells in mice (18). Another study found that AAV6 was highly efficient in transducing mouse islets in vitro and in vivo when using intraductal delivery. However, when delivered systemically, AAV8 proved to be the more robust serotype (19). Recently, tyrosine-phenylalanie (Y-F) mutant AAV8 vectors have been reported to achieve up to 10-fold improved gene transfer into mouse islets as compared with WT AAV8 (20). Interestingly, studies in rats found that AAV5 was the best capsid for islet transduction in this animal model (21). AAV2 is the serotype that has been described most frequently for transduction of human islets (22, 23). More recently, capsid variants AAV-DJ (24) and AAV-LK03 (25) were found to have better transduction efficiency on human islets as compared with AAV2 and AAV3B; however, a strong preference for α cells was observed for those capsids (26).
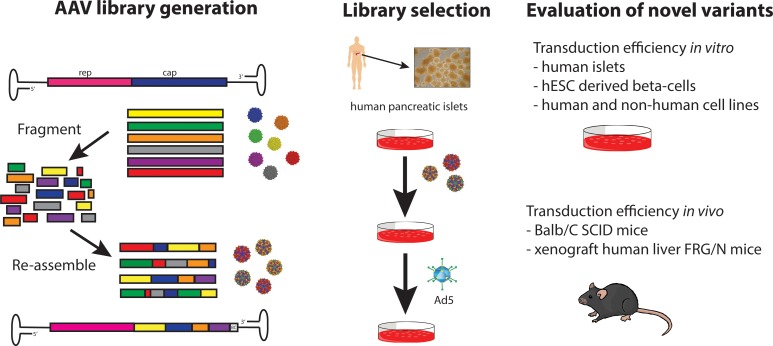
Even though different AAV capsids that are capable of transducing human islets have been described, the current low efficiencies limit their broad applicability to diabetes. In view of that, we sought to develop new capsid variants with further enhanced human islet cell transduction efficiency using directed evolution. Directed evolution by DNA shuffling is a powerful tool to mimic natural evolution in a vastly accelerated manner (27) and has yielded greatly improved variants in many different areas of research (28–41). Most importantly, no a priori knowledge about sequence-function relationships is needed, which stands in contrast to rational design strategies. In 2008, our group was the first to our knowledge to report the development of an improved AAV variant by subjecting a capsid shuffled AAV library to multiple rounds of selection on the target cell type (24). Since then, we and others have used this approach to identify AAV variants with improved properties (24, 25, 42–51).
In this project, we subjected capsid shuffled barcoded AAV libraries to multiple rounds of selection on primary human islets and analyzed enriched capsid variants for improved transduction efficiency. As previously described by others (52–54), the use of barcoded vectors allowed us to follow enrichment of chimeric variants by employing high-throughput sequencing (HTS). Among the candidates tested in our study, 3 chimeric variants were found to exhibit considerably improved transduction capacity of human islet cells — particularly of β cells. In addition, these variants showed improved transduction in other nonpancreatic cell types both in vitro and in vivo. Therefore, these capsids may be useful for various gene therapy applications targeting pancreatic islets, as well as other tissues relevant for other diseases.
Results
Evaluation of parental capsids for human islet transduction efficiency.
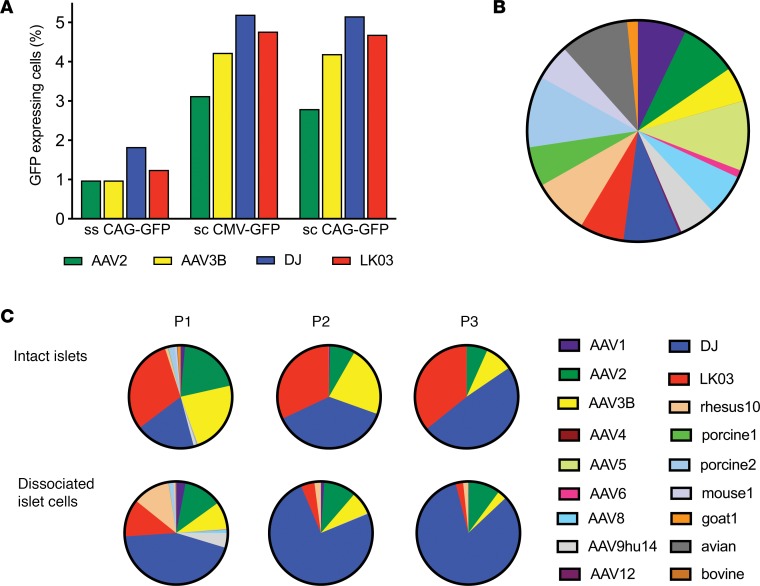
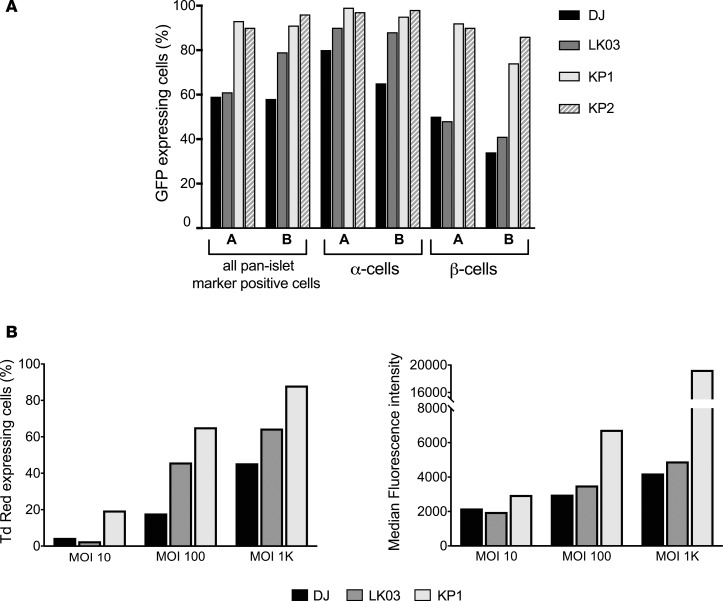
We first sought to confirm previous data showing that AAV-DJ and AAV-LK03 have higher human islet transduction efficiency than the closely related natural serotypes AAV2 and AAV3B (26). We transduced human islets with GFP-expressing rAAV vectors packaged with the different capsids and measured transduction efficiency using flow cytometry analysis (Figure 1A). Due to the limited availability of patient-derived islets, we were unable to run replicates in this and several other experiments in this study. As expected, AAV-DJ and AAV-LK03 had higher rates of transduction in human islets than AAV2 and AAV3B.
Figure 1. Evaluation of parental capsids for islet tropism.
(A) Dissociated islet cells were transduced with different vectors that had been packaged with AAV2, AAV3B, DJ, and LK03 capsids at a MOI of 1,000, and transduction efficiency was determined by flow cytometry 48 hours after transduction. Each rAAV was tested once. (B) Parental contribution in the 18-parent pool as analyzed using high-throughput sequencing of the barcodes. (C) Barcode sequences were amplified from viral genomes after passaging the 18-parent pool on human islets and were analyzed by high-throughput sequencing. Color coding of enriched parental capsids for passaging of the 18-parent pool is identical to that of the input parental pool.
This confirmatory result provided the basis for a validation study. For this experiment, we generated a control AAV pool containing parental AAVs that each carried a unique barcode (BC) sequence immediately downstream of the capsid coding sequence. After mixing equal copy numbers of each of the 18 parental AAVs, we purified the pool and analyzed its composition by HTS of the BCs (Figure 1B and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131610DS1). Although some of the parental AAVs were under- or overrepresented in the pool, the 18-parent pool was found to be useful in a pilot library screen as a first validation for the use of BC sequencing in a capsid selection screen. To evaluate which capsid was best at transducing human islets, we infected human intact — as well as dissociated — islets with the 18-parent pool and superinfected with human adenovirus 5 (Ad5) helpervirus to replicate AAVs prior to infection of the next selection round. Helpervirus-mediated replication ensured that only those AAV variants that had successfully undergone all steps involved in transduction of the target cell would be available for infection of the subsequent round. Moreover, this amplification of enriched AAVs greatly facilitated rescue of the capsid sequences by PCR. We used a 10-fold higher multiplicity of infection (MOI) for infection of intact (20K) than for dissociated (2K) islets, since we reasoned that cells located in the center of islets would be harder to infect than those on the periphery. Three consecutive rounds of selection were performed, and AAV replication (Supplemental Figure 5A), as well as composition of the viral pool (Figure 1C), was assessed at each round. For rounds 1 and 2 of selection, lower copy numbers of viral genomes were recovered after propagation on intact islets as compared with the input, whereas replication reached the highest levels at round 3. When a lower MOI of 2K was used to infect dissociated islet cells, the amount of virus obtained after islet culture was higher than the input at each round of selection. HTS of the BC sequences was used to assess the composition of AAVs at each round. Between 400,000 and 1,000,000 sequence reads were obtained for each sample. Certain parental AAVs had a clear advantage over others, as the diversity of the pool was already diminished after 1 round of selection, particularly when intact islets were used (Figure 1C). AAV2 and AAV3B, as well as shuffled variants AAV-DJ and AAV-LK03, were enriched after the first passage. After the third passage on intact islets, as expected, AAV-DJ and AAV-LK03 were equally represented, while AAV2 and AAV3B were present at lower concentrations. Passaging of the 18-parent pool on dissociated islet cells revealed early enrichment of AAV-DJ with lower proportions of AAV2, LK03, AAV3B, and AAV–rhesus 10. This experiment validated our selection screen approach, as it confirmed the results from previous transduction studies, showing that AAV-DJ and AAV-LK03 transduce primary human islets better than closely related serotypes AAV2 and AAV3B.
Characterization of highly diverse barcoded capsid shuffled AAV libraries.
We characterized the capsid shuffled barcoded libraries using various methods. The details are provided in Supplemental Results, as well as in Supplemental Figures 1–4 and Supplemental Table 2.
Selection of AAV capsids with human islet tropism.
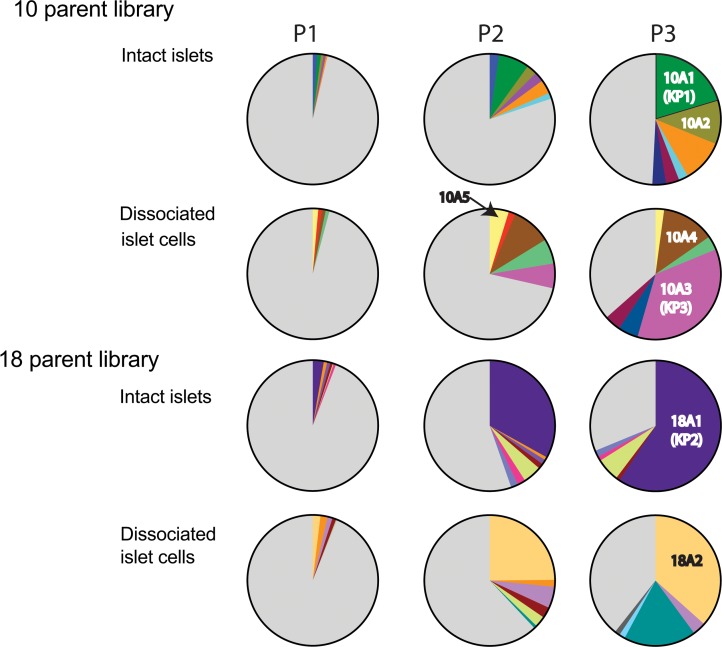
Both libraries were used to infect intact, as well as dissociated, islets and were replicated using Ad5 superinfection. After each round of selection, virus replication was assessed by quantitative PCR (qPCR) (Supplemental Figure 5A) and enrichment of variants was tracked by HTS of the BCs (Figure 2). The number of reads was between 400,000 and 800,000. In contrast to the 18-parent pool, we observed replication of the libraries at each round of selection, which might indicate that certain capsid sequences conferred improved tropism, transduction, and/or replication. Enrichment of distinct capsid variants was observed early in the selection process. Those are depicted as colored slices in the pie charts in Figure 2. A second set of library screens on intact islets using 2 different MOIs was performed in duplicate for the 10-parent library and in triplicate for the 18-parent library. Replication data for this library screen are shown in Supplemental Figure 5B, and the BC sequencing data are shown in Supplemental Figure 6. We observed similar levels of replication and enrichment during this second set of library screens. However, this time, 1 BC was found to be enriched in 2 independent screens of the 10-parent library (variant 10B5, highlighted in yellow), indicating that this particular variant may have a significant selection advantage in islets. A different BC was found to be enriched in 2 of the triplicate screens of the 18-parent library (18B2, highlighted in magenta).
Figure 2. Enrichment of distinct capsids during passaging.
Barcode sequences were amplified from viral genomes after each passage and were analyzed by high-throughput sequencing. Enriched variants are depicted in different colors, while all other variants are shown in gray. Enrichment of AAV capsid variants used for vectorization is indicated (10A1, 10A2, 10A3, 10A4 10A5, 18A1, 18A2).
Recovery of capsid sequences and evaluation of selected AAV capsid variants for improved transduction of primary human islets and β cells derived from human embryonic stem cells.
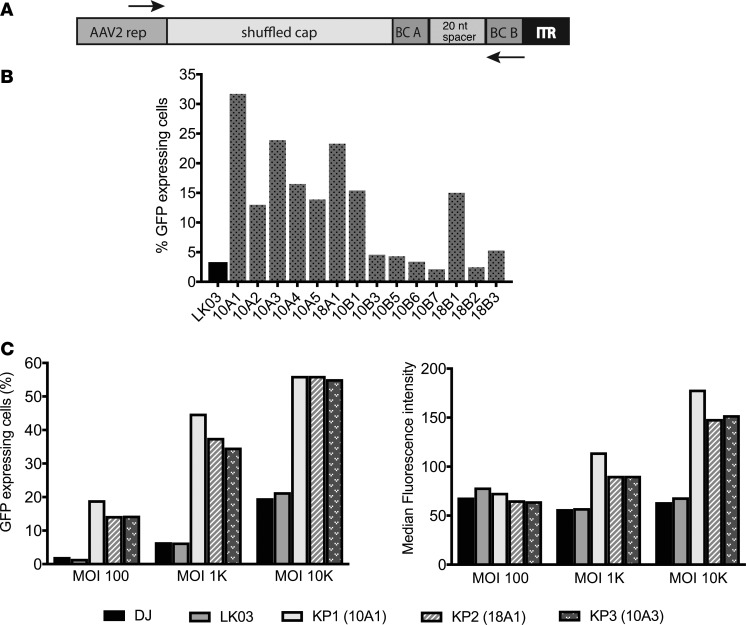
A total of 17 enriched capsid sequences were recovered from the different screens after 3 rounds of selection (see Figure 2 and Supplemental Figure 6 for enrichment data of those variants) using a forward primer upstream of cap and a BC sequence–specific reverse primer (Figure 3A), followed by TOPO cloning and sequencing. After performing sequencing analysis, it was found that all enriched capsids shared large sequences from AAV3B in the 3′ half, while the 5′ half was much more diverse (data not shown). All 17 capsid variants, as well as 1 of the control capsids (AAV-LK03), were used to package an AAV vector containing a CAG-GFP cassette. Three of the capsids (18A2, 10B2, and 10B4) failed to generate high-titer rAAV and were excluded from further testing. In an initial prescreen, crude cell lysate–derived rAAV was used to transduce dissociated islet cells using a low MOI. Due to the limited availability of human donor–derived islets, the experiment did not include biological replicates. Transduction efficiency was determined by flow cytometry analysis of GFP-expressing cells 2 days after transduction. While multiple capsid variant rAAVs exhibited higher transduction efficiency than AAV-LK03, others were only marginally improved or were not improved at all (Figure 3B). The 3 lead candidates (10A1, 18A1, and 10A3) were all derived from the first set of library screening and were renamed into KP1, KP2, and KP3, respectively. Dissociated islet cells were transduced with purified rAAVs packaged with the capsid variants, as well as with DJ and LK03 capsids, at 3 different MOIs, and transduction efficiency was analyzed by flow cytometry. As it was detected before, these 3 AAV variants were capable of transducing islet cells with improved efficiency when compared with the best parents (Figure 3C and Supplemental Figure 7A). In fact, these levels of transduction were achieved by AAV-DJ or AAV-LK03 only when a 10-fold–higher MOI was used.
Figure 3. Rescue of enriched capsid sequences and evaluation of selected capsids for islet transduction.
(A) The forward primer annealed to a sequence in the 3′ end of the rep gene; the reverse primer was specific to the sequence of the right barcode of the variant capsid to be amplified. (B) A self-complementary AAV-expressing GFP was packaged with LK03, as well as 12 capsid sequences, and islet cells were transduced using a low MOI of 1,000. Cells were sorted for GFP expression using FACS 48 hours later. Each rAAV was evaluated once. (C) Dissociated islet cells were transduced with CsCl gradient–purified scCAG-GFP rAAV preparations generated with the 2 best parental capsids, as well as the capsids that were the top transducers in the prescreen. Three different MOIs were used for transduction. Transduction efficiency is depicted both as the percentage of GFP+ cells (left graph) and the median fluorescence intensity within the GFP+ cell population (right graph). Results of a representative experiment that was performed twice are shown.
Next, we wanted to determine if rAAV packaged with the capsids transduced both α- and β cells with equal efficiency or if 1 cell population was being targeted preferentially. Due to limited islet availability, only 2 of the apparently most efficient AAV capsids were used in the study. In order to address this, islets were transduced with GFP-expressing vectors packaged into KP1, KP2, DJ, and LK03 capsids, and the different subpopulations were separated using specific antibody staining for α- and β cells. KP1 and KP2 remarkably outperformed DJ and LK03 in β cells but only modestly in α cells (Figure 4A). Importantly, these data also demonstrate that the AAV variants were capable of penetrating intact islets, as they transduced almost all of the α- and β cells when using high MOIs.
Figure 4. Analysis of transduction efficiency of the variants.
(A) GFP-expressing rAAV packaged with 2 of the variant capsids, as well as AAV-DJ and AAV-LK03 capsids, were used to transduce intact islets from 2 individual donors (donors A and B) at a MOI of 10,000, and α- and β cell–specific transduction was determined by surface staining followed by flow cytometry. The experiment was performed once. (B) Transduction efficiency of capsid KP1 for human embryonic stem cell–derived β cells. DJ, LK03, and KP1 capsids were used to package a Tomato Red vector, and hESC-derived mature β cells were transduced with the MOIs indicated. Intracellular staining for the β cell marker C-peptide was performed on day 6 after transduction, and cells were analyzed by flow cytometry. Transduction with MOI 100 was performed in a second independent experiment (Supplemental Figure 9); transduction at the other MOIs was performed only once.
β Cells derived from human embryonic stem cells (hESC) are currently being studied as a potential cell source for the treatment of patients with diabetes. Thus, we decided to test if our variants would also transduce these cells with high efficiency. Due to the limited availability of hESC and the labor intensity of the protocol for β cell generation that we had recently developed (55), we focused on the most powerful capsid variant for this study: KP1. Tomato Red–expressing vectors were used in this experiment, and cells were stained for the β cell marker C-peptide. The data for an experiment using 3 different MOIs for transduction are shown in Figure 4B, Supplemental Figure 7B, and Supplemental Figure 8. Similar to what had been observed in islet cells, transduction of these cells using AAV-KP1 was 5- to 10-fold more efficient as compared with AAV-DJ or AAV-LK03. The experiment was repeated using 1 MOI. Again, we observed improved transduction of AAV-KP1 as compared with AAV-DJ and AAV-LK03 in those cells (Supplemental Figure 9). Moreover, when immature hESC-derived β-like spheres were transduced with the 3 rAAVs, the cells transduced with rAAV packaged with KP1 capsid showed the highest rates of Tomato Red expression (Supplemental Figure 10).
Analysis of sequence determinants of the capsids.
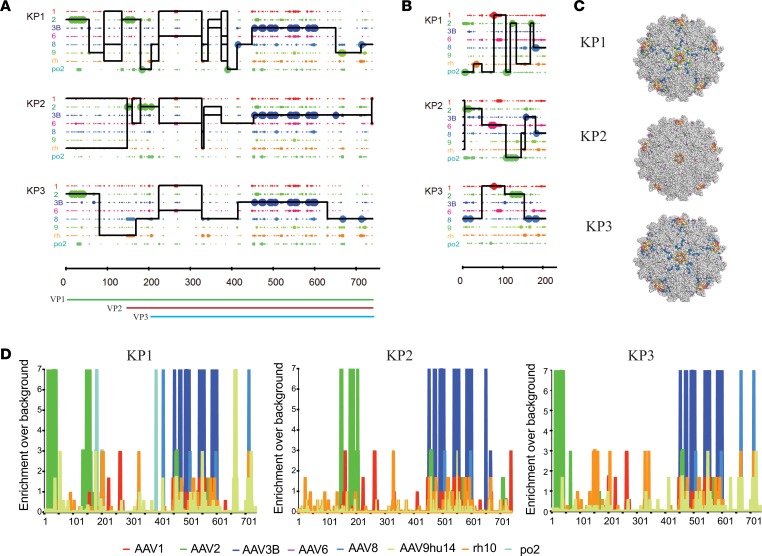
Figure 5A shows a Xover recombination analysis (56) of the 3 most improved capsid amino acid capsid sequences. While the N-terminal part of capsid KP1 contains stretches from several different parental sequences, capsids KP2 and KP3 are less diverse in this part. However, as seen for all capsids enriched during the islet screen, most of the C-terminal half of those capsids was derived from AAV3B, suggesting that this stretch may contain sequence determinants that are crucial for islet cell tropism. AAV3B is the parental that is most closely related to the 3 capsids, with 92% sequence identity to KP1 and KP3 and 95% identity to KP2 throughout the entire capsid sequence. According to recombination analysis of the nucleotide sequences (Supplemental Figure 11), the KP1 capsid sequence contains fragments from at least 7 of the 8 parental serotypes, while KP2 and KP3 contain fragments from at least 6 parents. The fact that all 3 improved capsids share 3 residues that are unique to AAV1 and AAV6 in the sequence stretch between amino acids 225 and 267 may indicate that these residues are important for human islet transduction (Figure 5A, Supplemental Figure 12, and Supplemental Table 3). Since assembly-activating protein (AAP) uses a different reading frame within the capsid gene, we also performed crossover analysis for this protein. We found that all 3 variants contained chimeric AAP sequences (Figure 5B and Supplemental Figure 13). Besides the crossover analysis, we performed enrichment analysis that confirmed strong selection pressure for certain amino acid residues in all 3 capsids (Figure 5D). In the N-terminus, the 2 most improved variants KP1 and KP3 showed a strong selection for AAV2 residues, while they share AAV8-derived residues in the C-terminus. KP2 capsid shows a strong enrichment of several AAV2 residues between positions 150 and 210. All 3 capsids have an arginine at the position that has been described to be the key HSPG binding site for AAV3B (position 597 in Supplemental Figure 12) (57). We also performed predictive 3-dimensional structural VP3 capsid mapping of the variants to reveal those residues that are displayed on the outside of the capsids and, thus, may contribute to improved islet transduction, possibly by interaction with cell surface receptors (Figure 5C). All amino acid residues that differ from the closest parental AAV3B and are shared among at least 2 of the variants are listed in Supplemental Table 3. The 3 capsids also share several non-AAV3B–derived residues within VP3 that are buried inside the capsid, as well as residues in the N-terminus of the capsid gene that encodes parts of VP1 and VP2. Those residues possibly also contribute to the observed strong transduction efficiency, as they may be involved in uncoating and other postentry steps. All but 1 of the surface-exposed tyrosine residues that have been associated with ubiquitination and subsequent proteasome-mediated degradation of AAV2 capsids are present in the variants (58). However, all variants have a phenylalanine at 1 of the positions where AAV2 contains a tyrosine, since this stretch is derived from AAV3B (position 504 in Supplemental Figure 12).
Figure 5. Amino acid sequence and structural composition of selected shuffled AAV capsid variants.
(A) Amino acid sequence mapping analysis of parental capsid fragment crossovers in vectorized shuffled capsids. Library parents are depicted in different colors as indicated on the left. Large dots represent 100% parental match (i.e., the position in question matches only 1 parent), and small dots represent more than 1 parental match (i.e., the position matches more than 1 parent) at each position. The solid line for each chimera represents the library parents identified within the sequence between crossovers. A set of thin horizontal parallel lines between crossovers indicates that multiple parents match at an equal probability. A vertical spike indicates a single position switch between parents. VP1, VP2, VP3, and AAP open reading frames are shown below. (B) Amino acid sequence mapping analysis of parental AAP fragment crossovers in vectorized shuffled capsids. (C) The residues different from AAV3B of shuffled variants were 3-dimensionally false-color mapped onto the crystal structure of AAV6 VP3. Light gray residues correspond to AAV3B amino acids, while colored residues indicate surface-exposed amino acids derived from other serotypes. With the exception of AAV3B, color coding is as in A and B. (D) Enrichment scores were calculated for each amino acid position in the sequence of each chimera by comparison of sequences from parental serotypes based on maximum likelihood. Library parents are depicted in different colors as shown.
Evaluation of transduction efficiency of the new AAV variants on a panel of diverse cells lines.
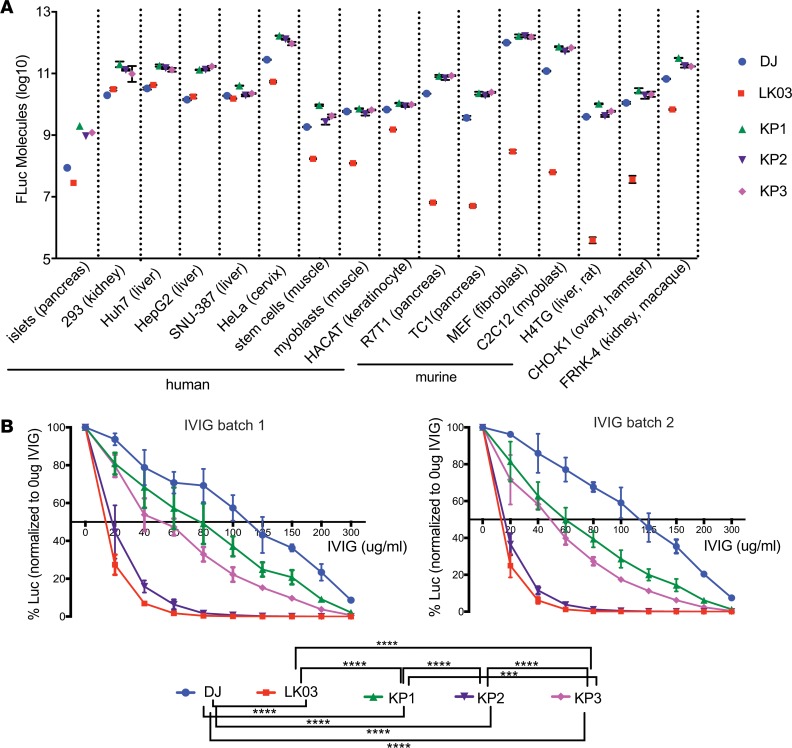
In order to evaluate if the AAV variants selected in human islets could also transduce other cell types, several primary cells — as well as cell lines derived from human and animal sources — were transduced with firefly luciferase (FLuc) rAAV vectors packaged with the 2 parental (AAV-DJ and AAV-LK03) and the 3 evolved AAV capsids (KP1, KP2, KP3) and were analyzed for transduction efficiency using a luciferase assay (Figure 6A). As had been shown previously (25), AAV-LK03 did not transduce murine cells efficiently, while AAV-DJ transduced all cell lines with high efficiency. Remarkably, the variants showed similar or slightly higher levels of transduction as compared with AAV-DJ on all cell types tested.
Figure 6. In vitro transduction experiments using rAAV-FLuc vectors.
(A) Transduction efficiency of the capsids, as well as AAV-DJ and AAV-LK03 capsids, on a variety of human- and nonhuman-derived cell types. Cells were transduced with the different capsid containing rAAVs that were packaging a FLuc expression cassette at a MOI of 1,000 in triplicate (with the exception of islet cells), and cell lysates were analyzed 48 hours after transduction in a luciferase activity assay. For primary human islet cells, results of 1 experiment that had been performed twice are shown. Two-fold dilutions of recombinant FLuc enzyme were used to prepare a standard curve, and raw luminescence units were calculated into luciferase molecules based on the standard curve. (B) Neutralization assay of rAAVs packaged with different capsids using dilutions of 2 different batches of pooled human immunoglobulin (IVIG). Huh-7 cells were transduced at a MOI of 100 with FLuc-expressing rAAVs that had been preincubated with different concentrations of IVIG for 1 hour at 37°C. Luciferase activity in cell lysates was measured 24 hours after transduction. Mean values of 5 replicates (obtained in 2 independent experiments) with SDs are shown for each sample. Experimental values were assessed via 2-way ANOVA using Tukey’s multiple comparisons test. Only statistically significant differences are indicated in the legend below the graph. ***P < 0.001, ****P < 0.0001.
Neutralization profile of the capsid variants.
One of the limitations of many AAV vectors is the fact that the majority of patients exhibit preexisting AAV neutralizing antibodies. Aiming to study the neutralization profile of our new variants, the capsid variants, as well as AAV-DJ and AAV-LK03, were analyzed for sensitivity toward neutralization by 2 different batches of pooled human immunoglobulin (Figure 6B). The experiment was performed in 7 biological replicates for each sample, split between 2 independent experiments. While AAV-DJ showed the highest resistance toward neutralization with ~120 μg/mL human i.v. immunoglobulin (IVIG) needed for 50% inhibition, AAV-LK03 was more sensitive, with 50% neutralization achieved with only ~15 μg/mL. AAV-KP2 had a neutralization profile only slightly better than that of AAV-LK03, while AAV-KP1 and AAV-KP3 needed more IVIG to be neutralized (~70 μg/mL and ~50 μg/mL for AAV-KP1 and AAV-KP3, respectively). In summary, when pooled IVIGs were used for neutralization, 2 of the variant capsids performed better than AAV-LK03 but less favorable than AAV-DJ.
In vivo biodistribution of the AAV-KP variants in mice.
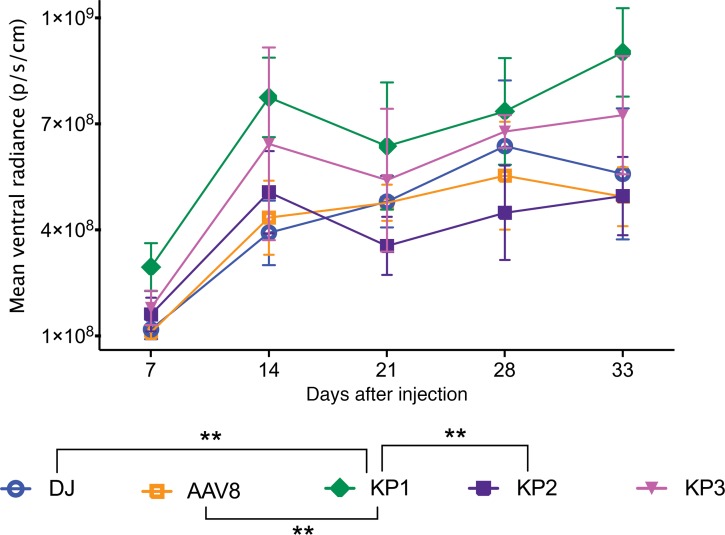
Preclinical rAAV studies are routinely performed in mice; thus, it is desirable that AAV capsids can be also used in this model prior to studies in human patients. As the capsid variants developed here had shown promise regarding transduction of a broad panel of different cell lines in vitro, we were interested to evaluate their transduction profile in vivo in mice. Mice were injected i.v. with FLuc rAAV packaged with AAV8 and AAV-DJ, as well as AAV-KP1, KP2, and KP3 capsids, and transgene expression was monitored over several weeks by live imaging (Figure 7 and Supplemental Figure 14). AAV8 was included, since AAV-LK03 does not transduce murine cells (25). As described previously (59), the majority of the injected AAVs was found to target the liver when delivering the virus i.v. While AAV-KP1 appeared to transduce mouse liver more rapidly than AAV-DJ, expression levels were only slightly higher once steady state expression was achieved at later time points. Thirty-five days after injection, several organs were harvested from each mouse and analyzed for luciferase expression. With the exception of liver, low levels of expression with high intragroup variation were detected (Supplemental Figure 15A). Vector genomes were quantified in the organs using qPCR. However, we were able to detect vector genome copies clearly above background only for the liver samples (Supplemental Figure 15B). Relative vector copy numbers correlated with the in vivo luciferase expression data.
Figure 7. In vivo transduction efficiency of rAAVs packaged with the AAV8 and AAV-DJ capsids.
Balb/C SCID mice were injected via tail vein with 2 × 1010 vg each FLuc-expressing rAAV, and luciferase expression in the livers was monitored over several weeks using live imaging after i.p injection of luciferin substrate. Four animals were injected for each group, with the exception of the AAV8 group, which contained 3 animals. The mean of each group’s mean ventral radiance is shown for each time point, with SDs indicated. One animal from the AAV8 group was omitted from analysis due to a failed substrate injection. Experimental values were assessed via 2-way ANOVA using Tukey’s multiple comparisons test. Only statistically significant differences are indicated in the legend below the graph. **P < 0.01.
Assessing functional human hepatocyte transduction in xenograft liver models in vivo.
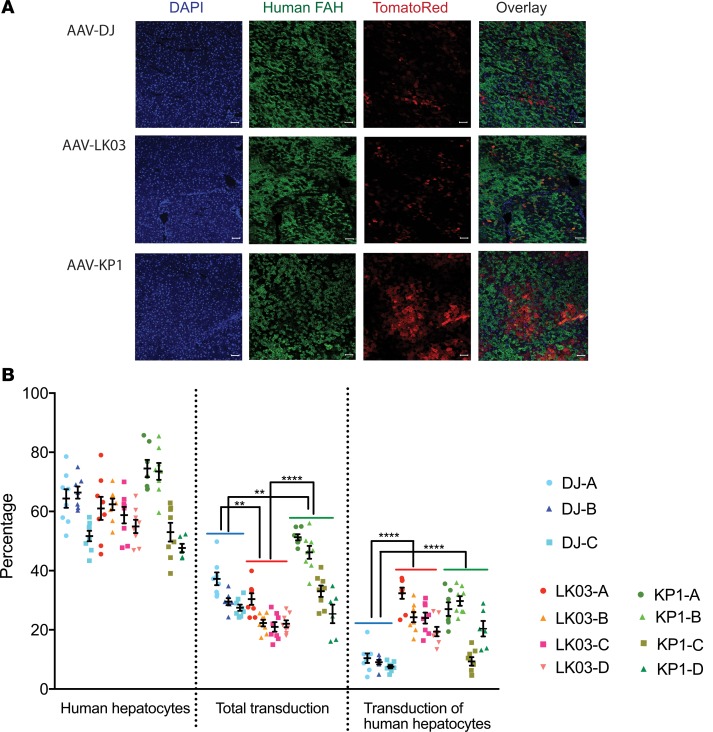
We transduced humanized Fah–/– Rag2–/– Il2rg–/– (FRG)xenograft mice to assess the functional human hepatic transduction capabilities of rAAV packaged with KP1, the most promising of our capsids, in an appropriate in vivo setting. Mice were highly repopulated with human hepatocytes, as shown by expression of high levels of human albumin (between 5.6 mg/mL and 8 mg/mL; data not shown). Humanized mice were administered Tomato Red–expressing rAAV packaged with DJ, LK03, or KP1 capsids at a dose of 1 × 1011 vg/mouse via the i.v. injection and assessed for expression of Tomato Red protein in human and mouse hepatocytes 14 days after AAV administration (Figure 8). We observed that AAV-LK03 had high transduction efficiency in human hepatocytes, but not in mouse hepatocytes, while transduction with AAV-DJ was not specific toward human cells. Those findings are not unexpected, as the LK03 capsid had been selected to be specific for transduction of primary human hepatocytes (25), while the DJ capsid was derived from an in vitro screen on human hepatoma cells in the presence of neutralizing antibodies (24) but has been reported to also transduce murine cells with high efficiency. AAV-KP1 was also found to transduce both human and mouse hepatocytes; however, levels were higher than those found with AAV-DJ. In 3 of the 4 AAV-KP1–injected mice, transduction efficiency for human hepatocytes was similar to that found in AAV-LK03–injected mice. However, total transduction was higher in AAV-KP1–injected mice, since — unlike AAV-LK03 — AAV-KP1 transduced both mouse and human cells.
Figure 8. Validation and quantification of human hepatocyte transduction in mice with humanized liver in vivo.
(A) Representative immunofluorescence images from livers of mice injected with ssAAV-Td Tomato Red at 1 × 1011 vg i.v. with varying capsid serotypes (3 mice for DJ; 4 mice each for LK03 and KP1). DAPI (blue), human-specific FAH (green), and Tomato Red (red) on liver sections. Scale bar: 50 μM. (B) Quantification of human hepatocyte repopulation levels, transduction efficiency for all hepatocytes, and transduction efficiency for human hepatocytes only. Each data point represents an area of interest for each mouse. A total of 6–9 areas of interest for each mouse were scanned and analyzed. The mean and SD for each mouse is indicated. Experimental values were assessed via 2-way ANOVA using Tukey’s multiple comparisons test. Only statistically significant differences between the groups are shown in the graphs. **P < 0.01, ****P < 0.000.
Discussion
In this work, we show for the first time to our knowledge that screening of highly diverse shuffled AAV capsid libraries that are tagged with unique short BCs yielded highly functional AAV capsids for gene transfer into human islets, as well as other clinically relevant cell types. The technology of tagging AAV capsids with individual BCs had previously been described for in vivo tracking of variants derived from small AAV libraries or parental pools and has been termed AAV BC sequences (52). For previous library screening studies in our lab that did not employ barcoded libraries, 4 or more rounds of selection were routinely performed (24, 25, 46, 47). However, in the current study, we found that 3 rounds of selection were sufficient to enrich and amplify variants with improved transduction of the target cell type to a level where they could be isolated reliably. In addition, combining short DNA BCs and single-molecule DNA sequencing approaches will enable us to optimize other important parameters, such as the best MOI to use during the selection process and the number of screening rounds necessary for efficient enrichment. When generating the AAV libraries by transfection, we were careful to not exceed 5,000 copies per cell, as this is the number that has been found to result in minimum cross-packaging while still yielding high-titer libraries (60). A recent report by the Dirk Grimm group in Heidelberg, Germany, laid to rest the concern that functionality of AAV capsid libraries generated for directed evolution studies might be severely compromised by inactivation of the assembly activating protein (AAP) in a large proportion of the chimeric variant pool (61). Although this small protein had previously been described to play an important role in virus assembly (62–65), it appears that AAV is strikingly tolerant toward recombination within the AAP coding sequence. Indeed, it has been described that natural AAV serotypes 4, 5, and 11 do not require AAP for successful capsid assembly (66). The AAP sequences of all the chimeras selected in the present studies are chimeric, and the rAAV titres obtained with the capsids were similar to or a little lower than those obtained with LK03.
When we subjected the libraries to stringent selection pressure on human islets, AAV variants with distinct capsid sequences were enriched after a few rounds of selection. Several of those variants showed improved transduction efficiency for human islets as compared with the best known capsid variant rAAVs to date — LK03 and DJ. The best performing capsid variants identified in the current study were at least 10 times more efficient at transducing human islet cells, particularly β cells. In addition, when 1 of the variants was tested for transduction efficiency of hESC-derived β cells, we were able to achieve similar levels of improvement. In addition, we confirmed prior observations that capsid variants AAV-DJ and AAV-LK03 were capable of transducing primary human islets with equal or better efficiency than AAV2 or AAV3B. While it is not clear which amino acid residues in the capsids are responsible for the improved islet phenotype, we noticed several features in the improved capsid sequences that may confer enhanced transduction of islet cells, as well as other cell types. It is possible that several surface-exposed tyrosine residues that were described to trigger degradation of the capsids (58) can be mutated to achieve further enhancement of transduction efficiency for the capsids.
Although the vectors described in this study mainly transduced the liver when delivered systemically, they could possibly be safely delivered in close proximity to islets using endoscopic retrograde cholangio-pancreatography (ERCP), a procedure that is routinely used in patients to examine the pancreatic and bile ducts (67). Several recent studies have reported retrograde pancreatic intraductal delivery for safe and effective administration of rAAV vectors into the pancreas of mice (5, 16, 68). The vectors generated in our study could be used for gene therapy of diabetes by delivering transcription factors or small hairpin RNAs (shRNA) into pancreatic islet cells using ERCP. The overexpression or inhibition of several transcription factors, such as Pdx1, Ngn3, MafA, Pax4, and Arx, was found to effectively convert pancreatic islet progenitors and committed islet α cells into β cells (7, 8, 10–15). Moreover, expression of other factors, such as Igf1 and Follistatin, has also shown promise in preserving β cell mass or protecting newly transplanted donor islets from apoptosis (4, 5).
When developing a vector for gene therapy, it is important to keep the goal of a clinical application in mind. Therefore, it is desirable to screen AAV capsid libraries in cells of human origin rather than in murine or other animal models. This has been clearly evidenced by a highly neurotropic AAV variant (AAV-PHP.B) that had recently been discovered through an in vivo library screen in mice (69) and, unfortunately, was recently found to exhibit this enhanced tropism in mice only (70, 71). However, for the purpose of preclinical testing, it is desirable to develop capsids that can transduce murine cells in addition to human cells, as this can accelerate the translation of different gene therapy approaches to the clinic. It is interesting that KP1-packaged rAAV — despite its similarity to AAV3B and AAV-LK03 — transduced mouse hepatocytes as well as human hepatocytes in the humanized liver mouse model. This humanized model was used previously to select for several highly human liver AAV transducing vectors (25, 47), one of which has been shown to provide robust human Factor VIII expression in an early human clinical trial (72, 73). However, all of the vectors selected in the previous screen showed poor transduction of mouse tissues or cells in vitro and in vivo. rAAV packaged with KP1 capsid showed similar transduction of human hepatocytes in the chimeric mouse model but, in contrast to AAV-LK03, also showed efficient transduction of mouse liver. This is intriguing because some researchers have suggested that the transduction of human hepatocytes in this mouse model was exaggerated because of the relatively low transduction of mouse cells (74). If the data obtained from the human hepatocytes that had been engrafted in the mice are influenced by the degree of mouse hepatocyte transduction, perhaps the KP1 variant will provide even more robust transduction when tested in human trials where the liver cell population will be more homogenous. At a minimum, because this vector transduces both mouse and human liver, a surrogate capsid for preclinical testing is not required. Ultimately, these capsids are good candidates for future study in human clinical liver–based gene therapy trials. In fact, because islet transduction will require localized delivery into the pancreatic duct, the KP variants could potentially be used in both applications.
The use of barcoded libraries for molecular evolution studies was found to be highly beneficial, as high-throughput analysis of the BCs of enriched variants is much more cost-effective and thorough than cloning and sequencing of the entire capsids. The libraries generated for this study are currently being evaluated in screens on a number of other target cells and may result in the discovery of other AAV capsid variants that are useful for other clinical gene therapy applications. The pool of parental barcoded AAVs proved to be a convenient and effective tool for validatation of the library selection parameters for the current study.
Methods
Generation of an AAV vector containing a library of unique BC sequences.
A WT AAV2 vector in which the capsid coding sequences had been replaced by a PacI and AscI containing linker fragment (provided by D. Grimm and S. Grosse, University of Heidelberg, Heidelberg, Germany) was used as the starting material for construction of the barcoded AAV library. Two unique restriction sites (AgeI and EagI) were introduced just downstream of the cap polyadenylation signal. BCs consisting of 2 stretches of 12 random nucleotides separated by a 20-nt long spacer sequence were generated as described previously (52). A detailed description of the BC library generation can be found in Supplemental Methods.
Generation of capsid shuffled barcoded AAV libraries.
DNAse I–mediated family shuffling was essentially performed as described previously (35, 54, 75). A detailed description of the method is provided in Supplemental Methods. Capsid sequences from 16 AAV serotypes (AAV1, AAV2, AAV3B, AAV4, AAV5, AAV6, AAV8, AAV9hu14, AAV12, AAV–rhesus 10, AAV–porcine 1, AAV–porcine 2, AAV-bovine, AAV–mouse 1, AAV-avian, AAV–goat 1), as well as from shuffled variants AAV-DJ and AAV-LK03 that had been selected in previous screens (24, 25), were used as parental sequences for the shuffling reactions. The capsid sequences had been obtained from various sources (D. Grimm, University of Heidelberg; Mark Kay lab, Vector Core, Stanford University).
AAV libraries were generated in HEK 293T cells using the calcium phosphate transfection method. AAV production is described in detail in Supplemental Methods.
Generation of barcoded parental AAV pools.
Capsids from all 18 parental AAVs were cloned into the BC library vector using PacI and AscI restriction sites. Each parental AAV contained a unique BC sequence, as confirmed by sequence analysis, and between 2 and 6 T225 with 293T cells were transfected with each parental AAV (37.5 μg AAV plasmid and 37.5 μg pAd5 helper plasmid per T225). Crude lysates of each barcoded parental cap AAV were generated, and 2.8 × 1012 vg of the 10 parents or 1.1 × 1012 vg of the 18 parents were mixed together to generate the 10-parent and the 18-parent pool, respectively. AAV pools were purified by double-CsCl gradient centrifugation.
Sequence contribution analysis of evolved AAV capsids.
Contigs were assembled using Sequencher 5.3 software and aligned using the Muscle multiple sequence alignment software (MacVector, Version 14.5.3). Xover 3.0 DNA/protein shuffling pattern analysis software (56) was used to generate parental fragment crossover maps of shuffled variants. Each parental serotype was color coded as indicated in the figures.
PacBio sequencing of AAV pools and libraries.
For the 10-parent library, as well as the 18-parent pool, a 2.4-kb fragment containing the capsid as well as the BC sequences was amplified using capF and QSeqRev from extracted viral genomes, loaded onto a 1% agarose gel, visualized by staining with SybrSafe, and gel-purified using a gel extraction kit (Qiagen). The 10-parent library was also assessed at the plasmid level prior to generating the AAV library using restriction enzymes (PacI and XbaI) to release the capsid sequences and was gel purified as described above for the amplified capsid sequences. Library preparation and Pacific Biosciences (PacBio) sequencing were performed at the University of Washington (Seattle, Washington, USA) PacBio Sequencing Service as described previously (47). Bioinformatic assessment of the sequence reads was performed as described (47).
False-colored structural capsid mapping.
Chimeric capsids (VP3 sequences only) were false-color mapped onto the AAV6 structure 4V86 (76) using PyMOL Version 2.3.0. Only surface-exposed amino acids that are different from the closest parental AAV3B are shown. With the exception of AAV3B, which is shown in gray, all mapped amino acid residue colors correspond to parental serotype colors used in the crossover and enrichment figures.
Conservation and enrichment calculations.
Amino acid conservation for each position was calculated using the alignment profile obtained with MacVector version 14.5.3. Average conservation values were calculated for stretches of 30 amino acid residues and were used to generate the graphs. Percent parental conservation was determined using an in-house algorithm that identifies the percentage of each parent on each aligned position in the shuffled library. The maximum square size indicates that 100% of variants share that amino acid from that parent at that position. All other square sizes are proportional to the percent of variants from 0%–100% that have that amino acid at that position from that parent. Enrichment scores were calculated for each amino acid position in the sequence of each chimera by comparison of sequences from parental serotypes based on maximum likelihood. Xover version 3.0 (56) was used to generate a crossover data analysis set for each chimera. Excel version 16.20 was used to convert those data into enrichment scores. Library parents are depicted in different colors as shown.
HTS of AAV BCs.
BC sequences were amplified with indexed primers (forward [F]: 5′ - AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T (I) CGC GCC ACT AGT AAT AAA C - 3′ and reverse [R]: 5′ - CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC T (I) TAG AGC AAC TAG AGT TCG - 3′, with the indices (I) containing between 4 and 6 nucleotides), gel-purified from 2% SybrSafe containing agarose gels, pooled (up to 30 samples), and sequenced on a MiSeq instrument. The number of PCR cycles was minimized to avoid amplification bias and was dependent on the concentration of input AAV genomes, as determined by qPCR using primers and a Taqman probe specific for the AAV2 rep gene. The following cycling conditions were used: 2 minutes at 98°C, 15–30 cycles of 15 seconds at 98°C, 15 seconds at 50°C, and 20 seconds at 72°C, with a final 15-minute extension at 72°C. Phusion Hot Start Flex (NEB) was used for all amplifications.
Cell culture conditions.
Cells and culture conditions are described in Supplemental Methods.
Selection of AAV libraries on human islets.
Islets were left in a 10-cm Petri dish with 10 mL complete media to recover overnight prior to AAV infection. Islets were infected either intact or were dissociated into single cell suspensions using Accumax prior to infection (1 mL Accumax per 1,000 islet equivalents [IEQ]). Approximately 300 IEQ or 1.7 × 105 dispersed islet cells were seeded in several wells of an ultralow attachment 24-well plate, infected with various MOIs of either the 18 parent AAV mix or the AAV libraries, and incubated in a 37°C incubator for 6 hours. After 2 PBS washes to remove leftover input virus, cells were superinfected with human Ad5 obtained from ATCC (catalog VR-5). For intact islets 8 × 107 PFU were used, for dissociated islet cells 4 × 107 PFU were added into 1 mL media per well. After 4 days at 37°C, the cells and supernatant were harvested, subjected to 3 freeze-thaw cycles, and incubated for 30 minutes at 65°C to inactivate Ad5. Cell debris was removed by centrifugation (2 minutes, 10,000 g), and viral genomes were isolated from 100-μL clarified supernatant for titration by qPCR using a rep primer-probe set. For subsequent rounds of passaging, similar MOIs as for the initial infections were used if sufficiently high titers were achieved. When titers were low, a maximum volume of 200 μL was used for infection.
Vector plasmids.
A self-complementary rAAV vector expressing GFP under control of a CAG promoter (pscAAV-CAG-GFP, Addgene, catalog 83279) was generated by replacing the CMV promoter in plasmid pscAAV- GFP (gift from John T. Gray, Addgene, catalog 32396) with the CAG promoter derived from pAAV-CAG-GFP (gift from Edward Boyden, Addgene, catalog 37825). A single-stranded rAAV vector expressing FLuc under control of the CAG promoter (pAAV-CAG-FLuc, Addgene, catalog 83281) was generated by replacing the GFP sequences in plasmid pAAV-CAG-GFP with FLuc sequences obtained from plasmid pAAV-EF1α-FLuc-WPRE-HGHpA (Addgene, catalog 87951). A single-stranded rAAV vector expressing codon diversified Tomato Red was a gift from Edward Boyden (pAAV-CAG-tdTomato, Addgene, catalog 59462).
Recovery and evaluation of enriched AAV capsid sequences.
Capsid sequences were amplified from viral genomes after the third round of selection using primer capF and a reverse primer containing the respective BC-specific sequence on its 3′ end. The method is described in detail in Supplemental Methods. The capsids were used to package a self-complementary CAG promoter–driven GFP expression vector by calcium phosphate triple transfection. For each T225 flask, 25 μg self-complementary (sc) CAG-GFP transfer vector, 25 μg packaging plasmid, and 25 μg pAd5 helper plasmid was used. Crude cell lysates were generated; rAAV titers were determined by qPCR using a GFP-specific primer-probe set and tested for transduction efficiency of dissociated human islet cells using a MOI of 1,000. Forty-eight hours after transduction, the reaggregated pseudo islets were dissociated into single-cell suspensions by incubation with Accumax followed by treatment with Dispase. The number of GFP-expressing cells was evaluated using a BD FACS Calibur cell analyzer (BD Biosciences), and FlowJo software Version 10 was used to analyze and graph data. Selected capsid variants were used to generate CsCl gradient purified vector preparations packaging different expression vectors.
Evaluation of cell type–specific transduction efficiency of capsid variants.
The cold transduction method was performed for those studies. Briefly, approximately 300 intact islets were resuspended in 100 µL CMRL-1066 (Thermo Fisher Scientific, 11530-037) with 2% FBS (Sigma-Aldrich, F2442), rAAV was added at an MOI of 10,000 (assuming 1,000 cells per islet), and the mixture was incubated on ice while gently rocking on a horizontal shaker in the cold room. After 2 hours, 1 mL prewarmed complete media (CMRL-1066 with 10 mM HEPES; Corning, 25060Cl), 0.5% human serum albumin (Sigma-Aldrich, 12668), 2% FBS, 10 mM nicotinamide (Sigma-Aldrich, N3376), 1% antimycotic-antibiotic (Thermo Fisher Scientific, 15240062), and 1% Glutamax (Thermo Fisher Scientific, 35050061) was added to each sample, and islets were incubated on 24-well ultralow attachment plates. Media was replaced after 2 days, and islets were harvested, dissociated, and analyzed by FACS as described previously using surface antibodies to subdivide into α- and β cells and non–α-/non–β cells (77).
Evaluation of rAAVs for transduction efficiency on hESC-derived β cells.
rAAVs were mixed with 800,000 GFP-high cells sorted from 20 spheres at an MOI of 10; 100; and 1,000 and reaggregated in Aggrewell-400 (Stemcell Technologies, 34415) in CMRL-1066 (Thermo Fisher Scientific, 11530-037) containing 10% FBS (Corning, 35011132), 1:100 Glutamax (Thermo Fisher Scientific, 35050-061), 1:100 NEAA (Thermo Fisher Scientific, 11140-050), 10 μm ALKi II (Cayman Chemical, 14794), 0.5 mM vitamin C (MilliporeSigma, 49752-10G), 1 μM T3 (MilliporeSigma, T6397), 1 mM N-acetyl Cysteine (MilliporeSigma, A9165), 10 μM zinc sulfate (MilliporeSigma, Z0251), and 10 μg /ml of heparin sulfate (MilliporeSigma, H3149). Media was replaced after 3 days, when the reaggregated enriched β-clusters (eBCs) were transferred into 6-well suspension plates. They were placed on orbital shakers at 100 rpm and further cultured for 3 days. Subsequently, the eBCs were dissociated, fixed, permeabilized, and stained with an anti–human C-peptide-488 monoclonal mouse antibody (Chemicon, catalog C-PEP-01, 1:200 dilution) and anti-RFP antibody (Rockland, catalog 600-401-379, 1:500 dilution) for analysis on LSRFortessa ×20 Dual, as described previously (78). Data were analyzed with FlowJo software. Anti–human C-peptide antibody was conjugated to FITC-488 in house using the Molecular Probes Antibody Labeling Kit (catalog A20181) according to manufacturer’s instructions. An Alexa Fluor 555–conjugated donkey anit–rabbit IgG antibody (Invitrogen, catalog A31572) was used to detect the anti-RFP antibody. Live images were taken using Leica DMI4000 B.
Evaluation of the variants for transduction efficiency on a variety of cell lines.
Capsid sequences of AAV-DJ and AAV-LK03, as well as the variants AAV-KP1, -KP2, and -KP3 were used to package a single stranded CAG-FLuc vector. rAAV preparations were double-CsCl purified and used to transduce a variety of human and mouse primary cells and cell lines at a MOI of 1,000 in triplicates. Except for the differentiated human muscle cells, all cells were seeded 1 day prior to transduction on 48-well plates so that they were about 60%–70% confluent at the time of transduction (seeding density of 20,000–80,000 per well, depending on size and proliferation rate). Cells were lysed and assayed for luciferase activity using the Luciferase Assay Kit (Promega) 48 hours after transduction. Purified recombinant luciferase protein (Promega) was used to generate a standard curve.
Neutralization assay.
Two different batches of pooled human immunoglobulin fractions (IVIG, Baxter) were used to evaluate the variants for sensitivity to neutralizing antibodies. Neutralization assays were essentially performed as described (79). Briefly, IVIG preparations were diluted in complement inactivated FBS and incubated for 1 hour at 37°C with 2 × 108 vector genomes of each ssCAG-FLuc vector packaged with the different capsids in a total volume of 100 μL. Huh7 cells that had been seeded on 48-well plates the day before (5 × 104 per well) were transduced with the virus-IVIG mixtures in triplicates (22.5 μL each well, corresponding to MOI of about 100), and luciferase activity in the cell lysates was determined 24 hours later.
Mice.
FRG female mice on a NOD-strain background (FRG/N) were housed and maintained in specific pathogen–free barrier facilities at OHSU. FRG/N mice were maintained on irradiated high-fat, low-protein mouse chow (Lab Diet, catalog 5LJ5) ad libitum to decrease flux through the tyrosine pathway. Beginning on the day of transplantation, FRG/N mice were maintained for 1 week on acidified water to prevent bacterial growth. The following week, mice were switched to 1 week of water supplemented with antimicrobial Sulfadiazine/Trimethoprim at 0.72 g/L (Equisul SDT, Aurora Pharmaceuticals; NADA, 141-360; NDC, 51072-020-00) and with 0.7 mol/L dextrose (Sigma-Aldrich) for palatability. Thereafter, FRG/N mice were cycled on and off 1 mg/L NTBC water as described (80). Thereafter, FRG/N mice were cycled on and off 1 mg/L NTBC water as described. Female Balb/C SCID mice between 6 and 8 weeks of age were purchased from The Jackson Laboratory (catalog 001803) for imaging studies.
Hepatocyte transplantation.
Donor human hepatocytes for transduction studies were acquired from BioreclamationIVT (lot QIE). Weanling FRG/N mice were preconditioned with administration of recombinant human adenovirus expressing urokinase (5 × 1010 PFU retroorbitally) 24 hours prior to transplant to promote human cell engraftment. Between 5 × 105 and 1 × 106 human hepatocytes were injected intrasplenically into anesthetized recipient FRG/N mice and cycled on/off NTBC to promote human hepatocyte engraftment and expansion. Broad-spectrum antibiotic (ceftiofur 4 mg/kg) was given by i.p. injection immediately prior to surgery and for 2 days following surgery. Six months after transplant, circulating human albumin levels as measure of engraftment were determined with the Bethyl Quantitative Human Albumin ELISA kit (catalog E88-129).
In vivo transduction experiments.
For evaluation of WT mouse liver transduction efficiency, white Balb/C SCID mice were injected with 2 × 1010 vector genomes of each CAG-FLuc vector via normodynamic i.v. lateral tail vein injections. AAV8 was used in place of LK03, as this capsid had previously been shown to be highly human specific. Mice were monitored for luciferase activity in the liver once a week by i.p. injection of 150 μg per g body weight D-Luciferin (Biosynth, catalog L-8220) and ventral luciferase readings using an Ami Imaging System. On day 35, mice were sacrificed, and various organs were recovered (liver, pancreas, heart, lung, spleen, brain, and kidney). Organs were homogenized in Passive lysis buffer (Promega) using a Bullet Storm Homogenizer, and luciferase activity from 1 mg each tissue sample was measured as described above. Genomic DNA was isolated from 10 mg of each tissue sample, and vector copy numbers were determined using qPCR. Primers for luciferase were FLuc forward (F): 5′ - CAC ATA TCG AGG TGG ACA TTA C - 3′ and FLuc reverse (R): 5′ - TG TTT GTA TTC AGC CCA TAG - 3′. Mouse actin primers (m-act F: 5′ - CCT GTA TGC CTC TGG TCG TA - 3′ and m-act R: 5′ - CCT CGT AGA TGG GCA CAG T - 3′) were used for normalization.
For evaluation of human hepatocyte transduction in vivo, humanized FRG/N mice (3 mice for DJ; 4 mice each for LK03 and KP1) were injected i.v. with 1 × 1011 ssCAG-Td Tomato Red vector genomes pseudotyped with DJ, LK03, or KP1 capsids and maintained on 1 mg/L NTBC during this 14-day transduction. Livers were harvested under inhalation isoflurane anesthesia. Liver tissue was cut into several 2 × 5–mm pieces from several lobes and fixed in 10× volume of 4% PFA for 5 hours at 25°C, protected from light. Fixed tissue was washed 1× in PBS and put through a sucrose cryoprotection and rehydration series (10% w/v sucrose for 2 hours at 25°C, 20% w/v sucrose overnight at 4°C, 30% w/v sucrose for 4 hours at 25°C). Liver pieces were rinsed in PBS, blotted dry, and mounted in cryomolds (Tissue-Tek, catalog 4557) with OCT (Tissue-Tek, catalog 4583) and frozen in a liquid nitrogen-cooled isopentane bath. Labeled cryomolds were wrapped in aluminum foil and placed at –80°C until sectioning.
Liver IHC.
Each liver sample with 4–5 lobes was cut in a microtome at 5 μM per section. Fixation and staining was essentially performed as described previously (47); a detailed description is provided in Supplemental Methods.
Sequences.
The complete nucleotide sequences for capsids KP1, KP2, and KP3 are available from GenBank at NCBI (accession numbers MN428626, MN428627, MN428628, respectively).
Statistics.
Statistical analyses were conducted with Prism v7.0d. Experimental values were assessed via 2-way ANOVA using Tukey’s multiple comparisons test. P < 0.05 was considered statistically significant.
Study approval.
The IACUCs of Stanford University and OHSU approved all mouse procedures.
Author contributions
KP, GDA, and MAK designed the experiments. KP, GDA, FG, MRT, YK, FZ, RS, JX, SN, QJL, JL, MH, MG, and MAK generated reagents and protocols, performed experiments, and analyzed data. KP and MAK wrote the manuscript and generated the figures. All authors reviewed, edited and commented on the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (U01DK089569 and RO1 AI116698). Human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program (IIDP) at City of Hope, NIH grant 2UC4DK098085, and the Stanford Islet Research Core. Work in MH’s laboratory was supported by NIH R01 (DK105831); YK was supported by a Kraft Family Postdoctoral Fellowship. We are deeply indebted to the islet donors and their families for making this study possible. We are grateful to Dirk Grimm and Stephanie Grosse (University of Heidelberg) for supplying this study with the AAV backbone vector and the majority of parental capsid sequences. We thank Javier Alcudia (Stanford Vector Core) for supplying us with several parental capsids and Hiroyuki Nakai (OHSU) for advice on BC library generation. We are indebted to Yan Hang, Heshan Peiris, and Robert Whitener (Stanford University) for help with islet culture, staining, and FACS. We thank Hak Kyun Kim (Stanford University) for help with figure generation, Francesco Puzzo for guidance on statistics, and Paul Valdmanis (University of Washington) for development of the conservation and enrichment analysis. This project was also supported by a NIH Shared Instrumentation Grant (S10-OD010580) from the National Center for Research Resources (NCRR), with significant contribution from Stanford’s Beckman Center, as well as the Stanford Small Animal Imaging Facility. The authors wish to acknowledge the Stanford Genomics Facility for performing HTS. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the various funding bodies or universities involved. Packaging plasmids of the new capsids described herein must be obtained through an MTA with Stanford University.
Version 1. 11/14/2019
Electronic publication
Footnotes
FG’s present address is: U.S. Food and Drug Administration, Silver Spring, Maryland, USA.
GDA’s present address is: Sangamo Therapeutics, Richmond, California, USA.
MRT’s present address is: Denali Therapeutics, South San Francisco, California, USA.
Conflict of interest: KP and MAK are named on patent applications for AAV variants used in this paper. MAK has equity interests in LogicBio Therapeutics. MG has equity interests in Yecuris Corp., Ambys Medicines, and LogicBio Therapeutics. MH owns stock in Encellin and Viacyte Inc., receives research support from Eli Lily, and holds roles as consultant and member of the scientific advisory board for Semma Therapeutics and Encellin.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(22):e131610.https://doi.org/10.1172/jci.insight.131610.
Contributor Information
Katja Pekrun, Email: kpekrun@stanford.edu.
Gustavo De Alencastro, Email: gus.alencastro@gmail.com.
Qing-Jun Luo, Email: qingjun.luo@gmail.com.
Jun Liu, Email: JUNLIU18@stanford.edu.
Youngjin Kim, Email: youngjin.kim@ucsf.edu.
Sean Nygaard, Email: nygaards@ohsu.edu.
Feorillo Galivo, Email: feorillo@gmail.com.
Feijie Zhang, Email: zfeijie@stanford.edu.
Ren Song, Email: rensong@stanford.edu.
Jianpeng Xu, Email: jxu6@stanford.edu.
Matthias Hebrok, Email: mhebrok@diabetes.ucsf.edu.
Markus Grompe, Email: grompem@ohsu.edu.
References
- 1. CDC. US Department of Health Human Services. Web Site. https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf Updated 2017. Accessed October 30, 2019.
- 2.Bruni A, Gala-Lopez B, Pepper AR, Abualhassan NS, Shapiro AJ. Islet cell transplantation for the treatment of type 1 diabetes: recent advances and future challenges. Diabetes Metab Syndr Obes. 2014;7:211–223. doi: 10.2147/DMSO.S50789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Meloche M, Verchere CB, Ou D, Mui A, Warnock GL. Improving islet engraftment by gene therapy. J Transplant. 2011;2011:594851. doi: 10.1155/2011/594851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C, et al. Overcoming Insulin Insufficiency by Forced Follistatin Expression in β-cells of db/db Mice. Mol Ther. 2015;23(5):866–874. doi: 10.1038/mt.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallol C, et al. AAV-mediated pancreatic overexpression of Igf1 counteracts progression to autoimmune diabetes in mice. Mol Metab. 2017;6(7):664–680. doi: 10.1016/j.molmet.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira A, et al. β-Cell replacement as a treatment for type 1 diabetes: an overview of possible cell sources and current axes of research. Diabetes Obes Metab. 2016;18 Suppl 1:137–143. doi: 10.1111/dom.12721. [DOI] [PubMed] [Google Scholar]
- 7.Collombat P, et al. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138(3):449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtney M, et al. The inactivation of Arx in pancreatic α-cells triggers their neogenesis and conversion into functional β-like cells. PLoS Genet. 2013;9(10):e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lima MJ, et al. Generation of Functional Beta-Like Cells from Human Exocrine Pancreas. PLoS ONE. 2016;11(5):e0156204. doi: 10.1371/journal.pone.0156204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Long-Term Correction of Diabetes in Mice by In Vivo Reprogramming of Pancreatic Ducts. Mol Ther. 2018;26(5):1327–1342. doi: 10.1016/j.ymthe.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao X, et al. Endogenous Reprogramming of Alpha Cells into Beta Cells, Induced by Viral Gene Therapy, Reverses Autoimmune Diabetes. Cell Stem Cell. 2018;22(1):78–90.e4. doi: 10.1016/j.stem.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Fava GE, Wang H, Mauvais-Jarvis F, Fonseca VA, Wu H. PAX4 Gene Transfer Induces α-to-β Cell Phenotypic Conversion and Confers Therapeutic Benefits for Diabetes Treatment. Mol Ther. 2016;24(2):251–260. doi: 10.1038/mt.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarthy H, et al. Converting Adult Pancreatic Islet α Cells into β Cells by Targeting Both Dnmt1 and Arx. Cell Metab. 2017;25(3):622–634. doi: 10.1016/j.cmet.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka TA, et al. Mafa Enables Pdx1 to Effectively Convert Pancreatic Islet Progenitors and Committed Islet α-Cells Into β-Cells In Vivo. Diabetes. 2017;66(5):1293–1300. doi: 10.2337/db16-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuyama K, et al. Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature. 2019;567(7746):43–48. doi: 10.1038/s41586-019-0942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez V, et al. In vivo genetic engineering of murine pancreatic beta cells mediated by single-stranded adeno-associated viral vectors of serotypes 6, 8 and 9. Diabetologia. 2011;54(5):1075–1086. doi: 10.1007/s00125-011-2070-3. [DOI] [PubMed] [Google Scholar]
- 17.Tonne JM, et al. Beta cell regeneration after single-round immunological destruction in a mouse model. Diabetologia. 2015;58(2):313–323. doi: 10.1007/s00125-014-3416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores RR, Zhou L, Robbins PD. Expression of IL-2 in β cells by AAV8 gene transfer in pre-diabetic NOD mice prevents diabetes through activation of FoxP3-positive regulatory T cells. Gene Ther. 2014;21(8):715–722. doi: 10.1038/gt.2014.45. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Widespread and stable pancreatic gene transfer by adeno-associated virus vectors via different routes. Diabetes. 2006;55(4):875–884. doi: 10.2337/diabetes.55.04.06.db05-0927. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, et al. Efficient Gene Delivery and Expression in Pancreas and Pancreatic Tumors by Capsid-Optimized AAV8 Vectors. Hum Gene Ther Methods. 2017;28(1):49–59. doi: 10.1089/hgtb.2016.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig AT, et al. Transduction of rat pancreatic islets with pseudotyped adeno-associated virus vectors. Virol J. 2009;6:61. doi: 10.1186/1743-422X-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flotte T, et al. Efficient ex vivo transduction of pancreatic islet cells with recombinant adeno-associated virus vectors. Diabetes. 2001;50(3):515–520. doi: 10.2337/diabetes.50.3.515. [DOI] [PubMed] [Google Scholar]
- 23.Goudy K, et al. Adeno-associated virus vector-mediated IL-10 gene delivery prevents type 1 diabetes in NOD mice. Proc Natl Acad Sci USA. 2001;98(24):13913–13918. doi: 10.1073/pnas.251532298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82(12):5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisowski L, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506(7488):382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z, et al. Strong Alpha Cell Preference of the AAV Strains That Best Transduce Human Pancreatic Islets In Vitro. Molecular Therapy. 2017;25(5S1):47 [Google Scholar]
- 27.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91(22):10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apt D, et al. Tetravalent neutralizing antibody response against four dengue serotypes by a single chimeric dengue envelope antigen. Vaccine. 2006;24(3):335–344. doi: 10.1016/j.vaccine.2005.07.100. [DOI] [PubMed] [Google Scholar]
- 29.Chang CC, et al. Evolution of a cytokine using DNA family shuffling. Nat Biotechnol. 1999;17(8):793–797. doi: 10.1038/11737. [DOI] [PubMed] [Google Scholar]
- 30.Christians FC, Scapozza L, Crameri A, Folkers G, Stemmer WP. Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat Biotechnol. 1999;17(3):259–264. doi: 10.1038/7003. [DOI] [PubMed] [Google Scholar]
- 31.Crameri A, Dawes G, Rodriguez E, Silver S, Stemmer WP. Molecular evolution of an arsenate detoxification pathway by DNA shuffling. Nat Biotechnol. 1997;15(5):436–438. doi: 10.1038/nbt0597-436. [DOI] [PubMed] [Google Scholar]
- 32.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391(6664):288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 33.Gafvelin G, et al. Hypoallergens for allergen-specific immunotherapy by directed molecular evolution of mite group 2 allergens. J Biol Chem. 2007;282(6):3778–3787. doi: 10.1074/jbc.M607938200. [DOI] [PubMed] [Google Scholar]
- 34.Ness JE, et al. DNA shuffling of subgenomic sequences of subtilisin. Nat Biotechnol. 1999;17(9):893–896. doi: 10.1038/12884. [DOI] [PubMed] [Google Scholar]
- 35.Pekrun K, et al. Evolution of a human immunodeficiency virus type 1 variant with enhanced replication in pig-tailed macaque cells by DNA shuffling. J Virol. 2002;76(6):2924–2935. doi: 10.1128/JVI.76.6.2924-2935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell SK, et al. Breeding of retroviruses by DNA shuffling for improved stability and processing yields. Nat Biotechnol. 2000;18(12):1279–1282. doi: 10.1038/82391. [DOI] [PubMed] [Google Scholar]
- 37.Raillard S, et al. Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chem Biol. 2001;8(9):891–898. doi: 10.1016/S1074-5521(01)00061-8. [DOI] [PubMed] [Google Scholar]
- 38.Soong NW, et al. Molecular breeding of viruses. Nat Genet. 2000;25(4):436–439. doi: 10.1038/78132. [DOI] [PubMed] [Google Scholar]
- 39.Stutzman-Engwall K, et al. Semi-synthetic DNA shuffling of aveC leads to improved industrial scale production of doramectin by Streptomyces avermitilis. Metab Eng. 2005;7(1):27–37. doi: 10.1016/j.ymben.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Wright A, et al. Diverse plasmid DNA vectors by directed molecular evolution of cytomegalovirus promoters. Hum Gene Ther. 2005;16(7):881–892. doi: 10.1089/hum.2005.16.881. [DOI] [PubMed] [Google Scholar]
- 41.Zhang JH, Dawes G, Stemmer WP. Directed evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proc Natl Acad Sci USA. 1997;94(9):4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asuri P, Bartel MA, Vazin T, Jang JH, Wong TB, Schaffer DV. Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol Ther. 2012;20(2):329–338. doi: 10.1038/mt.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray SJ, et al. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol Ther. 2010;18(3):570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16(7):1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 45.Maguire CA, et al. Directed evolution of adeno-associated virus for glioma cell transduction. J Neurooncol. 2010;96(3):337–347. doi: 10.1007/s11060-009-9972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulk NK, et al. Bioengineered Viral Platform for Intramuscular Passive Vaccine Delivery to Human Skeletal Muscle. Mol Ther Methods Clin Dev. 2018;10:144–155. doi: 10.1016/j.omtm.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulk NK, et al. Bioengineered AAV Capsids with Combined High Human Liver Transduction In Vivo and Unique Humoral Seroreactivity. Mol Ther. 2018;26(1):289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siu JJ, et al. Improved gene delivery to adult mouse spinal cord through the use of engineered hybrid adeno-associated viral serotypes. Gene Ther. 2017;24(6):361–369. doi: 10.1038/gt.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tervo DG, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92(2):372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci USA. 2009;106(10):3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang L, Li J, Xiao X. Directed evolution of adeno-associated virus (AAV) as vector for muscle gene therapy. Methods Mol Biol. 2011;709:127–139. doi: 10.1007/978-1-61737-982-6_8. [DOI] [PubMed] [Google Scholar]
- 52.Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun. 2014;5:3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davidsson M, Diaz-Fernandez P, Schwich OD, Torroba M, Wang G, Björklund T. A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing. Sci Rep. 2016;6:37563. doi: 10.1038/srep37563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrmann AK, et al. A Robust and All-Inclusive Pipeline for Shuffling of Adeno-Associated Viruses. ACS Synth Biol. 2019;8(1):194–206. doi: 10.1021/acssynbio.8b00373. [DOI] [PubMed] [Google Scholar]
- 55.Nair GG, et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat Cell Biol. 2019;21(2):263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang W, Johnston WA, Boden M, Gillam EM. ReX: A suite of computational tools for the design, visualization, and analysis of chimeric protein libraries. BioTechniques. 2016;60(2):91–94. doi: 10.2144/000114381. [DOI] [PubMed] [Google Scholar]
- 57.Lerch TF, Chapman MS. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B) Virology. 2012;423(1):6–13. doi: 10.1016/j.virol.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong L, et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci USA. 2008;105(22):7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palaschak B, Herzog RW, Markusic DM. AAV-Mediated Gene Delivery to the Liver: Overview of Current Technologies and Methods. Methods Mol Biol. 2019;1950:333–360. doi: 10.1007/978-1-4939-9139-6_20. [DOI] [PubMed] [Google Scholar]
- 60.Nonnenmacher M, van Bakel H, Hajjar RJ, Weber T. High capsid-genome correlation facilitates creation of AAV libraries for directed evolution. Mol Ther. 2015;23(4):675–682. doi: 10.1038/mt.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrmann AK, et al. Impact of the Assembly-Activating Protein on Molecular Evolution of Synthetic Adeno-Associated Virus Capsids. Hum Gene Ther. 2019;30(1):21–35. doi: 10.1089/hum.2018.085. [DOI] [PubMed] [Google Scholar]
- 62.Sonntag F, et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol. 2011;85(23):12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107(22):10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naumer M, et al. Properties of the adeno-associated virus assembly-activating protein. J Virol. 2012;86(23):13038–13048. doi: 10.1128/JVI.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grosse S, et al. Relevance of Assembly-Activating Protein for Adeno-associated Virus Vector Production and Capsid Protein Stability in Mammalian and Insect Cells. J Virol. 2017;91(20):e01198-17. doi: 10.1128/JVI.01198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Earley LF, et al. Adeno-associated Virus (AAV) Assembly-Activating Protein Is Not an Essential Requirement for Capsid Assembly of AAV Serotypes 4, 5, and 11. J Virol. 2017;91(3):e01980-16. doi: 10.1128/JVI.01980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hibberts F, Barnes E. The use of endoscopic retrograde cholangio pancreatography. Nurs Times. 2003;99(20):26–27. [PubMed] [Google Scholar]
- 68.Quirin KA, et al. Safety and Efficacy of AAV Retrograde Pancreatic Ductal Gene Delivery in Normal and Pancreatic Cancer Mice. Mol Ther Methods Clin Dev. 2018;8:8–20. doi: 10.1016/j.omtm.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deverman BE, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34(2):204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hordeaux J, Wang Q, Katz N, Buza EL, Bell P, Wilson JM. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol Ther. 2018;26(3):664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuzaki Y, et al. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci Lett. 2018;665:182–188. doi: 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- 72.Doshi BS, Arruda VR. Gene therapy for hemophilia: what does the future hold? Ther Adv Hematol. 2018;9(9):273–293. doi: 10.1177/2040620718791933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.George L, et al. Spk-8011: preliminary results from a phase 1/2 dose escalation trial of an investigational AAV-mediated gene therapy for hemophilia A. Blood. 2017;130:604 [Google Scholar]
- 74.Wang L, et al. Comparative Study of Liver Gene Transfer With AAV Vectors Based on Natural and Engineered AAV Capsids. Mol Ther. 2015;23(12):1877–1887. doi: 10.1038/mt.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kienle E, et al. Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling. J Vis Exp. 2012;(62):3819. doi: 10.3791/3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie Q, Lerch TF, Meyer NL, Chapman MS. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420(1):10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dorrell C, Abraham SL, Lanxon-Cookson KM, Canaday PS, Streeter PR, Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res. 2008;1(3):183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 78.Russ HA, et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34(13):1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meliani A, Leborgne C, Triffault S, Jeanson-Leh L, Veron P, Mingozzi F. Determination of anti-adeno-associated virus vector neutralizing antibody titer with an in vitro reporter system. Hum Gene Ther Methods. 2015;26(2):45–53. doi: 10.1089/hgtb.2015.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Azuma H, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25(8):903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.