Abstract
Broadly neutralizing Abs targeting the HA stem can provide broad protection against different influenza subtypes, raising the question of how best to elicit such Abs. We have previously demonstrated that vaccination with pandemic live-attenuated influenza vaccine (pLAIV) establishes immune memory for HA head-specific Abs. Here, we determine the extent to which matched versus mismatched LAIV-inactivated subunit vaccine (IIV) prime-boost vaccination elicits stem-specific memory B cells and Abs. We vaccinated African green monkeys with H5N1 pLAIV-pIIV or H5N1 pLAIV followed by seasonal IIV (sIIV) or with H5N1 pLAIV alone and measured Abs and HA-specific B cell responses. While we observed an increase in stem-specific memory B cells, head-specific memory B cell responses were substantially higher than stem-specific responses and were dominant even following boost with mismatched IIV. Neutralizing Abs against heterologous influenza viruses were undetectable. Head-specific B cells from draining lymph nodes exhibited germinal center markers, while stem-specific B cells found in the spleen and peripheral blood did not. Thus, although mismatched prime-boost generated a pool of stem-specific memory B cells, head-specific B cells and serum Abs substantially dominated the immune response. These findings have implications for including full-length native HA in prime-boost strategies intended to induce stem-specific Abs for universal influenza vaccination.
Keywords: Infectious disease, Vaccines
Keywords: Influenza
Priming and boosting with mismatched influenza vaccines elicited more HA head- than stem-specific memory B cells and did not induce broadly cross-neutralizing antibodies.
Introduction
Influenza causes considerable morbidity and mortality worldwide. The error-prone nature of the viral RNA polymerase and the segmented RNA genome endow influenza A viruses (IAVs) with the ability to rapidly evade host Ab responses induced by prior infection or vaccination through antigenic drift and shift. Creating vaccines that provide broad and prolonged protection from a range of IAVs would be a critical advance in controlling seasonal and pandemic influenza.
Abs induced by influenza virus infection and licensed vaccines mostly target the viral surface glycoprotein, the HA, which has two domains: the head and stem. The globular head domain contains the receptor binding site that is responsible for virus attachment to terminal sialic acid residues on host cells and is the target of neutralizing Abs. However, the HA head is under strong selection for mutations that allow the virus to escape from neutralizing Abs (1, 2). Conversely, the stem domain is highly conserved and is involved in membrane fusion during virus entry into cells. The HAs of the 18 known IAV subtypes cluster phylogenetically into groups 1 and 2 (3). However, stem-specific Abs and B cells are much less common than head-specific Abs in both humans and animal models.
Several vaccine strategies have been explored to induce stem-specific Abs, including headless HA constructs and chimeric HAs (reviewed in ref. 4). In the latter case, the idea is to sequentially vaccinate with chimeric HAs that share stem domains but have diverse head domains and, thus, selectively boost the stem Ab response. Another potential approach, which utilizes currently licensed and available vaccines, is to prime with a given subtype of a live-attenuated influenza vaccine (LAIV) and boost with an inactivated subunit vaccine (IIV) derived from a different influenza A subtype.
LAIVs are currently licensed for the prevention of seasonal influenza infections in children and adults. LAIVs bearing the 6 internal protein genes of the A/Ann Arbor/6/60 cold-adapted (AA ca) donor virus and the HA and neuraminidase (NA) genes from influenza viruses with pandemic potential have been evaluated in preclinical models (5–12) and in phase I clinical trials (13–17). Seasonal LAIVs and pandemic LAIVs (pLAIVs) are attenuated because their replication is restricted to the upper respiratory tract; both have been shown to be safe in humans. Although the ability of pLAIVs to induce a primary Ab response in humans is limited (13–17), they induce long-term immune memory and, when coupled with matched pIIV boost, generate a robust, rapid, and broadly neutralizing Ab response against heterologous viruses within the same subtype (18–20).
We recently established an African green monkey (AGM) IAV model and established that vaccination recapitulates the clinical phenomena observed during prime-boost human vaccination with pLAIV-pIIV (12, 21). The AGM model allowed us to dissect the HA-specific Ab and B cell responses to matched pLAIV-pIIV vaccination in the peripheral blood and lymphoid tissues. We observed HA-specific B cells being produced in the mediastinal lymph node (MLN) following intranasal pLAIV priming and distinct peripheral expansion of HA-specific plasmablasts and memory B cells in the axillary lymph nodes and spleen following intramuscular pIIV boost.
Here, we use the AGM model to determine to what extent mismatched LAIV and IIV vaccines with a conserved HA stem but antigenically distinct HA heads specifically boost HA stem-specific Abs.
Results
Low levels of stem-specific class-switched memory B cells are generated following primary immunization with H5N1 pLAIV.
There is great interest in devising strategies to induce B cells and potent neutralizing Abs against the highly conserved HA stem (22). It is clear from animal studies that high stem-specific Ab titers are necessary for protection from challenge infection (23). To date, robust titers of HA head-specific neutralizing Abs have been achieved by prime-boost vaccination strategies with H5 or H7 pLAIV/pIIVs in humans (18–20).
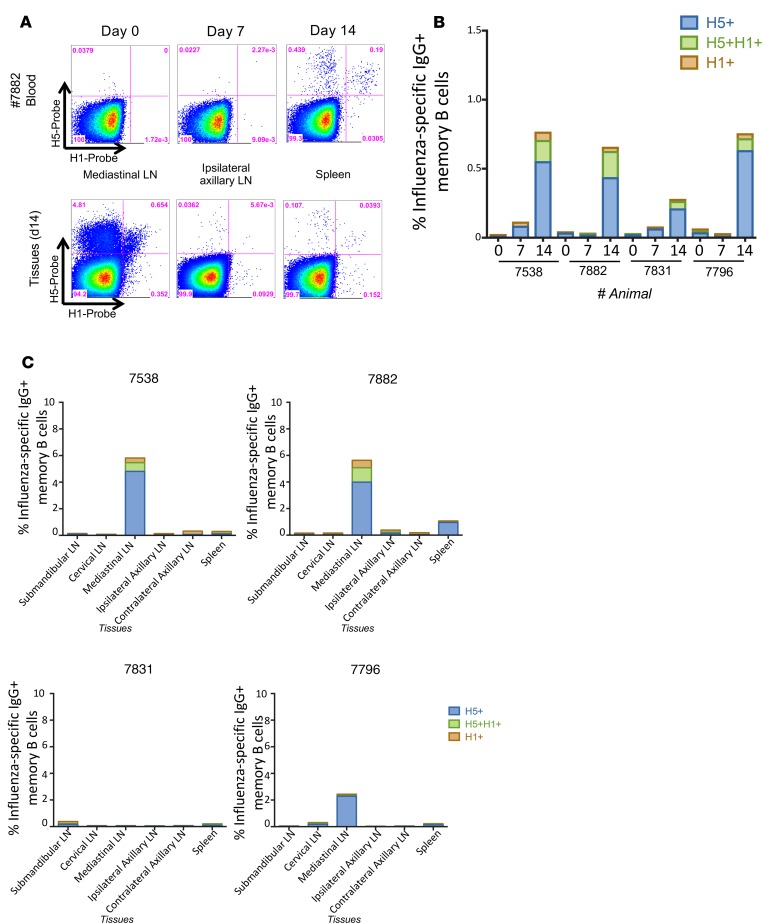
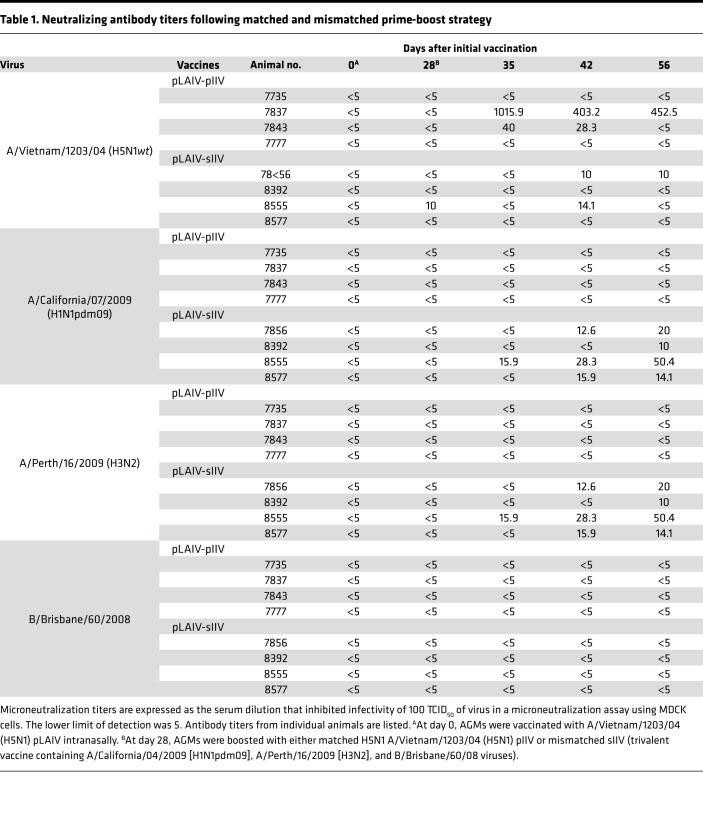
Exploring the immunologic basis for these clinical observations in AGMs, we recently reported that intranasal administration of H5N1 pLAIV followed by matched pIIV boost generates a robust H5-specific B cell response and H5-specific neutralizing Ab response. Further, we found that H5-specific memory B cells are generated in the MLN following pLAIV (21). We did not, however, assess the ability of pLAIV to generate a broad stem-specific memory B cell pool that can be recalled upon pIIV boost. To test this, we used recombinant influenza HA probes (HAΔSA), previously used to track and measure HA-specific memory B cell responses (24–26), to assess the frequency of H5+ head-specific and H5+H1+ double-positive stem-specific memory B cells following primary pLAIV vaccination (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129035DS1). Using a fluorescent stabilized stem immunogen (27), we determined that most, if not all, H5+H1+ double-positive cells bound the HA stem while the H5+ single-positive cells did not (Supplemental Figure 1B). AGMs vaccinated with H5N1 pLAIV showed a small but detectable population of H5+H1+ double-positive stem-specific memory B cells in the peripheral blood (Figure 1, A and B) and MLNs at day 14 (Figure 1C), but, as previously reported (21), the frequency of H5+ head-specific memory B cells was markedly higher in both the peripheral blood and MLNs at day 14 after pLAIV (0.05%–0.19% and 0.21%–0.64% for stem- and head-specific frequencies in the MLN, respectively). Consistent with these findings and our previous reports (12, 21), serum-neutralizing Abs were not detected at days 14 and 28 against either A/Vietnam/1203/04 (H5N1) or A/California/07/09 (H1N1pdm09) viruses following administration of H5N1 pLAIV (Table 1). These data show that immunodominance for the HA head is established early following pLAIV vaccination in the MLN memory B cell pool.
Figure 1. HA-specific memory B cells following primary H5N1 pLAIV vaccination.
(A) Representative plots of H5-specific IgG+ memory B cells (CD19+CD20+CD27+IgG+ from animal 7882) from peripheral blood at day 0, 7, and 14 and mediastinal lymph nodes, ipsilateral axillary lymph nodes, and spleen at day 14. (B) Frequency of H5+ or H1+ head-specific or H5+H1+ double-positive stem-specific memory B cells in peripheral blood at day 0, 7, and 14 following primary H5N1 pLAIV vaccination. (C) Frequency of H5+ or H1+ head-specific or H5+H1+ double-positive stem-specific memory B cells in submandibular, cervical, mediastinal, and ipsilateral and contralateral axillary lymph nodes (LN) and spleen on day 14.
Table 1. Neutralizing antibody titers following matched and mismatched prime-boost strategy.
Both head-specific and stem-specific memory B cells are expanded following matched H5N1 pIIV boost.
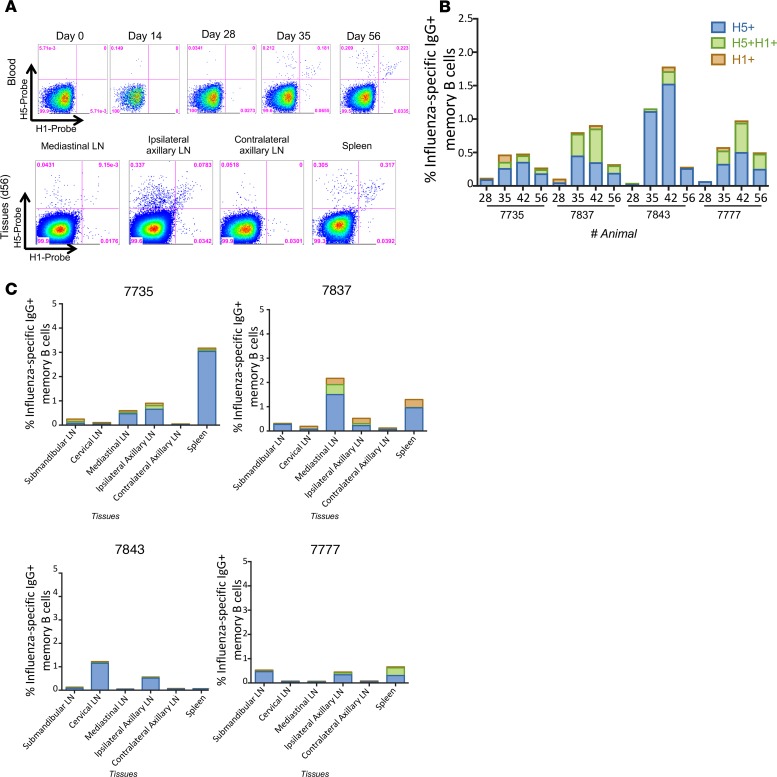
Prior studies have demonstrated that seasonal influenza vaccines as well as prime-boost strategies with pandemic influenza vaccines in humans expand pools of stem-specific memory B cells, leading to an increase in circulating stem-specific memory B cells and Ab titers (18–20). To determine whether matched H5N1 prime-pIIV boost expands both H5+ head-specific and H5+H1+ double-positive stem-specific memory B cells generated by primary pLAIV vaccination, we boosted AGMs at day 28 after pLAIV with a dose of H5N1 pIIV and measured both memory B cells and serum Abs (21). This revealed an increase in the frequency of H5+ head-specific and H5+H1+ double-positive stem-specific memory B cells in the peripheral blood at days 35–56 (Figure 2, A and B). Some monkeys (7837 and 7777, Figure 2, A and B) had far more stem-specific B cells than others. Analysis of tissues on day 56 after pIIV boost showed a lower frequency of stem-specific compared with head-specific memory B cells in all monkeys (Figure 2C).
Figure 2. HA-specific memory B cells following matched IIV boost vaccination.
(A) Representative plots of H5-specific IgG+ memory B cells (CD19+CD20+CD27+IgG+, animal 7777) from peripheral blood at day 0, 14, 28, 35, and 56 and mediastinal lymph nodes (LN), ipsilateral axillary lymph nodes, contralateral axillary lymph nodes, and spleen at day 56 after H5N1 pLAIV, which is 28 days after pIIV boost. (B) Frequency of H5+ or H1+ head-specific or H5+H1+ double-positive stem-specific memory B cells in peripheral blood at day 28, 35, 42, and 56 following H5N1 pLAIV-pIIV vaccination. (C) Frequency of H5+ or H1+ head-specific or H5+H1+ double-positive stem-specific memory B cells in submandibular, cervical, mediastinal, ipsilateral and contralateral axillary lymph nodes and spleen at day 56 after H5N1 pLAIV, which is 28 days after pIIV boost.
Despite the presence of both H5+ head-specific and H5+H1+ double-positive stem-specific memory B cells, neutralizing Ab titers were only detected in 2 of 4 AGMs against the H5N1wt virus (animal numbers 7837 and 7843), and none of the AGMs developed Ab against the heterologous A/California/07/09 (H1N1pdm09) virus, following matched H5N1 pLAIV-pIIV vaccination (Table 1). These data indicate that H5N1 pLAIV-pIIV administration can temporarily boost stem-specific memory B cells, but this strategy does not generate a robust stem-specific neutralizing Ab response.
Mismatched H5N1 pLAIV-sIIV preferentially induces memory B cells and a serologic response to the HA head.
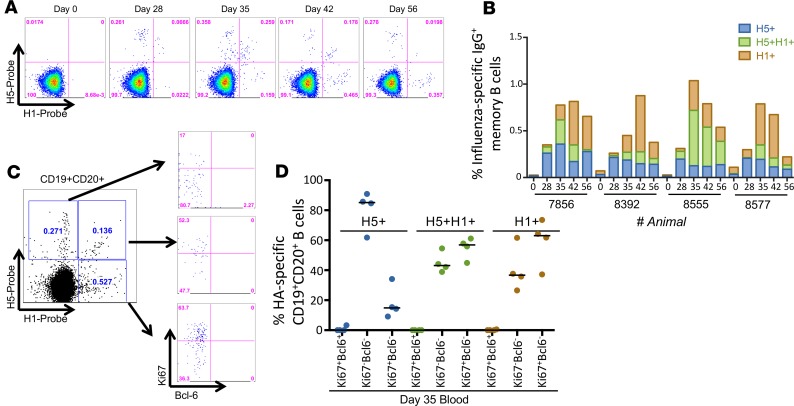
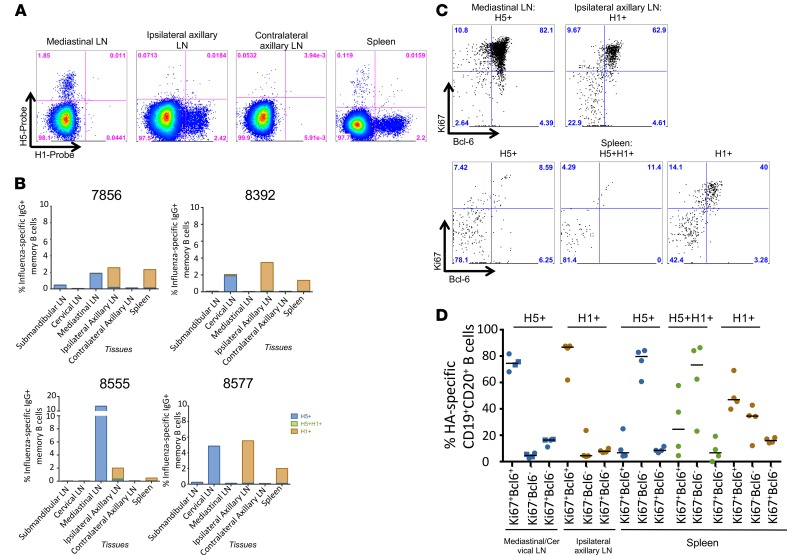
Studies have shown that prime-boost vaccination with divergent influenza viruses or chimeric influenza HA proteins can boost HA stem Ab titers and provide robust protection from heterologous virus challenge in animal models (4). To investigate whether prime-boost vaccination with mismatched seasonal IIV (sIIV) following H5N1 pLAIV preferentially boosts cross-reactive memory B cells to the HA stem, we vaccinated 4 AGMs with an H5N1 pLAIV followed by sIIV containing A/California/07/09 (H1N1pdm09), A/Perth/16/2009-like virus (H3N2), and B/Brisbane/60/2008 viruses. Consistent with our previous findings, we detected H5+ head-specific and H5+H1+ double-positive stem-specific memory B cells in the peripheral blood at day 28 following H5N1 pLAIV (Figure 3A), and we observed a distinct increase in H5+H1+ double-positive stem-specific memory B cells 7 days after the sIIV boost (day 35 after pLAIV) (Figure 3B). There was no significant difference between the percentage of H5+H1+ IgG-specific B cells in animals that were boosted with pIIV or sIIV. However, we observed a significant difference in the percentage of H5+ IgG-specific B cells at day 42 in pIIV-boosted animal (P = 0.0178) and in the percentage of H1+ IgG-specific B cells at days 35, 42, and 56 in sIIV-boosted animals (P = 0.0027, P < 0.0001, and P = 0.024, respectively). Surprisingly, we observed an increase in H1+ head-specific memory B cells at day 7 after sIIV, with H5+ head-specific and H1+ head-specific memory B cells being the predominant populations in the peripheral blood of 3 of the 4 animals by day 56 after pLAIV (or day 28 after sIIV boost) (Figure 3B).
Figure 3. HA-specific memory B cells following mismatched prime-boost vaccination in peripheral blood.
(A) Representative plots of H5-specific IgG+ memory B cells (CD19+CD20+CD27+IgG+) from peripheral blood at day 0, 28, 35, 42, and 56 after pLAIV followed by mismatched sIIV vaccination. (B) Frequency of H5+ or H1+ head-specific or H5+H1+ double-positive stem-specific memory B cells in peripheral blood at day 0, 28, 35, 42, and 56 after H5N1 pLAIV followed by sIIV boost in 4 separate animals. (C) Representative plot of Ki67 and Bcl-6 expression of CD19+CD20+ B cells from peripheral blood that are H5+, H5+H1+, or H1+ probe positive at day 35 (day 7 after boost) after pLAIV-sIIV. (D) Analysis of Ki67 and Bcl-6 expression on H5+, H5+H1+, or H1+ probe–positive B cells from 4 separate animals at day 35 after pLAIV, which is day 7 after sIIV boost.
To determine whether these cells were recent germinal center (GC) emigrants, we measured GC B cell surface marker Ki67 and Bcl-6 expression at day 35 after pLAIV (day 7 after sIIV boost) (ref. 21 and Figure 3C). We found Ki67+Bcl-6– and Ki67–Bcl-6– phenotypes among both H5+H1+ double-positive stem-specific and H1+ head-specific B cells, whereas H5+ head-specific B cells were predominantly Ki67–Bcl6– (Figure 3D). Curiously, by day 56 after pLAIV (day 28 after sIIV boost), we observed very few H5+H1+ double-positive stem-specific memory B cells in any of the lymphoid organs sampled, including the spleen, axillary lymph nodes, and MLNs (Figure 4, A and B). We observed increased proportions of antigen-specific memory B cells in relevant draining lymph nodes, with H5+ head-specific memory B cells in the mediastinal and cervical lymph nodes, as previously observed (21), and H1+ head-specific memory B cells in the ipsilateral axillary lymph nodes and spleen (Figure 4, A and B). Analysis of GC markers in these distinct sites showed large frequencies of Ki67+Bcl-6+ GC-B cells in the mediastinal/cervical lymph nodes and ipsilateral axillary lymph nodes for H5+ head-specific and H1+ head-specific B cells, respectively (Figure 4, C and D). The low frequencies of H1+H5+ double-positive stem-specific B cells in the lymph nodes made Ki67+Bcl-6+ quantification difficult; however, moderate frequencies of both Ki67+Bcl6+ cells and Ki67–Bcl6– cells were detectable in the spleen. H5+ head-specific B cells were highly enriched for the Ki67–Bcl6– phenotype in the spleen while the H1+ head-specific B cells displayed a combination of Ki67+Bcl6+ and Ki67–Bcl6– phenotypes. The relatively lower frequency of H1+H5+ double-positive stem-specific GC B cells compared with H1+ or H5+ head-specific B cells in the lymph nodes is consistent with a limited ability of stem-specific B cells to survive GC selection.
Figure 4. HA-specific memory B cells following mismatched prime-boost vaccination in lymphoid tissues.
(A) Representative plots of H5-specific IgG+ memory B cells (CD19+CD20+CD27+IgG+) from mediastinal, ipsilateral axillary, contralateral axillary lymph nodes (LN) and spleen at day 56 after pLAIV, which is day 28 after sIIV boost vaccination. (B) Frequency of H5+, H5+H1+, or H1+ memory B cells in submandibular, cervical, mediastinal, ipsilateral and contralateral axillary lymph nodes and spleen. (C) Representative plot of Ki67 and Bcl-6 expression of CD19+CD20+ B cells from mediastinal or ipsilateral axillary lymph nodes that are H5+, H5+H1+, or H1+ probe positive at day 56 after pLAIV (or day 28 after sIIV boost). (D) Analysis of Ki67 and Bcl-6 expression on H5+, H5+H1+, or H1+ probe–positive B cells in submandibular, cervical, mediastinal, ipsilateral and contralateral axillary lymph nodes and spleen at day 56 after pLAIV (or day 28 after sIIV boost) in 4 separate animals.
Low titers of serum-neutralizing Ab against both A/California/07/09 (H1N1pdm09) and A/Perth/16/2009 (H3N2) components of sIIV were detected at day 56 after pLAIV (day 28 after sIIV boost) in AGMs that received the mismatched pLAIV-sIIV vaccines. Neutralizing Abs against B/Brisbane/60/2008 were not detected (Table 1). In contrast, there was no detectable neutralizing activity against heterologous group 1 IAVs, including older seasonal H1N1 (A/New Caledonia/20/1999 and A/Brisbane/59/2007), H2N2 (A/Japan/305/1957), and H6N1 (A/Teal/HK/W132/1997) viruses (Supplemental Table 1). Taken together, the data show that priming with H5N1 pLAIV followed by sIIV boost can generate a pool of stem-specific memory B cells; however, they are lower in frequency than head-specific memory B cells and do not generate sufficient circulating Abs to cross-neutralize heterologous and heterosubtypic influenza viruses.
Discussion
In this study, we tested the hypothesis that sequential immunization with mismatched LAIV and IIV would focus the Ab response on the HA stem. This hypothesis was based on 4 important prior observations: sequential administration of homologous pLAIV and pIIV in humans elicits a robust neutralizing Ab response against heterologous viruses within the same subtype (18–20); these serologic findings were reproduced in AGMs, which would therefore serve as a useful model to investigate human immunity (21); HA stem memory B cell and Ab responses are elicited in previously primed individuals by infection (28) or vaccination (29) with heterologous influenza viruses that bear a novel HA head but with a shared stem; and mismatched influenza viruses bearing chimeric HAs with distinct head domains on a conserved stem elicited HA stem Abs in animal models (30).
Despite this compelling evidence, we found that the mismatched pLAIV-sIIV strategy fails to generate broadly cross-reactive neutralizing Abs against IAV subtypes, reflecting the fact that stem-specific memory B cell responses are clearly subdominant to the HA head-specific response.
Following administration of H5- or H1-containing vaccines, irrespective of which lymphoid tissues were sampled, we observed limited elicitation of H5+H1+ double-positive stem-specific B cells compared with the HA head-specific response. The dominance of HA head-specific Abs is consistent with the serological dominance of head Ab observed in humans (31), ferrets (32), and mice (33, 34). Although the mechanistic basis for HA stem subdominance remains unclear (35), a study in mice suggests that under typical immunization conditions, HA-head specific naive B cells outcompete stem-specific naive B cells for limiting amounts of antigens based on the higher avidity of the B cell receptors for head versus stem epitopes (36).
Our data indicate that the mechanism underlying this phenomenon is independent of viral antigen (H5 versus H1), vaccine vector (LAIV versus IIV), or immunological site of initiation of humoral immunity.
Consistent with our previous report (21), we found that intranasally administered H5 pLAIV initiated H5-specific immunity that was highly localized to the local draining mediastinal and cervical lymph nodes, with many H5-specific B cells exhibiting the Ki67+Bcl6+ phenotype characteristic of active GC reactions. In contrast, intramuscular administration of sIIV elicited potent H1-specific GC B cells in the draining axillary lymph node and spleen. Interestingly, both H1 and H5 memory B cells were readily detected in the peripheral blood, as were serum-neutralizing responses against H5N1 and H1N1 viruses, indicating that all lymphoid tissues were equally capable of initiating humoral immunity. Notably, there was marked segregation of the B cell responses in the lymphoid tissues of animals receiving sequential LAIV/IIV immunizations, with little evidence of seeding GC or memory B cell responses at distal lymphoid sites. These highly localized responses suggest that vaccine antigen is sequestered in lymphoid tissues draining the immunization site, presumably making it difficult for vaccine delivered at a different site to boost HA stem responses. Although a similar phenomenon may occur in naive human subjects (e.g., children), it is likely that adults, who have been exposed to influenza viruses repeatedly throughout their lifetime will have HA stem-specific memory B cells that can be recruited into draining lymphoid tissues following vaccination.
Finally, there are a number of potential explanations for why the mismatched H5N1-H1N1 vaccination strategy in the AGM model failed to recapitulate the dominant HA stem responses reported in humans following the emergence of the 2009 pandemic H1N1 virus (24, 29, 37). First, a majority of the HA stem Ab responses in humans following exposure to heterologous influenza viruses are derived from a unique single germline family (VH1-69) (25, 38–40), which displays pan–Group 1–binding activity and uses a noncanonical CDR-H2–dependent recognition of HA (41). The anti-stem Ab repertoire may be limited in AGMs, which, typical of nonhuman primates, lack VH1-69 germline analogs. Second, adult humans have experienced multiple exposures to influenza viruses via infection and vaccination while AGMs are influenza naive. It is possible that the establishment of HA stem memory requires repeated exposure, a longer interval between exposures, or administration of several antigenically diverse, drifted influenza HA immunogens. Studies in children would provide valuable insights into the differences between human and AGM immunity. In any event, a more complex influenza exposure history might be required for monkeys to recapitulate human anti-stem Ab responses. Finally, the exquisite compartmentalization of the LAIV and IIV responses to their respective draining lymph nodes might mean these 2 vaccine platforms elicit spatially distinct responses that are not suitable for driving expansion of an HA stem response.
Methods
Animal study design.
Samples from 2 studies were included in this analysis. We used stored PBMC and serum samples from a previously described study (21) in which 24 influenza-naive male and female adult AGMs (Chlorocebus aethiops) were inoculated intranasally with 1 × 107 TCID50 in 0.4 ml (0.2 ml in each naris) of H5N1 pLAIV (A/Vietnam/1203/2004) using a mucosal atomization device (Teleflex). Twenty-eight days later, 12 of the H5N1 pLAIV-immunized AGMs and the 12 mock-vaccinated AGMs were given a single dose of H5N1 pIIV (45 μg) intramuscularly in the right deltoid muscle, and the remaining 12 H5N1 pLAIV-immunized AGMs were given a single dose of Leibovitz-15 (L15) diluent as a sham control for pIIV. Blood (sodium heparin tubes) and serum samples (serum separator tubes) were collected at days 0, 7, 14, 28, 32, 35, 42, and 56 from the start of the study. At days 14, 35, and 56, 4 AGMs from each group were euthanized and tissue samples, including cervical lymph nodes, submandibular lymph nodes, MLNs, ipsilateral and contralateral axillary lymph nodes, spleen, and bone marrow (EDTA tubes) were collected and processed as previously described (21). Only the animals that received H5N1 pLAIV followed by H5N1 pIIV are included in this paper.
Additionally, 4 influenza naive AGMs were inoculated intranasally with 1 × 107 TCID50 in 0.4 ml (0.2 ml in each naris) of H5N1 pLAIV (A/Vietnam/1203/2004), as previously described (21); 28 days later, they were given a single dose of trivalent seasonal inactivated vaccine 2011–2012 sIIV (15 μg per HA, Fluzone). Blood (sodium heparin tubes) and serum samples (serum separator tubes) were collected at days 0, 28, 35, 42, and 56 from the start of the study. Animals were euthanized at day 28 after boost (or day 56), and tissues were collected and processed as above.
Viruses and vaccine.
As previously described, the HA and NA genes of the cold-adapted pLAIV vaccine viruses were derived from avian influenza A/Vietnam/1203/2004 (H5N1), with a deletion of the multibasic cleavage site engineered into the H5 HA (11). The pLAIV and pIIV vaccine viruses were generated by reverse genetics (20). Viruses were propagated in 10- to 11-day-old embryonated chicken eggs. Virus was titered in Madin-Darby canine kidney (MDCK) cells maintained in modified Eagle’s medium with 10% heat-inactivated fetal calf serum (Hyclone) and L-glutamine (Gibco). The monovalent subvirion vaccine influenza rgA/Vietnam/1203/2004 H5N1 pIIV (A/Vietnam/1203/2004) vaccine and trivalent split-virion seasonal influenza vaccine containing 15 μg HA from A/California/07/09 (H1N1), A/Victoria/210/2009 (H3N2, A/Perth/16/2009-like virus), and B/Brisbane/60/2008 (Fluzone 2011–2012 season) were obtained through the NIH Biodefense and Emerging Infections Resources Repository, National Institute of Allergy and Infectious Diseases (NIAID), NIH (catalog nos. NR-4143 and NR-36747).
Microneutralization assay.
Heat-inactivated sera (56°C for 1 hour) were tested for neutralizing activity against wild-type influenza A/Vietnam/1203/2004 (H5N1) virus, A/California/07/2009 (H1N1pdm09), A/Perth/16/2009 (H3N2), B/Brisbane/60/2008, A/Brisbane/59/2007 (H1N1), A/New Caledonia/20/1999 (H1N1), A/Japan/305/1957 (H2N2), and A/Teal/HK/W132/1997 (H6N1) viruses in a microneutralization assay as previously described (42). Briefly, 2-fold dilutions of heat-inactivated serum were tested for the presence of Abs capable of neutralizing the infectivity of 100 TCID50 of virus in MDCK cells (ATCC). The serum dilution that completely prevented cytopathic effect in 50% of the wells on day 4 was calculated by the Reed-Muench formula (43).
HA-specific B cell phenotyping and sorting.
HA probes were previously generated and validated for use in characterizing HA-specific B cells in PBMC samples (21, 24). HA-specific memory B cells and plasmablasts were identified using cryopreserved PBMC or processed tissue samples using recombinant H5 HA (A/Vietnam/1203/2004) conjugated to streptavidin APC (Life Technologies, catalog no. S-32362) and H1 HA (A/California/07/2009) conjugated to streptavidin PE and stabilized stem-HA conjugated to streptavidin BV785 (44). Cell viability was determined using Aqua LIVE/DEAD (Life Technologies, catalog no. L34957). AGM PBMCs were stained with the following Abs: CD14-BV510 (Biolegend, catalog no. 301842, clone M5E2), CD19-ECD (Beckman Coulter, catalog no. IM2708U, clone J3-119), CD20-BV605 (Biolegend, catalog no. 302334, clone 2H7), CD27-BV650 (Biolegend, catalog no. 302828, clone 0323), CD38-AF680 (provided by Margaret Beddall, Vaccine Research Center, NIAID), IgG-BV421 (BD, catalog no. 562581, clone G18-145), IgD-FITC (Southern Biotech, catalog no. 2030-02), IgM-PerCP-Cy55 (BD, catalog no. 561285, clone G20-127), CD21-PE-Cy5 (BD, catalog no. 551064, clone B-ly4), CXCR5-PE-Cy7 (eBioscience, catalog no. 25-9185-42, clone MU5UBEE). To identify H5-specific GC B cells, additional staining was performed for intracellular Ki67-PerCP-Cy5.5 (BD, catalog no. 561284, clone B56) and Bcl-6-APC-Cy7 (BD, catalog no. 563581, clone K1129-19) using the FoxP3/Transcription Factor staining buffer set (eBioscience) as previously described (44). A minimum of 2 million total events was collected on a custom LSR II instrument (BD Immunocytometry Systems), and analysis was performed using FlowJo Software 9.7.6 (TreeStar). Flow cytometric data were subsequently analyzed using FlowJo v9.9.5.
Statistics.
Statistical analyses used Prism GraphPad, version 7. Two-tailed Student’s t test, Mann-Whitney U test, and two-way ANOVA Bonferroni’s multiple comparison tests were performed for analyses between groups. Wilcoxon matched-pair rank tests were performed for analyses within each group. A P value of less than 0.05 was considered significant.
Study approval.
Animal studies were carried out in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) of the NIH, the Office of Animal Welfare, and the United States Department of Agriculture. All animal work was approved by the NIAID Division of Intramural Research Animal Care and Use Committees in Bethesda, Maryland, USA (protocol no. LID3E). The animal facility is accredited by the American Association for Accreditation of Laboratory Animal Care.
Author contributions
SJ, SFA, AKW, ABM, and KS designed the study. SJ and SFA performed assays and analysis. KS, SFA, AKW, and JWY wrote the paper. All authors reviewed and approved the submission.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of NIAID, NIH. SJ was supported by a National Health and Medical Research Council Australia Early Career Fellowship (APP1072127).
Version 1. 11/14/2019
Electronic publication
Funding Statement
Division of Intramural Research
Footnotes
KS’s current address is: WHO Collaborating Centre for Reference and Research on Influenza, The Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(22):e129035.https://doi.org/10.1172/jci.insight.129035.
Contributor Information
Sinthujan Jegaskanda, Email: jegaskanda8@gmail.com.
Sarah F. Andrews, Email: sarah.andrews2@nih.gov.
Adam K. Wheatley, Email: a.wheatley@unimelb.edu.au.
Jonathan W. Yewdell, Email: jyewdell@mail.nih.gov.
Adrian B. McDermott, Email: adrian.mcdermott@nih.gov.
Kanta Subbarao, Email: kanta.subbarao@influenzacentre.org.
References
- 1.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286(5446):1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 2.Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol. 1999;16(11):1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 3.Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krammer F. The Quest for a Universal Flu Vaccine: Headless HA 2.0. Cell Host Microbe. 2015;18(4):395–397. doi: 10.1016/j.chom.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Chen GL, Lamirande EW, Yang CF, Jin H, Kemble G, Subbarao K. Evaluation of replication and cross-reactive antibody responses of H2 subtype influenza viruses in mice and ferrets. J Virol. 2010;84(15):7695–7702. doi: 10.1128/JVI.00511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, et al. Generation and characterization of a cold-adapted influenza A H9N2 reassortant as a live pandemic influenza virus vaccine candidate. Vaccine. 2003;21(27-30):4430–4436. doi: 10.1016/S0264-410X(03)00430-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, et al. Development of a high-yield live attenuated H7N9 influenza virus vaccine that provides protection against homologous and heterologous H7 wild-type viruses in ferrets. J Virol. 2014;88(12):7016–7023. doi: 10.1128/JVI.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, et al. Evaluation of live attenuated influenza a virus h6 vaccines in mice and ferrets. J Virol. 2009;83(1):65–72. doi: 10.1128/JVI.01775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph T, et al. A live attenuated cold-adapted influenza A H7N3 virus vaccine provides protection against homologous and heterologous H7 viruses in mice and ferrets. Virology. 2008;378(1):123–132. doi: 10.1016/j.virol.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min JY, et al. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J Virol. 2010;84(22):11950–11960. doi: 10.1128/JVI.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suguitan AL, et al. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3(9):e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka Y, et al. African green monkeys recapitulate the clinical experience with replication of live attenuated pandemic influenza virus vaccine candidates. J Virol. 2014;88(14):8139–8152. doi: 10.1128/JVI.00425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karron RA, et al. A live attenuated H9N2 influenza vaccine is well tolerated and immunogenic in healthy adults. J Infect Dis. 2009;199(5):711–716. doi: 10.1086/596558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karron RA, et al. Evaluation of two live attenuated cold-adapted H5N1 influenza virus vaccines in healthy adults. Vaccine. 2009;27(36):4953–4960. doi: 10.1016/j.vaccine.2009.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talaat KR, et al. A live attenuated H7N3 influenza virus vaccine is well tolerated and immunogenic in a Phase I trial in healthy adults. Vaccine. 2009;27(28):3744–3753. doi: 10.1016/j.vaccine.2009.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talaat KR, et al. An open-label phase I trial of a live attenuated H2N2 influenza virus vaccine in healthy adults. Influenza Other Respir Viruses. 2013;7(1):66–73. doi: 10.1111/j.1750-2659.2012.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talaat KR, et al. An open label Phase I trial of a live attenuated H6N1 influenza virus vaccine in healthy adults. Vaccine. 2011;29(17):3144–3148. doi: 10.1016/j.vaccine.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babu TM, et al. Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine. 2014;32(50):6798–6804. doi: 10.1016/j.vaccine.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobhanie M, et al. Evaluation of the Safety and Immunogenicity of a Candidate Pandemic Live Attenuated Influenza Vaccine (pLAIV) Against Influenza A(H7N9) J Infect Dis. 2016;213(6):922–929. doi: 10.1093/infdis/jiv526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talaat KR, et al. A live attenuated influenza A(H5N1) vaccine induces long-term immunity in the absence of a primary antibody response. J Infect Dis. 2014;209(12):1860–1869. doi: 10.1093/infdis/jiu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jegaskanda S, et al. Intranasal Live Influenza Vaccine Priming Elicits Localized B Cell Responses in Mediastinal Lymph Nodes. J Virol. 2018;92(9):e01970-17. doi: 10.1128/JVI.01970-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowe JE. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Potential for a Universal Influenza Vaccine. Cold Spring Harb Perspect Biol. 2018;10(7):a029496. doi: 10.1101/cshperspect.a029496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120(5):1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittle JR, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol. 2014;88(8):4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheatley AK, et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J Immunol. 2015;195(2):602–610. doi: 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews SF, et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol. 2017;2(13):eaan2676. doi: 10.1126/sciimmunol.aan2676. [DOI] [PubMed] [Google Scholar]
- 27.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21(9):1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 28.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellebedy AH, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci USA. 2014;111(36):13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87(12):6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui J, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. 2011;52(8):1003–1009. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchenbaum GA, Carter DM, Ross TM. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J Virol. 2016;90(2):1116–1128. doi: 10.1128/JVI.02372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman MO, Bennink JR, Yewdell JW, Herrin BR. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. Elife. 2015 doi: 10.7554/eLife.07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angeletti D, et al. Defining B cell immunodominance to viruses. Nat Immunol. 2017;18(4):456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7(316):316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angeletti D, et al. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc Natl Acad Sci USA. 2019;116(27):13474–13479. doi: 10.1073/pnas.1816300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avnir Y, et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 2014;10(5):e1004103. doi: 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avnir Y, et al. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep. 2016;6:20842. doi: 10.1038/srep20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pappas L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516(7531):418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- 41.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe T, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27(3):493–497. [Google Scholar]
- 44.Havenar-Daughton C, et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci USA. 2016;113(10):2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.