ABSTRACT
Bacteriophages employ small proteins to usurp host molecular machinery, thereby interfering with central metabolic processes in infected bacteria. Generally, phages inhibit or redirect host transcription to favor transcription of their own genomes. Mechanistic and structural studies of phage-modulated host transcription may provide inspirations for the development of novel antibacterial substances.
KEWORDS: Antimicrobial peptides, bacteriophage antibacterial proteins, structural biology, transcription regulation in bacteria
Introduction
The bacterial transcription machinery is a proven target for antibiotics [1]. Besides RNA polymerase (RNAP) itself, bacteria also harbor numerous transcription regulatory factors that are essential. While sequence and structural similarities exist between bacterial and eukaryotic RNAPs, the molecular machinery comprising the transcription regulatory layers is very distinct between the different domains of life. Indeed, only the NusG/Spt5-family of transcription elongation factors appears to be universally conserved [2–4]. Thus, transcription regulatory factors or transcription complexes that are modified by specific regulatory factors may represent highly attractive targets for the development of novel antimicrobial substances. However, as most of these factors do not possess easy-to-monitor enzymatic activities, targeting this regulatory layer by traditional inhibitor screening strategies is difficult. In light of an increasing number of high-resolution 3D structures of transcription factors and their complexes becoming available, in silico screening methodologies may offer some remedy [5].
Bacteriophages provide large reservoirs of unique proteins that modulate diverse bacterial molecular machineries to enable successful propagation of the phages [6, 7]. Many of these phage-derived protein modulators do not exhibit any homology to known proteins from organisms in all three kingdoms of life. Often such effector molecules target host molecular machineries associated with essential metabolic pathways, eventually leading to the complete shutdown of the host metabolism and killing of the host bacteria. The essentiality of host molecular machinery is one of the major criteria to be considered when selecting a potential drug target, and identification and characterization of novel phage effector proteins could thus lead to the identification of novel, attractive drug targets in pathogens.
With an estimated number of more than a billion bacterial species on the planet [8] and more than ten phage species on average estimated to infect each microbial species [9], phages might harbor bactericidal proteins for pathogenic bacterial species in numbers that dwarf the size of chemical libraries presently in use. Phages, therefore, might offer a rich resource for discovering novel molecules that interfere with the metabolism of otherwise difficult to control human pathogens. Indeed, mycobacteriophage endolysins lyse the complex peptidoglycans of mycobacteria [10], and the gp52 protein of mycobacteriophage Fruitiloop has been shown to exert its toxicity by affecting mycobacterial cell wall biosynthesis [11]. Recently, a mycobacteriophage genomics approach has been initiated to identify novel mycobacteriophage factors with bactericidal properties [12].
Antimicrobial peptides (AMPs) are oligopeptides comprising around 5 to 50 amino acid residues. Due to the emergence of multi-drug resistant bacteria and due to a dearth in novel antibiotics, AMPs are presently garnering renewed attention [13, 14]. In addition to their bactericidal activities, phage-derived protein modulators are typically small (≤20kDa) [6], rendering them attractive templates for designing new bactericidal peptides or peptidomimetics. Detailed biochemical and structural analyses of their interactions with their host targets might enable the definition of small regions of the phage proteins that embody target-binding properties independent of the remaining parts of the proteins, which might be further developed into effective AMPs.
As a general strategy, phages hijack or subvert parts of the host transcription machinery [15]. In modulating host transcription, phages often target not only host RNAP but also transcription regulatory factors [16, 17]. Thus, studying the structural basis of phage-derived mechanisms to interfere with bacterial transcription may uncover hitherto unexplored transcription-modulatory strategies and, thus, may inspire the development of novel antibacterial compounds that target bacterial transcription, including the transcription regulatory layer. High-resolution macromolecular structures, as can be obtained via macromolecular crystallography, may serve as templates for medicinal chemists to devise small peptides, peptidomimetics or even non-protein small molecules, which could possess inhibitory prowess. Indeed, high-resolution crystal structures of a transcription factor complex have recently been used to rationally design small molecules that interfere with protein–protein interactions in this complex [18, 19]. Recent revolutionary developments in imaging of biomacromolecular complexes at the atomic level using single-particle cryo-electron microscopy (cryoEM) [20] also provide deep insights into macromolecular interactions. With additional technological and computational improvements expected in the foreseeable future, cryoEM may provide additional templates for inhibitor development, in particular, high-resolution structures of complete transcription complexes.
Phage-mediated modulation of transcription initiation
Bacteria use a conserved, multi-subunit RNAP (core subunit composition α2ββ’ω) to transcribe their genomes [21]. The core enzyme associates with one of the several σ-factors to initiate transcription specifically at promoter sequences [22]. After transcription initiation, RNAP forms a stable elongation complex (EC) with the DNA and nascent RNA. The EC can be modified by transcription elongation factors, such as N-utilization substance (Nus) A or G [23]. RNAP dissociates from the template only in response to certain signals, called terminators [24]. There are two major modes of transcription termination in bacteria. In intrinsic termination, a GC rich inverted repeat that forms an RNA hairpin, followed by a stretch of consecutive uridinylates induces RNAP to pause and subsequently disengage from the template [25, 26]. The predominant mode of factor-dependent termination relies on a hexameric, NTP-dependent RNA translocase/helicase, ρ [27, 28].
Traditionally, transcription has been regarded to be regulated predominantly during the initiation phase. Thus perhaps not surprisingly, several phage modulators of bacterial transcription initiation have been discovered [29]. The study of their functional mechanisms revealed an astonishing diversity of how phages can subvert the function of host σ factors. For instance, enterobacterial phage T4 proteins AsiA and MotA activate phage middle genes [30]. AsiA binds conserved region 4 of the primary E. coli σ-factor, σ70, preventing its canonical interaction with −35 promoter elements and enabling subsequent MotA binding. Upon binding, AsiA undergoes a conformational change and engages upstream DNA [31]. MotA binds to a conserved DNA element (MotA box) that replaces the −35 element in middle promoters as well as to AsiA-remodeled σ region 4 [31]. Thus, AsiA and MotA cooperate to substitute for σ70 interactions with a − 35 promoter element during middle gene expression. For late gene expression, T4 proteins gp33 and gp55 form a “composite” σ factor that acts in cooperation with the T4 sliding clamp gp45 to recognize the single −10 elements of the late promoters [32]. The gp39 and gp76 proteins of Thermus phage P23-45 redirect host RNAP to late phage genes. In the crystal structures of a gp39- [33] and gp39/gp76-holoenzyme complexes [34], the globular part of p39 binds to the β flap at the base of the flap tip, while a C-terminal helix interacts with σ region 4, displacing σ region 4 bound to the flap tip. As a consequence, σ region 4 can no longer interact with promoter −35 regions, leading to inhibition of transcription from the −10/-35 class of promoters, but not of transcription from an extended −10 class of phage middle/late promoters [33]. Inhibition of transcription of host genes is supported by the phage gp76 protein, which binds deep inside the active site cleft of RNAP and to a linker connecting σ regions 3 and 4, obstructing accommodation of the melted template DNA strand and thus hindering the conversion of a closed to an open transcription initiation complex. Most likely, initiation of phage genes remains efficient due to the higher affinity of RNAP to extended −10 regions, which may overcome gp76-mediated inhibition [34]. As another example, protein P7 of Xanthomonas oryzae phage Xp10 directly binds to the β and β’ subunits of RNAP in a manner that induces σ70 displacement [35] and that locks the RNAP clamp in a closed conformation that inhibits loading of promoter DNA into the RNAP active site cleft [36]. Finally, enterobacterial phage T7 encodes its own RNAP and benefits from shutting off host RNAP-dependent transcription. The gp2 protein of T7 phage binds the 1.1 domain of σ70 and the RNAP β’ subunit, thereby locking σ70 domain 1.1 in the RNAP active site channel [37].
However, it is now well established that the transcription elongation and termination phases also are highly regulated by both intrinsic signals on the template DNA/product RNA and by trans-acting, extrinsic protein transcription factors [38]. These signals and factors, among others, cause RNAP to frequently pause during elongation, offering windows of opportunity for other regulatory mechanisms to take effect [23, 38, 39], or they can modulate the strength of terminators [24, 25]. Indeed, it has been the investigation of lambdoid phages that led to the discovery of host-encoded transcription elongation factors, termination factor ρ as well as phage-derived factors that modulate the behavior of RNAP during elongation and termination [40–42].
Phage-mediated modulation of transcription elongation and termination
Lambdoid phages are known for a long time to employ strategies that modulate host RNAP pausing and termination functions during transcription of the phage genomes [40–44]. For example, most of these phages employ N and Q proteins to facilitate the switch from immediate-early to delayed-early gene expression and to support the expression of late genes, respectively, during their lytic life cycles (Figure 1). To this end, N and Q proteins interact with host RNAP, transcription factors, RNA and/or DNA, stably modifying ECs to confer pause- and termination resistance on RNAP and thus allowing it to read through intra- and intergenic terminators, even if the terminators are located far downstream of the site at which N or Q originally load onto the EC (processive anti-termination) [40–44].
Figure 1.
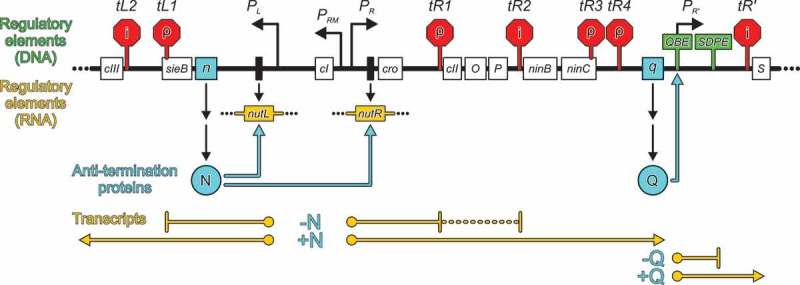
Biological activities of lambdoid phage N and Q proteins.
Scheme of part of the phage λ genome (thick black line) containing early and late control regions. The sizes of regions and elements, and their positions, are not drawn to scale. Open boxes with names – protein-coding regions; narrow black boxes, nut site DNA; black-angled arrows, promoters; red stop signs, intrinsic (“i”) and ρ-dependent (“ρ”) terminators; green signs, regulatory regions active as DNA (QBE and SDPE); dark yellow boxes, nut regulatory regions active as RNA; cyan spheres, anti-termination proteins; cyan angled arrows, sites of recruitment of anti-termination proteins to ECs; dark yellow lines, transcripts. Scheme adapted from [43] with changes.
Processive anti-pausing and anti-termination by n proteins
N recognizes signal sequences in untranslated regions of nascent phage RNA, so-called N-utilization (nut) sites (Figure 1), comprising a linear boxA element and a boxB stem-loop structure. N binds boxB and RNAP and recruits Nus factors A, B, E (equivalent to ribosomal protein S10) and G, building up a complex ribonucleoprotein (a “modifying” RNP) on the surface of RNAP that stays associated with RNAP during the entire transcription elongation process by an RNA looping mechanism [42, 43, 45].
Recently, the group of one of the authors (M.C.W.) and collaborators reported a crystal structure of an λN-Nus factor-nut RNA complex [46] and a high-resolution cryoEM structure of a complete λN-based transcription anti-termination complex (λN-TAC), comprising RNAP, template DNA, product RNA with a nut site, all Nus factors and the λN protein [17] (Figure 2a). N proteins are intrinsically unstructured, ~110-residue polypeptides. The structural analyses revealed that in the λN-TAC, λN only locally adopts the regular secondary structure and remains highly elongated, which enables it to contact many sites on RNAP, the Nus factors and the nascent RNA (Figure 2b). It thereby implements a multi-pronged strategy to suppress transcription pausing as well as intrinsic and factor-dependent termination.
Figure 2.
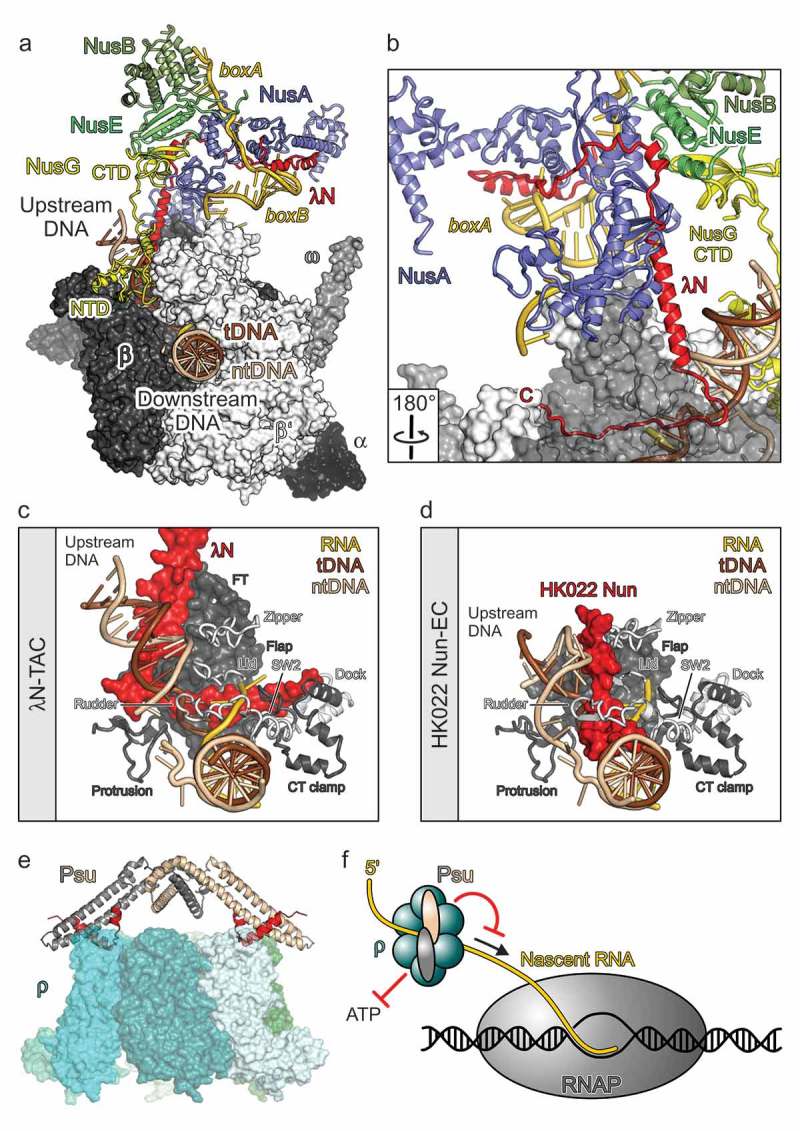
Molecular models of phage factors modulating host transcription elongation and termination.
(a) A Single-particle cryoEM structure of an λN-TAC [17]. RNAP subunits in surface representation. Nucleic acids, Nus factors, and λN in cartoon representation. (b) λN remains highly extended in the λN-TAC, allowing short peptides along its sequence to interact with spatially widely distributed regions on nascent RNA, Nus factors and RNAP. RNAP subunit β as semi-transparent surface. Rotation symbol – view relative to (a). (c) Interaction of the C-terminal region of λN with nucleic acids and various elements of RNAP in and around the active site cleft (β elements: flap, FT – flap tip, protrusion, CT clamp – C-terminal clamp; β’ elements: zipper, lid, rudder, SW2 – switch 2, dock). View as in (a). (d) HK022 Nun interacting with nucleic acids and RNAP elements [60]. Same orientation of RNAP as in (d). (e) presumed mode of action of Psu [16]. By inhibiting ρ’s ATPase, Psu will hinder the translocation of ρ along the nascent RNA toward RNAP (arrow). Red symbols – inhibition. (f) Docking model of the phage P4 Psu protein interacting with E. coli transcription termination factor ρ [62]. (c) and (d) adapted from [17].
Hairpin-stabilized pausing and intrinsic termination are disfavored by multiple strategies aimed at preventing pause-stabilizing or termination hairpins from invading the RNAP RNA exit tunnel: λN repositions NusA on RNAP, such that it can no longer stabilize exit tunnel-invading hairpins and instead may support their unfolded state. In addition, λN conformationally remodels RNAP exit tunnel elements, and its C-terminal residues line the RNA exit tunnel, constricting the tunnel and physically blocking its invasion by RNA secondary structures. Moreover, λN organizes NusA and S10/NusE regions such that they present a binding site for the C-terminal domain of NusG, which is thereby sequestered and prevented from supporting the activity of termination factor ρ. Furthermore, λN and NusG line opposite flanks of the upstream DNA duplex; λN thereby apparently supports the otherwise weak intrinsic DNA re-annealing and RNAP anti-backtracking activities of NusG. Most remarkably, the C-terminal 25 residues of λN traverse the RNAP active site cleft, stringing mobile RNAP elements, which are repositioned during pausing and presumably also during termination [47, 48], together, thus keeping RNAP in an anti-paused, processive conformation (Figure 2c).
Notably, λN employs diverse, short peptide segments to implement its many anti-pausing/anti-termination strategies, each of which may in principle lend itself to the design of novel interfering substances. The present resolution of the λN-TAC structure (3.7 Å) most likely does not suffice to serve as a reliable template for detailed modeling studies with the aim to derive new modulators. Mild crosslinking [49] and/or imaging a co-transcriptionally assembled complex may still offer room for improvement. Moreover, different lambdoid phages encode different N proteins, which may lend themselves to even higher-resolution structural analyses. Of particular interest may be a C-terminally extended N polypeptide of phage H-19B [50]. Biochemical analyses by the group of one of the authors (R.S.) have revealed that H-19B N repositions NusA and prevents ρ function [51, 52], and suggested that H-19B N may even directly interact with the RNAP active site region [50]. Thus, a detailed structural analysis may uncover yet additional strategies of RNAP modulation in the H-19B case.
RNA exit tunnel modulation as a widespread strategy to counteract pausing and termination
Structural modulation of the RNA exit tunnel and surrounding elements to prevent invasion by pause/termination-enhancing RNA hairpins, as well as prevention of NusA-mediated stabilization of such hairpins, appear to be widespread strategies employed by phages to regulate their gene expression. Again, phages have evolved surprisingly diverse molecular mechanisms to achieve these tasks. One alternative to the N-based strategy is exemplified by lambdoid phage Q proteins [44, 45]. Q recognizes a Q-binding DNA element (QBE) located between the −35 and −10 elements of the phage late gene promoter, which is followed by a σ-dependent pause element (SDPE) and a terminator (Figure 1). Q loads onto the σ-modified, paused EC and, upon pause escape, remains associated with the EC, persistently suppressing RNAP pausing and termination. Recent cryoEM structures of Q-loading complexes, based on the Q protein of phage 21 (Q21), revealed that two Q21 molecules recognize direct repeats of the QBE [53, 54]. In the loading complex, σ remains anchored to the paused EC via σ regions 2 and 3, but due to the presence of >10 nucleotides of initial RNA, the σ region 3–4 linker and σ region 4 are displaced. Besides binding the β’ dock domain (occupied by σ region 4 in transcription initiation complexes) and the αI-β interface, the Q21 protomer bound at the upstream QBE (Q21u) uses a helix and neighboring linkers to form a ring-like structure (the “Q torus” [54]) around the mouth and inside of the RNA exit tunnel, which extends and constricts the RNA exit tunnel. The Q21 protomer bound at the downstream QBE (Q21d) additionally contacts the β flap tip helix in a manner mutually exclusive with σ region 4-β flap tip interactions in initiation complexes. A structure of the Q21-loaded complex revealed that Q21u maintains its RNAP interactions after pause escape, while σ and Q21d are displaced [54]. This structure also showed that single-stranded RNA can be threaded through the Q21u torus, while nucleation, propagation and exit tunnel penetration of RNA hairpins are prevented [54].
P7 protein of phage Xp10 provides yet another example of exit tunnel modulation. Recent cryoEM structures of P7-modified ECs without and with NusA revealed that P7 can bind between a short N-terminal helix of β’, the β’ dock domain and the C-terminal region of β at the mouth of the RNA exit tunnel, thereby restricting the local diameter of the exit tunnel and preventing accommodation of an RNA hairpin [36]. Moreover, P7 in a P7/NusA-modified EC lines a concave surface of the NusA N-terminal and S1 domains [36]. Thus, P7 exploits the very surfaces of NusA that are normally used to stabilize exit tunnel-invading hairpins for its own stable binding to RNAP, essentially converting NusA from a pause/termination-supporting factor to an anti-pausing/anti-termination factor [36].
Transcription arrest by the HK002 nun proteins
Lambdoid phage HK022 resorts to a different strategy to implement transcription anti-termination for delayed-early gene expression. Here, a cis-acting, bi-lobed RNA structure, the polymerase-utilization (put) site, in the untranslated regions of the phage RNA directly binds to the β’ Zinc-finger domain of RNAP and confers pause/termination resistance [55] 56]. HK022 also encodes an N-related protein, Nun [57]. Presumably due to the availability of the put element, Nun evolved to have a diametrically opposite function to other N proteins: It responds to the same nut sites as N and recruits the same set of host Nus factors to RNAP, but induces pre-mature transcription arrest [57–59], likely to prevent super-infection by other lambdoid phages. A cryoEM structure of an HK022 Nun-arrested EC has been elucidated [60]. Only the C-terminal 23 residues of Nun on RNAP could be imaged, the rest of the protein remained unresolved due to its intrinsically unstructured nature and high flexibility in the absence of the Nus factors and nut RNA. The structure revealed how Nun, similar to λN, inserts its C-terminal region into the interior of RNAP, but entering along a different flank of upstream DNA, where no natural crevices are available to accommodate the protein without distorting RNAP (Figure 2d). As a consequence, Nun distorts and displaces several RNAP elements and wedges into the nucleic acid network, inhibiting nucleic acid movement inside RNAP (Figure 2d). Thus, this C-terminal region of Nun provides a highly attractive template for the design of novel RNAP-inhibitory substances. It will be interesting to see in the future how other regions of Nun interact with the Nus factors and whether these interactions augment the transcription inhibitory potential of the protein.
Anti-ρ activity of the Psu protein
Another interesting phage-derived transcription modulator is the capsid protein, Psu, of enterobacterial phage P4. Psu is an antagonist of the conserved bacterial transcription termination factor, ρ [16]. The group of one of the authors (R.S.) demonstrated that Psu inhibits ρ ATPase activity but does not prevent the binding of RNA to ρ’s primary and secondary RNA binding sites [16]. Thus, Psu presumably interferes with ρ-mediated transcription termination by inhibiting 5ʹ-to-3ʹ translocation of ρ on the mRNA (Figure 2e). Together with collaborators, the Sen lab also unraveled the crystal structure of Psu, showing that the protein adopts a novel fold that supports the formation of an unusual, knotted dimer [61]. Based on this structure, the known structure of E. coli ρ and the mapping of interacting residues on Psu and ρ, a docking model of dimeric Psu on a closed ρ hexamer was constructed (Figure 2f) [62, 63]. The biochemical data and the docking model revealed that Psu most likely uses a C-terminal α helix and neighboring residues to contact two ρ subunits on opposite sides of the ring. Importantly, the Sen group demonstrated that Psu can inhibit the ATPase and transcript release activities of ρ proteins from diverse pathogenic bacteria in vitro and that overproduction of Psu was bactericidal [64]. Novel AMPs could be designed based on the C-terminal helices of Psu that directly contact ρ. The rational design of Psu-derived anti-ρ peptides or peptidomimetics would strongly benefit from the elucidation of a high-resolution experimental structure of a Psu-ρ complex.
Conclusions
Ample examples have been documented for how phages employ small proteins to target all phases of transcription of their hosts. It is to be expected that with more phages being discovered and studied, more such mechanisms, as well as variations of known mechanisms, will be revealed. Notably, in many cases, not only the host RNAP but also the host transcription factors that regulate the various phases of transcription are key targets of the phage proteins. While these transcription factors are typically not conserved in eukaryotes, they are widely distributed in bacteria and in many species they are essential, rendering them “naturally selected” (“chosen” by phages) drug targets. With improved techniques for molecular docking and design, high-resolution structures of phage-derived transcription modulators in the course of their action may provide valuable assets for guiding the rational development of novel, phage-informed, transcription-targeting antibacterial substances.
Funding Statement
Work in the authors’ laboratories was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 740-3, SFB 958-2, RTG 2473-1) to M.C.W. and the Departments of Biotechnology (DBT) and Science and Technology (DST) (DST-SERB) government of India (EMR/2015/001620 and BT/PR27969/BRB/10/1662/2018) to R.S.
Acknowledgments
We are grateful to members of our labs for critical reading of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Ma C, Yang X, Lewis PJ.. Bacterial transcription as a target for antibacterial drug development. Microbiol Mol Biol Rev. 2016;80:139–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yakhnin AV, Babitzke P.. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol. 2014;18:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Svetlov D, Shi D, Twentyman J, et al. In silico discovery of small molecules that inhibit RfaH recruitment to RNA polymerase. Mol Microbiol. 2018;110:128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B, et al. Learning from bacteriophages - advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci. 2012;13:699–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nechaev S, Severinov K. The elusive object of desire–interactions of bacteriophages and their hosts. Curr Opin Microbiol. 2008;11:186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dykhuizen D. Species numbers in bacteria. Proc Calif Acad Sci. 2005;56:62–71. [PMC free article] [PubMed] [Google Scholar]
- [9].Rohwer F. Global phage diversity. Cell. 2003;113:141. [DOI] [PubMed] [Google Scholar]
- [10].Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7:1147–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ko CC, Hatfull GF. Mycobacteriophage Fruitloop gp52 inactivates Wag31 (DivIVA) to prevent heterotypic superinfection. Mol Microbiol. 2018;108:443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Singh S, Godavarthi S, Kumar A, et al. A mycobacteriophage genomics approach to identify novel mycobacteriophage proteins with mycobactericidal properties. Microbiol. 2019;165:722–736. [DOI] [PubMed] [Google Scholar]
- [13].Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel). 2013;6:1543–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Parai D, Dey P, Mukherjee SK. Antimicrobial Peptides: an Approach to Combat Resilient Infections. Curr Drug Discov Technol. 2019. doi: 10.2174/1570163816666190620114338. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- [15].Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. [DOI] [PubMed] [Google Scholar]
- [16].Pani B, Banerjee S, Chalissery J, et al. Mechanism of inhibition of Rho-dependent transcription termination by bacteriophage P4 protein Psu. J Biol Chem. 2006;281:26491–26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krupp F, Said N, Huang YH, et al. Structural basis for the action of an all-purpose transcription anti-termination factor. Mol Cell. 2019;74:143–157. [DOI] [PubMed] [Google Scholar]
- [18].Cossar PJ, Abdel-Hamid MK, Ma C, et al. Small-molecule inhibitors of the NusB-nuse protein-protein interaction with antibiotic activity. ACS Omega. 2017;2:3839–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cossar PJ, Ma C, Gordon CP, et al. Identification and validation of small molecule modulators of the NusB-NusE interaction. Bioorg Med Chem Lett. 2017;27:162–167. [DOI] [PubMed] [Google Scholar]
- [20].Kuhlbrandt W. The resolution revolution. Science. 2014;343:1443–1444. [DOI] [PubMed] [Google Scholar]
- [21].Darst SA. Bacterial RNA polymerase. Curr Opin Struct Biol. 2001;11:155–162. [DOI] [PubMed] [Google Scholar]
- [22].Helmann JD. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol Microbiol. 2019;112:335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Belogurov GA, Artsimovitch I. Regulation of Transcript Elongation. Annu Rev Microbiol. 2015;69:49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of Bacterial Transcription Termination: all good things must end. Annu Rev Biochem. 2016;85:319–347. [DOI] [PubMed] [Google Scholar]
- [26].Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3ʹ-end chronicles. J Mol Biol. 2011;412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mitra P, Ghosh G, Hafeezunnisa M, et al. Rho protein: roles and mechanisms. Annu Rev Microbiol. 2017;71:687–709. [DOI] [PubMed] [Google Scholar]
- [28].Kriner MA, Sevostyanova A, Groisman EA. Learning from the leaders: gene regulation by the transcription termination factor Rho. Trends Biochem Sci. 2016;41:690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tabib-Salazar A, Mulvenna N, Severinov K, et al. Xenogeneic regulation of the bacterial transcription machinery. J Mol Biol. 2019. pii: S0022-2836(19)30085-3. doi: 10.1016/j.jmb.2019.02.008. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- [30].Hinton DM. Transcriptional control in the prereplicative phase of T4 development. Virol J. 2010;7:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shi J, Wen A, Zhao M, et al. Structural basis of sigma appropriation. Nucleic Acids Res. 2019;47:9423–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nechaev S, Kamali-Moghaddam M, Andre E, et al. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2004;101:17365–17370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tagami S, Sekine S, Minakhin L, et al. Structural basis for promoter specificity switching of RNA polymerase by a phage factor. Genes Dev. 2014;28:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ooi WY, Murayama Y, Mekler V, et al. A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation. Nucleic Acids Res. 2018;46:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu B, Shadrin A, Sheppard C, et al. A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement. Nucleic Acids Res. 2014;42:4294–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].You LL, Shi J, Shen LQ, et al. Structural basis for transcription antitermination at bacterial intrinsic terminator. Nat Commun. 2019;10:3048. doi: 10.1038/s41467-019-10955-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bae B, Davis E, Brown D, et al. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma(70) domain 1.1. Proc Natl Acad Sci USA. 2013;110:19772–19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang J, Landick R. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci. 2016;41:293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gottesman ME, Weisberg RA. Little Lambda, who made thee? Microbiol Mol Biol R. 2004;68:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Weisberg RA, Gottesman ME. Processive antitermination. J Bacteriol. 1999;181:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–406. [DOI] [PubMed] [Google Scholar]
- [43].Goodson JR, Winkler WC. Processive Antitermination. Microbiol Spectr. 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Roberts JW, Yarnell W, Bartlett E, et al. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. [DOI] [PubMed] [Google Scholar]
- [45].Nudler E, Gottesman ME. Transcription termination and anti-termination in E. Coli. Genes Cells. 2002;7:755–768. [DOI] [PubMed] [Google Scholar]
- [46].Said N, Krupp F, Anedchenko E, et al. Structural basis for lambdaN-dependent processive transcription antitermination. Nat Microbiol. 2017;2:17062. [DOI] [PubMed] [Google Scholar]
- [47].Kang JY, Mishanina TV, Bellecourt MJ, et al. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol Cell. 2018;69:802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo X, Myasnikov AG, Chen J, et al. Structural basis for NusA stabilized transcriptional pausing. Mol Cell. 2018;69:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kastner B, Fischer N, Golas MM, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. [DOI] [PubMed] [Google Scholar]
- [50].Cheeran A, Kolli NR, Sen R. The site of action of the antiterminator protein N from the lambdoid phage H-19B. J Biol Chem. 2007;282:30997–31007. [DOI] [PubMed] [Google Scholar]
- [51].Muteeb G, Dey D, Mishra S, et al. A multipronged strategy of an anti-terminator protein to overcome Rho-dependent transcription termination. Nucleic Acids Res. 2012;40:11213–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mishra S, Sen R. N protein from lambdoid phages transforms NusA into an antiterminator by modulating NusA-RNA polymerase flap domain interactions. Nucleic Acids Res. 2015;43:5744–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shi J, Gao X, Tian T, et al. Structural basis of Q-dependent transcription antitermination. Nat Commun. 2019;10:2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yin Z, Kaelber JT, Ebright RH. Structural basis of Q-dependent antitermination. Proc Natl Acad Sci USA. 2019;116:18384–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Komissarova N, Velikodvorskaya T, Sen R, et al. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol Cell. 2008;31:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sen R, King RA, Weisberg RA. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol Cell. 2001;7:993–1001. [DOI] [PubMed] [Google Scholar]
- [57].Robert J, Sloan SB, Weisberg RA, et al. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987;51:483–492. [DOI] [PubMed] [Google Scholar]
- [58].Hung SC, Gottesman ME. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995;247:428–442. [DOI] [PubMed] [Google Scholar]
- [59].Hung SC, Gottesman ME. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 1997;11:2670–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kang JY, Olinares PD, Chen J, et al. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLife. 2017;6:e25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Banerjee R, Nath S, Ranjan A, et al. The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer. J Biol Chem. 2012;287:44667–44675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ranjan A, Sharma S, Banerjee R, et al. Structural and mechanistic basis of anti-termination of Rho-dependent transcription termination by bacteriophage P4 capsid protein Psu. Nucleic Acids Res. 2013;41:6839–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Pani B, Ranjan A, Sen R. Interaction surface of bacteriophage P4 protein Psu required for complex formation with the transcription terminator Rho. J Mol Biol. 2009;389:647–660. [DOI] [PubMed] [Google Scholar]
- [64].Ghosh G, Reddy J, Sambhare S, et al. A bacteriophage capsid protein is an inhibitor of a conserved transcription terminator of various bacterial pathogens. J Bacteriol. 2018;200:e00380–17. [DOI] [PMC free article] [PubMed] [Google Scholar]


