Colonial animals reconcile selection among organisms and colonies by limiting the evolutionary potential of organism.
Abstract
The evolution of modular colonial animals such as reef corals and bryozoans is enigmatic because of the ability for modules to proliferate asexually as whole colonies reproduce sexually. This reproductive duality creates an evolutionary tension between modules and colonies because selection operates at both levels. To understand how this evolutionary conflict is resolved, we compared the evolutionary potential of module- and colony-level traits in two species of the bryozoan Stylopoma, grown and bred in a common garden experiment. We find quantitatively distinct differences in the evolutionary potential of modular and colony traits. Contrary to solitary organisms, individual traits are not heritable from mother to daughter modules, but colony traits are strongly heritable from parent to offspring colonies. Colony-level evolution therefore dominates because no evolutionary change can accumulate among its modules.
INTRODUCTION
One is tempted to view the problem of modular animal evolution (1) through the lens of the evolution of castes in eusocial insects (2–5) or the evolution of individuality (6–8), but neither approach has borne fruit (9, 10). Social insects impede the evolutionary potential of members by limiting their ability to proliferate and evolve within the context of the insect colony (11, 12), whereas colonial animals have no such restrictions. Buss (6) hypothesized that the evolutionary tension between clonal cell lineages and multicellular bodies is broken by a sequestered germ line that polices the evolutionary potential of members. However, modular colonial animals do not have sequestered germ lines at the cellular level (13–16), and their colony-level sexual reproductive specialists tend to be dispersed around the colony and to originate from a number of independent clonal lineages (17–19). Consequently, modular colonial animals do not have the ability to enforce limitations on the propagation of novel variants (9, 20).
In contrast, we can understand how modular colonial animals resolve the evolutionary tension between modules and colonies through an examination of the way that phenotypic variation is channeled within a colony from module to module and propagated from parent to offspring colonies to produce the phenotype (Fig. 1). Evolution by natural selection results from a multiplicative interaction between selection, phenotypic variation, and the heritability of phenotypes from parent to offspring (21–27). If any one of these three attributes is absent (effectively zero), then no evolution is possible. Animal colonies somehow remain coherent entities in the face of the potential for selection among modules to undermine them. Hence, the pattern of heritable variation among modules within colonies and between parent and offspring colonies is the key to understanding the organizational stability of modular colonies because these features control the evolutionary potential of traits even in the presence of active selection or passive drift.
Fig. 1. A Stylopoma colony grows by adding modules and the population of Sylopoma colonies increase by the settling of sexually produced larvae.
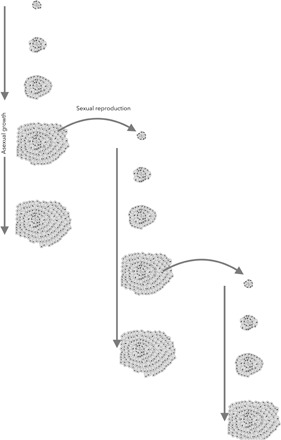
Modules proliferate by asexual clonal budding. The growth of a single colony is illustrated in this series along with the numeric increase in the number of colonies. New colonies are founded from sexually derived larvae that disperse from a mother colony. The first sexually derived ancestral set of zooids is shown in the top left. Time runs from top to bottom so that by the end of this series, this colony has expanded markedly from the clonal proliferation of its modules, and the populations of colonies have also expanded from the sexual production of offspring colonies.
Here, we examine whether traits at the modular and colonial levels have the potential to evolve by natural selection by measuring the heritability between mother and daughter modules (i.e., individual animal bodies, termed zooids, within a colony) and parent and offspring colonies (figs. S1 and S2). Using two generations of colonies of two species of the cheilostome bryozoan genus Stylopoma, we measured heritability as the phenotypic parent-offspring covariance among twelve characters consisting of eight modular characters and four colony-level characters (a depiction of the characters and methods are included in full in the Supplementary Materials). Modular characters include measurements of their dimensions, and colony traits include the numbers, relative positions, and orientations of modules within multimodular complexes, which Beklemishev (1) termed “cormidia” (see Fig. 2 and the Supplementary Materials for bryozoan basics and details of these traits). Stylopoma colonies were raised in a common garden breeding experiment to produce offspring of known maternity and environmental influence (28, 29). These colonies where originally studied by Jackson and Cheetham (28) to investigate whether or not skeletal characteristics are sufficient to rigorously identify biological species in the fossil record. They were subsequently used in a quantitative genetics analysis where the among-zooid variability was used to infer the magnitude of heritable genetic variation (29–31). However, that approach neglected the hierarchical structure that nests zooids within colonies, effectively conflating patterns of variability among zooids for heritability between colonies. Here, we used measurements of at least three modules of each of 82 mother colonies and 326 daughter colonies to calculate parent-offspring phenotypic covariance between mother and daughter modules within colonies and also for colonial traits expressed between parent and offspring colonies.
Fig. 2. We measure the evolutionary potential of these twelve traits.
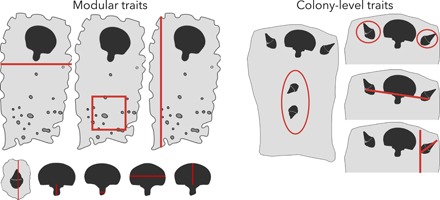
Module-level traits are expressed by single modules. These traits are quantitative measures, lengths, widths, and densities. Colony-level traits are expressed by multiple modules and are measures of their relative positions, orientations, and numbers of modules. See the Supplementary Materials for a full description of these traits.
Unlike additive genetic variance, parent-offspring phenotypic covariance can be negative or larger than one because it is based on the linear regression of offspring onto parent phenotypes (25). The advantage of using parent-offspring phenotypic covariance is that its sign (positive or negative) provides a qualitative measure of evolutionary potential that is fully independent of the pattern of selection. Positive covariance works with selection. Random covariance (those near zero) neutralizes selection. In addition, negative covariance opposes selection and leads to evolutionary outcomes opposite of those that selection favors.
RESULTS AND DISCUSSION
The outcome is unexpectedly clear. Module-level traits are heritable neither between parent and offspring modules within a colony nor between parent and offspring colonies (Fig. 3). In contrast, two of four colony-level traits, which involve the position and organization of multiple members, are positively heritable across colony generations (Fig. 3). The rest of the colony-level traits have low evolutionary potential because they are invariant among colonies and across generations not because they show no similarity between parent and offspring.
Fig. 3. The evolutionary potential for eight modular and four colony-level traits in two species of the bryozoan Stylopoma.
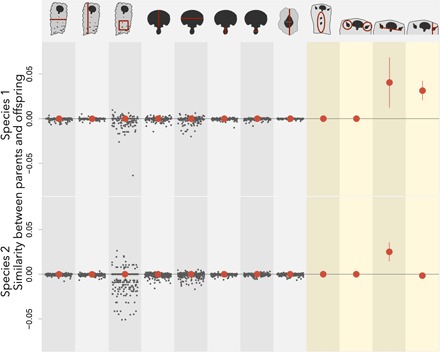
The parent-offspring phenotypic covariance for each trait is shown in columns. We calculate the heritability of member-level traits within each colony (indicated by gray circles jittered horizontally to avoid plotting overlap). The median heritability across colonies is shown by the large red circles. For colony-level traits, only a single estimate of heritability is possible; thus, the dispersion around the heritability estimate is indicated by vertical bars. Colony traits without bars do not vary among colonies.
The lack of heritability of modular traits means that there is no propagation of modular characteristics within a colony. The pattern of higher heritability of colony-level traits across colony generations acts to crystalize evolution at the colony level because only colonial traits have the evolutionary potential to respond to natural selection.
This result contrasts with typical patterns of heritability observed in solitary organisms where metric traits commonly have positive heritability much greater than those we observe within Stylopoma colonies (23). Furthermore, the quantitative separation of modular traits has proven to be inadequate to distinguish species in Stylopoma (32, 33) and in other bryozoans (34).
Differential proliferation among modules is likely to be ubiquitous within colonies. Consequently, Stylopoma’s pattern of inheritance transforms what could be an evolutionary problem into an ecological asset. Encrusting species, like Stylopoma, must cope with many and varied enemies and with an unpredictable world as they grow and occupy space (35–39), and the ability for a Stylopoma colony to grow in any direction without accumulating evolutionary change allows them to thrive in the face of intense and unpredictable biological overgrowth interactions.
Variation in evolutionary potential across traits together with the proliferation of both modules and colonies implies that the emergence of the colony as a level of evolution is under developmental control rather than a result of environmental influence. The presence of developmental machinery for both colony and modular traits is necessary to produce an evolutionary response to natural selection at any level. Furthermore, Stylopoma colonies achieve and maintain their integration without resorting to the policing of modules.
Broadly speaking, there are two alternative modes of development to maintain colony integration: policing and cultivation. Eusocial insects, such as ants and bees, use policing to maintain the integrity of their colonies by limiting the reproductive potential of members (40, 41). However, modular colonies, like those made by bryozoans, take a diametrically opposing strategy. Colonies use the developmental mode of cultivation to take advantage of an individual module’s ability to proliferate without propagating morphological errors or accumulating character change. Like a farmer cultivating crops, colonies provide conditions and support that help the modules to thrive and vary within broad limitations. The advantage of the mode of cultivation is that the module’s capability of proliferation can be channeled into colony development, increasing growth rates and the potential for internal complexity without the risk of evolutionary conflict, as any conflict that may arise can only last a single generation. Moreover, the cultivation mode of development may not be limited to only modular animal colonies. It could occur in any group with members that proliferate. During their origin before the Cambrian explosion, metazoans may have used developmental cultivation to guide their cellular constituents from independently evolving lineages into the complex developmental ferment that they now have.
MATERIALS AND METHODS
Experimental design
Here, we use colonies that were grown in a previous breeding experiment. The specimens were used previously to tell whether skeletons are enough to tell species apart (28), whether selection or random change drove differentiation between species (29), and a phylogenetic estimate of the tempo of speciation (32). The scientific versatility of these specimens is due to the controlled way that they were grown and bred. From wild-caught colonies, two generations of colonies with known maternity were born. Furthermore, offspring colonies grown in a common garden experiment, allowing generations of scientists to tease apart the complex processes involved in phenotypic macroevolution.
The experimental design was modified from one developed by Maturo (42). This experiment was designed to strictly limit the number of possible paternal colonies that could fertilize maternal colonies and then to use the limited dispersal ability of Stylopoma larvae to know the maternal colonies that gave rise to offspring colonies. The experiment was conducted at the Smithsonian field station just east of San Blas Point on the Caribbean coast of the Republic of Panamá. Using this field station allowed offspring colonies to be grown in a common garden with a shared environment that allows the quantification of the impact of the environment on phenotypic expression. Bryozoans were collected from one to five sites, depending on species, between Holandes Cays and Isla Grande in depths of <1 to >40 m and were maintained in running sea water for usually no more than 1 day before use. Maternal colonies of two Stylopoma species were collected from the localities shown on this map. Colonies were collected in the 1980s from the following localities: Isla Grande, the west and eastern shores of Ulaksukan, West Palina, and southwest and northeast Aguadargana.
As described in Jackson and Cheetham (28) and Cheetham et al. (29), corals with Stylopoma colonies containing embryos were collected from these Caribbean localities. These coral substrata were cleaned of other organisms and isolated in transport chambers made from plastic food containers with sides cut open and replaced by plankton nets. Filtered sea water was run through the top of the chambers and exited through the nets in the walls. A single maternal colony was kept in each container under a piece of bare coral for daughter colonies to settle. After 5 to 10 days, the coral substratum with newly settled F1 colonies were removed and attached to concrete blocks on a sandy bottom about −0.5 m below low water at the Smithsonian field station just east of San Blas Point on the Caribbean coast of the Republic of Panamá (Fig. 4). Every month, the condition of the colonies was assessed, and the colonies and substrata were cleaned of other organisms.
Fig. 4. Map indicating the location of the Smithsonian San Blas field station in Panamá.
Quantifying evolutionary potential
If traits have the capability to respond to natural selection, then they have evolutionary potential. The trait may or may not experience selection, yet traits have the potential to evolve if they vary and if the trait is inherited with some fidelity from parent to offspring. The two components of variation and heritability together define the evolutionary potential. Without variation in a trait, there can be no evolution, because without phenotypic variation, there can be no variation in fitness. If heritability is zero, then it means that there is no degree of similarity between parents and offspring. Then, likewise, there can be no evolution by natural selection no matter the strength of selection or the amount of phenotypic variation. This is because, with zero heritability, offspring are free to take any form, and therefore, the change in frequencies of forms relative to their fitnesses is suppressed by the inability for forms to propagate across generations. A trait with heritability maintains its phenotype over time, much like a thrown snowball that maintains its identity as it flies through the air. A trait without heritability changes its phenotype overtime unpredictably, just as throwing a handful of sand spreads through the air. As soon as it leaves your hand it spreads out, increasingly losing its coherence the further it flies from your hand.
Multivariate Price’s theorem
The evolutionary logic above forms the basis of quantitative genetics and breeding by artificial selection. These fields have given us powerful mathematical tools to understand just how evolution proceeds given patterns of selection, variation, and heritability (22–25). These tools derive part of their power because they are not reductionists; they deal with phenotypic evolution at the phenotypic level. There is no need to dig lower into genetic levels of explanation for them to work. Thus, the methods do not offer a complete understanding of the mechanism of evolutionary change at all levels, nevertheless their success in agriculture and evolutionary biology underline their utility. For our purposes, we want to know how hierarchically organized Stylopoma colonies evolve, given the simultaneous proliferation of asexually and sexually produced zooids.
To see why this works, let us use a formality of Price’s theorem. Price’s theorem describes the evolutionary response to selection of a trait or set of covarying traits, given the structure of variation and heritability of those traits. Price’s theorem defines the evolutionary potential of traits in terms of two matrices: C, the heritability matrix, which measures the similarity of traits between parents and offspring. The diagonal values within the heritability matrix can be calculated as the variation in the offspring phenotype multiplied by the linear regression of parent phenotype onto offspring phenotype. The off-diagonal elements measure the co-heritability between two traits, for example, how similar egg size in a bird is to the egg number that her chicks are able to produce. These interactions can be strong if, as in some birds, egg size and egg numbers have a strong inverse relationship that persists over generations. The phenotypic covariance matrix, P, describes the amount of variation of all traits and the covariance between them. The product of C and the inverse of P define the evolutionary potential of traits. Selection, βw,ϕ, is defined as the linear regression of fitness (w) on phenotypes (ϕ). The response to selection is measured as the change in the average phenotype, denoted , and is determined by the product of evolutionary potential and selection (25)
| () | 1 |
Because this equation is a simple product of three terms, if any of the three factors selection (βw,ϕ), variation (P), or heritability (C) is equal to zero, then there will be no response to selection.
Hierarchical expansion
The equation above is very simple and it is another way to write the Breeder’s equation. For solitary organisms such as cattle, chickens, or beans, it summarizes the evolutionary processes involved adequately. However, it, as written, only gets half the story for colonial organisms like Stylopoma. It either partially describes colony-level evolution or it partially describes zooid level evolution. We want both. In the general form of Price’s theorem, there is an additional term, , which represents the expected change in the mean phenotype due to processes within the parts
| () | 2 |
It may help to use a paleontological example to think about this term (43–45). Species can evolve over time. New species may also change phenotypes during speciation so that they differ more or less from their ancestor. There is even a process of selection at the species level that acts by differential rates of extinction and speciation. The term, , is the average amount of evolution within all species—and in colonies, it is the change in phenotype due to biased changes among zooid members. Hamilton (46) and Price (47) were among the first to realize that Price’s theorem can be hierarchically expanded such that this last additive term is equivalent to a lower level of selection. The way this works is to notice that the change in the average traits within an entity has the same units as the change in the average traits among entities
| () | 3 |
If we rewrite Eq. 2 with this recursive evolutionary level in mind, then we can combine the evolution of wholes with the evolution of constituent parts
| () | 4 |
This looks more complex than it is because of the notation keeping track of whole colonies and their zooid parts. However, in words, Eq. 4 shows that the evolution of whole colonies is due to the product of the evolutionary potential of traits and selection at the colony level and the product of the evolutionary potential of traits and selection at the zooid level.
Unlike previous hierarchical expansions of Price’s theorem (26, 46–48), we do not assume that fitness at the colony level is a direct function of fitness at the zooid level. The standard assumption would be that average fitness of zooids is equal the average fitness of colonies. However, we know, from observed life-history patterns of Stylopoma and other bryozoans, that colony fitness is not a simple function of zooid fitness (49). As discussed in the context of the natural history of Stylopoma, the rate of production of ovicells in a colony is not related to the growth rate of the colony. Ovicells can be rare in fast-growing large colonies and common in small slow-growing colonies just as often as ovicells can be common in fast growing colonies (20, 50, 51).
Therefore, selection may occur at both the colony level and the zooid level as colonies beget colonies sexually and zooids beget zooids asexually. Given the importance of sexual and asexual modes of reproduction in these colonies, we should assume that selection is rampant at both levels. This fact brings the importance of evolutionary potential at each level into stark focus. What is the pattern of and cause of evolutionary potential at both the colony and the zooid level? If colony traits are variable and heritable, then they can respond to colony-level selection. Likewise, if zooid traits are variable and heritable, then they too have the potential to evolve by natural selection. There may be a conflict between these two levels of selection or they may be aligned.
Quantifying evolutionary potential in Stylopoma
As noted above, covariances can be calculated as the variance of a traits multiplied by the linear regression of traits. This even works in the case of a single trait, because its variance can be multiplied by the linear regression of the trait on itself. Because a linear regression of a trait with itself will always be equal to 1, the product of a variance value multiplied by 1 is equal to the variance value.
Heritability is of special interest because it is the product of the variance of a trait and its change over generations. This means that the heritability of traits is an efficient feature to investigate because the heritabilities of each trait automatically incorporate a measure of the variance of traits. Thus, we can understand the evolutionary potential of a trait by only looking at its heritability. This is true because there are two ways that heritability can be equal to zero. First, if there is no variation, then heritability will be equal to 0, and as a consequence, the equivalent element of the P matrix will also be equal to 0. The second way heritability will be equal to 0 is if the linear regression of parent and offspring phenotypes is equal to 0.
Measuring heritability
Figure 2 presents the heritability values for traits and pairwise combinations. Heritability is the phenotypic covariance between parent and offspring. Using the algebraic shortcut, we breakdown that covariance into a variance and a linear regression. So, for a single trait, the heritability (h) between parent (ϕp) and offspring (ϕo) is equal to
| () | 5 |
In Stylopoma, and all other bryozoans, clonal lineages are aligned in a linear chain. The distal end of a parent is where the offspring buds out and forms. At the growing margin of the colony, there will be many clonal lineages, each contributing a new generation of zooid. For our analysis, we compare phenotypes of zooids along individual clonal chains of parents and offspring.
For our heritability measure, we compare parents to offspring. As a consequence, the offspring in one generation will be the parent in the next. So, the phenotype of many individual zooids will be used twice in the calculation of heritability. For example, the width measurement of the zooid contributes to both PO1 and PO2, first as an offspring and second as a parent. Each comparison, PO(i,j), represents a coordinate on a scatter plot comparing parent to offspring phenotypes (fig. S1). It is from this scatter plot that the heritability is calculated. We then measure the heritability using the linear regression of parent on offspring phenotypes. In fig. S1, this linear component of heritability is shown by the solid regression line. Heritability for colony level traits (fig. S2) was calculated for parent-offspring pairs.
Supplementary Material
Acknowledgments
We thank J. Sanner and D. Erwin. This paper is dedicated to the memory of O. Coleman: “I don’t want them to follow me. I want them to follow themselves, but to be with me.” Funding: This work was funded by the Abbott and Springer Fellowships from the Department of Paleobiology, National Museum of Natural History, Smithsonian Institution. The original breeding experiments conducted in 1989 to 1990 were supported by the Smithsonian Institution Scholarly Studies Program and by the Smithsonian Tropical Research Institute. Author contributions: J.B.C.J. collected the specimens and designed the breeding experiment. A.H.-C. ran the breeding experiment. C.S. designed the analysis and analyzed the data. C.S. and J.B.C.J. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/2/eaaw9530/DC1
Supplementary Materials and Methods
Fig. S1. A small Stylopoma colony consisting of many thousands of members.
Fig. S2. A closeup of a Stylopoma colony showing a clonal lineage of autozooids.
Fig. S3. Polymorphic zooids of Stylopoma.
Fig. S4. A closeup of a Stylopoma colony consisting autozooids, three types of avicularia, and an ovicell.
Fig. S5. Evolutionary potential as measured by the heritability of traits between parent and offspring zooids.
Fig. S6. Evolutionary potential as measured by the heritability of traits between parent and offspring colonies.
Table S1. Zooid measurements of maternal Stylopoma colonies.
Table S2. Zooid measurements of offspring Stylopoma colonies.
REFERENCES AND NOTES
- 1.W. Beklemishev, Principles Of Comparative Anatomy Of Invertebrates 1: Promorphology (University of Chicago Press, 1969). [Google Scholar]
- 2.Wilson E. O., The origin and evolution of polymorphism in ants. Q. Rev. Biol. 28, 136–156 (1953). [DOI] [PubMed] [Google Scholar]
- 3.Wilson E. O., The ergonomics of caste in the social insects. Am. Nat. 102, 41–66 (1968). [Google Scholar]
- 4.T. J. M. Schopf, in Animal Colonies: Development and Function Through Time, R. S. Boardman, A. H. Cheetham, W. A. Oliver, Eds. (Dowden, Hutchinson, and Ross, 1973), pp. 247–294. [Google Scholar]
- 5.Harvell C. D., The evolution of polymorphism in colonial invertebrates and social insects. Q. Rev. Biol. 69, 155–185 (1994). [Google Scholar]
- 6.L. W. Buss, The Evolution of Individuality (Princeton Univ. Press, 1987). [Google Scholar]
- 7.J. Maynard Smith, E. Szathmáry, The Major Transitions In Evolution (Oxford Univ. Press, 1995). [Google Scholar]
- 8.R. E. Michod, Darwinian Dynamics: Evolutionary Transitions in Fitness and Individuality (Princeton Univ. Press, 1999). [Google Scholar]
- 9.C. Simpson, in The Major Transitions in Evolution Revisited, B. Calcott, K. Sterelney, Eds. (MIT Press, 2011), pp. 199–226. [Google Scholar]
- 10.Simpson C., Jackson J. B. C., Herrera-Cubilla A., Evolutionary determinants of morphological polymorphism in colonial animals. Am. Nat. 190, 17–28 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Wilson E. O., Hölldobler B., Eusociality: Origin and consequences. Proc. Natl. Acad. Sci. U.S.A. 102, 13367–13371 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson E. O., One giant leap: How insects achieved altruism and colonial life. Bioscience 58, 17–25 (2008). [Google Scholar]
- 13.P. D. Nieuwkoop, L. A. Sutasurya, Primordial Germ Cells in the Invertebrates: From Epigenesis to Preformation (Cambridge Univ. Press, 1981). [Google Scholar]
- 14.Buss L. W., Evolution, development, and the units of selection. Proc. Natl. Acad. Sci. U.S.A. 80, 1387–1391 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackstone N. W., Jasker B. D., Phylogenetic considerations of clonality, coloniality, and mode of germline development in animals. J. Exp. Zool. Mol. Dev. Evol. 297, 35–47 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Extavour C. G., Akam M., Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 130, 5869–5884 (2003). [DOI] [PubMed] [Google Scholar]
- 17.A. N. Ostrovsky, Evolution of Sexual Reproduction in Marine Invertebrates: Example of Gymnolaemate Bryozoans (Springer, 2013). [Google Scholar]
- 18.Church S. H., Siebert S., Bhattacharyya P., Dunn C. W., The histology of Nanomia bijuga (Hydrozoa: Siphonophora). J. Exp. Zool. B Mol. Dev. Evol. 324, 435–449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siebert S., Goetz F. E., Church S. H., Bhattacharyya P., Zapata F., Haddock S. H. D., Dunn C. W., Stem cells in Nanomia bijuga (Siphonophora), a colonial animal with localized growth zones. EvoDevo 6, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson C., The evolutionary history of division of labour. Proc. Biol. Sci. 279, 116–121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewontin R. C., The units of selection. Annu. Rev. Ecol. Syst. 1, 1–18 (1970). [Google Scholar]
- 22.Lande R., Arnold S. J., The measurement of selection on correlated characters. Evolution 37, 1210–1226 (1983). [DOI] [PubMed] [Google Scholar]
- 23.D. S. Falconer, T. F. C. Mackay, Introduction To Quantitative Genetics (Longmans Green, ed. 4, 1996). [Google Scholar]
- 24.M. Lynch, B. Walsh, Genetics And Analysis Of Quantitative Traits (Sinauer Sunderland, 1998), vol. 1. [Google Scholar]
- 25.S. H. Rice, Evolutionary Theory: Mathematical and Conceptual Foundations (Sinauer Associates, Sunderland, 2004). [Google Scholar]
- 26.S. Okasha, Evolution and the Levels of Selection (Oxford: Oxford Univ. Press, 2006). [Google Scholar]
- 27.P. Godfrey-Smith, Darwinian Populations And Natural Selection (Oxford Univ. Press, 2009). [Google Scholar]
- 28.Jackson J. B. C., Cheetham A. H., Evolutionary significance of morphospecies: A test with cheilostome bryozoa. Science 248, 579–583 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Cheetham A. H., Jackson J. B., Hayek L.-A. C., Quantitative genetics of bryozoan phenotypic evolution. I. Rate tests for random change versus selection in differentiation of living species. Evolution 47, 1526–1538 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Cheetham A. H., Jackson J. B., Hayek L.-A. C., Quantitative genetics of bryozoan phenotypic evolution. II. Analysis of selection and random change in fossil species using reconstructed genetic parameters. Evolution 48, 360–375 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Cheetham A. H., Jackson J. B., Hayek L.-A. C., Quantitative genetics of bryozoan phenotypic evolution. III. Phenotypic plasticity and the maintenance of genetic variation. Evolution 49, 290–296 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Jackson J. B., Cheetham A. H., Phylogeny reconstruction and the tempo of speciation in cheilostome Bryozoa. Paleobiology 20, 407–423 (1994). [Google Scholar]
- 33.Tilbrook K. J., Indo-West Pacific species of the genus Stylopoma Levinsen, 1909 (Bryozoa: Cheilostomea). Zool. J. Linn. Soc. 131, 1–34 (2001). [Google Scholar]
- 34.Herrera-Cubilla A., Jackson J. B., Phylogeny of genus Cupuladria (Bryozoa, Cheilostomata) in the Neogene of tropical America. J. Paleo. 88, 851–894 (2014). [Google Scholar]
- 35.Jackson J. B. C., Overgrowth competition between encrusting cheilostome ectoprocts in a Jamaican cryptic reef environment. J. Anim. Ecol. 48, 805–823 (1979). [Google Scholar]
- 36.J. B. C. Jackson, Morphological strategies of animals, in Biology and Systematics of Colonial Organisms, G. Larwood, B. R. Rosen, Eds. (Academic Press, 1979), pp. 499–555. [Google Scholar]
- 37.Jackson J. B. C., Winston J. E., Ecology of cryptic coral reef communities. I. Distribution and abundance of major groups of encrusting organisms. J. Exp. Mar. Biol. Ecol. 57, 135–147 (1982). [Google Scholar]
- 38.Palumbi S. R., Jackson J., Ecology of cryptic coral reef communities. II. Recovery from small disturbance events by encrusting bryozoa: The influence of “host” species and lesion size. J. Exp. Mar. Biol. Ecol. 64, 103–115 (1982). [Google Scholar]
- 39.F. K. McKinney, J. B. C. Jackson, Bryozoan Evolution (University of Chicago Press, 1991), pp. 243. [Google Scholar]
- 40.Queller D. C., Relatedness and the fraternal major transitions. Philos. Trans. R. Soc.Lond. B Biol. Sci. 355, 1647–1655 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbot P., Abe J., Alcock J., Alizon S., Alpedrinha J. A. C., Andersson M., Andre J.-B., van Baalen M., Balloux F., Balshine S., Barton N., Beukeboom L. W., Biernaskie J. M., Bilde T., Borgia G., Breed M., Brown S., Bshary R., Buckling A., Burley N. T., Burton-Chellew M. N., Cant M. A., Chapuisat M., Charnov E. L., Clutton-Brock T., Cockburn A., Cole B. J., Colegrave N., Cosmides L., Couzin I. D., Coyne J. A., Creel S., Crespi B., Curry R. L., Dall S. R. X., Day T., Dickinson J. L., Dugatkin L. A., Mouden C. El, Emlen S. T., Evans J., Ferriere R., Field J., Foitzik S., Foster K., Foster W. A., Fox C. W., Gadau J., Gandon S., Gardner A., Gardner M. G., Getty T., Goodisman M. A. D., Grafen A., Grosberg R., Grozinger C. M., Gouyon P.-H., Gwynne D., Harvey P. H., Hatchwell B. J., Heinze J., Helantera H., Helms K. R., Hill K., Jiricny N., Johnstone R. A., Kacelnik A., Kiers E. T., Kokko H., Komdeur J., Korb J., Kronauer D., Kümmerli R., Lehmann L., Linksvayer T. A., Lion S., Lyon B., Marshall J. A. R., McElreath R., Michalakis Y., Michod R. E., Mock D., Monnin T., Montgomerie R., Moore A. J., Mueller U. G., Noë R., Okasha S., Pamilo P., Parker G. A., Pedersen J. S., Pen I., Pfennig D., Queller D. C., Rankin D. J., Reece S. E., Reeve H. K., Reuter M., Roberts G., Robson S. K. A., Roze D., Rousset F., Rueppell O., Sachs J. L., Santorelli L., Schmid-Hempel P., Schwarz M. P., Scott-Phillips T., Shellmann-Sherman J., Sherman P. W., Shuker D. M., Smith J., Spagna J. C., Strassmann B., Suarez A. V., Sundström L., Taborsky M., Taylor P., Thompson G., Tooby J., Tsutsui N. D., Tsuji K., Turillazzi S., Úbeda F., Vargo E. L., Voelkl B., Wenseleers T., West S. A., West-Eberhard M. J., Westneat D. F., Wiernasz D. C., Wild G., Wrangham R., Young A. J., Zeh D. W., Zeh J. A., Zink A., Inclusive fitness theory and eusociality. Nature 471, E1–E4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.F. J. S. Maturo Jr., Offspring variation from known maternal stocks of Parasmittina nitida (Verrill), in Living and Fossil Bryozoa, G. P. Larwood, Eds. (Academic Press, 1973), pp. 577–584. [Google Scholar]
- 43.Simpson C., Species selection and driven mechanisms jointly generate a large-scale morphological trend in monobathrid crinoids. Paleobiology 36, 481–496 (2010). [Google Scholar]
- 44.Simpson C., Species selection and the macroevolution of coral coloniality and photosymbiosis. Evolution 67, 1607–1621 (2013). [DOI] [PubMed] [Google Scholar]
- 45.C. Simpson, The measurement of species selection on evolving characters (2017); bioRxiv 176438.
- 46.W. D. Hamilton, Innate social aptitudes of man: An approach form evolutionary genetics, in Biosocial Anthropology, R. Fox, Ed. (Wiley, 1975), pp. 133–155. [Google Scholar]
- 47.Price G. R., Extension of covariance selection mathematics. Ann. Hum. Genet. 35, 485–490 (1972). [DOI] [PubMed] [Google Scholar]
- 48.Maynard Smith J., Group selection. Q. Rev. Biol. 51, 277–283 (1976). [Google Scholar]
- 49.Herrera A., Jackson J., Hughes D., Jara J., Ramos H., Life-history variation in three coexisting cheilostome bryozoan species of the genus Stylopoma in Panamá. Mar. Biol. 126, 461–469 (1996). [Google Scholar]
- 50.J. B. C. Jackson, S. P. Wertheimer, Patterns of reproduction in five common species of Jamaican reef-associated bryozoans, in Bryozoa: Ordovician to Recent, C. Nielsen, G. P. Larwood, Eds. (Dredensborg, Denmark: Olsen and Olsen, 1985), pp. 161–168. [Google Scholar]
- 51.A. Herrera, J. Jackson, Life history variation among “dominant” encrusting cheilostomate Bryozoa, in Bryozoans In Space And Time, D. Gordon, A. Smith, J. Grant-Mackie, Eds. (NIWA, 1996), pp. 117–123. [Google Scholar]
- 52.Cheetham A. H., Sanner J., Taylor P. D., Ostrovsky A. N., Morphological differentiation of avicularia and the proliferation of species in mid-Cretaceous Wilbertopora Cheetham, 1954 (Bryozoa: Cheilostomata). J. Paleo. 80, 49–71 (2006). [Google Scholar]
- 53.W. C. Banta, in Degrees of colony dominance in stenolaemate and gymnolaemate Bryozoa, R. S. Boardman, A. H. Cheetham, W. A. Oliver Jr., Eds. (Dowden, Hutchinson, and Ross, 1973), pp. 295–303. [Google Scholar]
- 54.R. S. Boardman, A. H. Cheetham, W. A. Oliver Jr., Animal colonies: Development and Function Through Time (Dowden, Hutchinson, and Ross, 1973). [Google Scholar]
- 55.L. W. Buss, Hebitat salection, directional growth and spatial refuges: Why colonial animals have more hiding places, in Biology And Systematics Of Colonial Organisms, G. Larwood, B. R. Rosen, Eds. (Academic Press, 1979), pp. 459–497. [Google Scholar]
- 56.Carter M., Gordon D., Gardner J. P. A., Polymorphism and vestigiality: Comparative anatomy and morphology of bryozoan avicularia. Zoomorphology 129, 195–211 (2010). [Google Scholar]
- 57.Hughes D. J., Jackson J. B. C., Do constant environments promote complexity of form?: The distribution of bryozoan polymorphism as a test of hypotheses. Evolution 44, 889–905 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Hughes T. P., Jackson J. B. C., Population dynamics and life histories of foliaceous corals. Ecol. Monogr., 141–166 (1985). [Google Scholar]
- 59.Jackson J. B. C., Competition on marine hard substrata: The adaptive significance of solitary and colonial strategies. Am. Nat. 111, 743–767 (1977). [Google Scholar]
- 60.Jackson J. B. C., Ecology of cryptic coral reef communities. III. Abundance and aggregation of encrusting organisms with particular reference to cheilostome Bryozoa. J. Exp. Mar. Biol. Ecol. 75, 37–57 (1984). [Google Scholar]
- 61.J. B. C. Jackson, L. W. Buss, R. E. Cook, Population Biology And Evolution Of Clonal Organisms (Yale Univ. Press London, 1985). [Google Scholar]
- 62.J. B. C Jackson, A. Herrera-Cubilla, 2000. Adaptation and constraint as determinants of zooid and ovicell size among encrusting ascophoran cheilostome Bryozoa from opposite sides of the Isthmus of Panama, in Proceedings of the International Bryozoology Association Conference, Republic of Panama, A. Herrera-Cubilla, J. B. C. Jackson, Eds. (Smithsonian Tropical Research Institute), 26 to 31 January 1998, pp. 249–258. [Google Scholar]
- 63.J. S. Ryland, Bryozoans (Hutchinson & Co, 1970), pp. 175. [Google Scholar]
- 64.L. Silén, Polymorphism in marine bryozoans, in Biology of Bryozoans, R. Woollacott, R. Zimmer, Eds. (Academic Press, 1977), pp. 184–232. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/2/eaaw9530/DC1
Supplementary Materials and Methods
Fig. S1. A small Stylopoma colony consisting of many thousands of members.
Fig. S2. A closeup of a Stylopoma colony showing a clonal lineage of autozooids.
Fig. S3. Polymorphic zooids of Stylopoma.
Fig. S4. A closeup of a Stylopoma colony consisting autozooids, three types of avicularia, and an ovicell.
Fig. S5. Evolutionary potential as measured by the heritability of traits between parent and offspring zooids.
Fig. S6. Evolutionary potential as measured by the heritability of traits between parent and offspring colonies.
Table S1. Zooid measurements of maternal Stylopoma colonies.
Table S2. Zooid measurements of offspring Stylopoma colonies.


