Abstract
We present an easy-to-use integrated software suite, DIA-NN, that exploits deep neural networks and new quantification and signal correction strategies for the processing of data-independent acquisition (DIA) proteomics experiments. DIA-NN improves the identification and quantification performance in conventional DIA proteomic applications, and is particularly beneficial for high-throughput applications, as it is fast and enables deep and confident proteome coverage when employed in combination with fast chromatographic methods.
Proteomics provides the functional links between the genome and metabolome of a cell, and is rapidly gaining importance within both personalised medicine and the emerging field of data-driven biology1–4. The generation of data is hampered, however, by the inherent complexity of the proteome. In mass spectrometry-based (bottom-up) proteomics, this complexity leads to stochasticity in peptide detection, reducing the proteomic sampling depth5,6. A popular solution to these issues is to decrease sample complexity by proteome or peptidome pre-fractionation. Extensive fractionation promotes excellent proteome coverage, but at the cost of the time and resources as well as the introduction of added variability between samples.
Alternatively, data-independent acquisition (DIA) approaches, such as SWATH-MS7,8, have been developed to reduce stochastic elements. In DIA, rather than selecting the most abundant precursor ions (i.e. peptides bearing a specific charge) for further analysis in a data-dependent manner, the mass spectrometer is configured to cycle through a predefined set of precursor isolation windows, thus consistently fragmenting all the precursors within the mass range of interest (reviewed by Ludwig et al9). DIA workflows show high reproducibility, and recent developments have demonstrated that they can achieve higher proteomic depth in single injections, compared to conventional data-dependent proteomic methods, at least when recorded on Orbitrap-type mass spectrometers10–12.
The computational processing of DIA datasets, however, remains challenging due to their inherent complexity. A key difficulty originates from the fact that in DIA each precursor gives rise not just to a single spectrum, but to a set of chromatograms corresponding to numerous fragment ions generated by collision-induced dissociation. In addition, these chromatograms are often highly multiplexed due to interferences from co-fragmenting precursors. These interferences are further amplified in situations where short chromatographic gradients are coupled with complex samples, limiting the application of DIA-MS in high-throughput workflows. Given that increased sample throughput reduces batch effects and speeds up research, thus enabling studies on large cohorts of samples, it is imperative that such challenges are overcome.
To meet these challenges, we have developed DIA-NN, a software suite to process highly complex DIA data. DIA-NN utilizes deep neural networks to distinguish real signals from noise, as well as new quantification and interference correction strategies. The DIA-NN pipeline is fully automated (Fig. 1A; all procedures are described in detail in Methods) and includes both an intuitive graphical interface as well as a command line tool, with results being reported in a simple text format. DIA-NN does not require the presence of retention time standards, instead performing retention time alignment using endogenous peptides. DIA-NN also performs automatic mass correction and automatically determines such search parameters as the retention time window and the extraction mass accuracy. This eliminates the lengthy and laborious process of optimising the processing workflow for each particular data set.
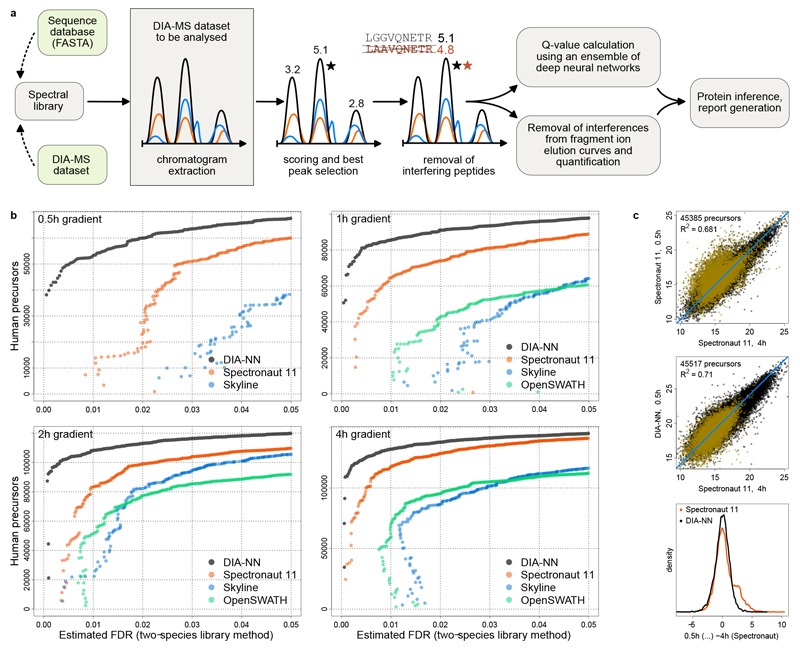
Fig. 1. DIA-NN workflow and its performance on conventional and short chromatographic gradients.
a, Schematic: DIA-NN workflow. Chromatograms are extracted for each precursor ion and all its fragment ions (the chromatograms are shown schematically, with different colours corresponding to different fragments). Putative elution peaks are then scored, and the ‘best’ peak (marked with a star) is selected. Potentially interfering peptides are then detected and removed. The precursor-peak matches obtained allow to calculate q-values using an ensemble of deep neural networks as well as remove interferences from the fragment elution curves. b, Identification performance of DIA-NN when processing technical repeat injections of a HeLa tryptic digest analysis (QExactive HF, 0.5h - 4h gradient lengths12). Precursor identification numbers are plotted against the FDR, estimated using a two-species compound human-maize spectral library method12 (Methods). Each point on the graph corresponds to a decoy (maize) precursor, its x-axis value reflecting the estimated FDR at the respective score threshold and its y-axis value being the number of identified target (human) precursors at this threshold. The 0.5h acquisition was not analysed with OpenSWATH for technical reasons. c, Log2-quantities of precursors reported for both the 0.5h acquisition – among top 50000 by Spectronaut (top panel) or DIA-NN (middle panel), and the 4h acquisition (among top 100000 by Spectronaut). R2 values were calculated using linear regression with unity slope. Precursors identified exclusively by either Spectronaut (8379 total) or DIA-NN (8511 total) at 0.5h (i.e. those precursors, identifications of which are not supported by the other tool at the same gradient) are highlighted in yellow. For these, the distribution densities of the differences (centered) between the 0.5h log2-quantities reported by Spectronaut or DIA-NN and 4h log2-quantities reported by Spectronaut (bottom panel) were plotted.
The DIA-NN workflow starts with a peptide-centric approach13, based on a collection of precursor ions (with several fragment ions from each precursor annotated), which can be provided separately (in a spectral library) or automatically generated by DIA-NN in silico from a protein sequence database (library-free mode). DIA-NN then generates a library of negative controls (i.e. decoy precursors13,14), extracts a chromatogram for each target or decoy precursor and identifies putative elution peaks, comprised of the precursor and fragment ion elution profiles in the vicinity of the putative retention time of the precursor. Each of the elution peaks is then described by a set of scores that reflect peak characteristics, including co-elution of fragment ions, mass accuracy or similarity between observed and reference (library) spectra. In total, DIA-NN calculates 73 peak scores in the various steps of the workflow (see Supplementary Note 1 for details of the scoring system). The best candidate peak is then selected per precursor using iterative training of a linear classifier, which allows to calculate a single discriminant score for each peak.
While being highly sensitive15, this peptide-centric search alone would lead to false identifications and unreliable quantification, as a single putative elution peak in the data could be used as detection evidence for several precursors that share one or more fragments with close m/z values. One solution to this problem is to draw upon the advantages of spectrum-centric approaches, wherein for each spectrum the single best matching precursor is selected16. DIA-NN evaluates the degree of interference between multiple precursors initially matched to the same retention time, and, if it is deemed significant enough, only reports the ones best supported by the data as identified, improving the identification performance at strict FDR thresholds (see Supplementary Note 2 for a benchmark).
A key step in DIA-MS workflows is to assign statistical significance to the identified precursors, typically in the form of precursor q-values. To calculate the q-values, all target and decoy precursors need to be assigned a single discriminant score each, based on the characteristics of the respective candidate elution peaks. For this step, DIA-NN relies on deep neural networks (DNNs). DNNs encompass a group of artificial intelligence methods, that have been developed extensively towards the analysis of complex data of heterogeneous nature17. In DIA-NN, an ensemble of feed-forward fully-connected DNNs (with 5 tanh-activated hidden layers and a softmax output layer) is trained for one epoch to distinguish between the target and decoy precursors using cross-entropy as the loss function. For each precursor, the set of scores corresponding to the respective elution peak is used as neural network input. Subsequently, each trained network, when provided with a set of scores as input, yields a quantity that reflects the likelihood that this set originated from a target precursor. These quantities, calculated for all the precursors and averaged across the networks, are then used as discriminant scores to obtain the q-values (see Methods for details on the DNNs implementation; we demonstrate that DNNs enable comprehensive proteome coverage at strict FDR thresholds, see Supplementary Note 2 for a benchmark).
Additionally, DIA-NN includes an algorithm for detection and removal of interferences from tandem-MS spectra. For each putative elution peak, DIA-NN selects the fragment least affected by interference (as the one with the elution profile best correlated with the elution profiles of the other fragments). Its elution profile is then considered representative of the true elution profile of the peptide. Comparison of this profile with the elution profiles of other fragments allows to subtract interferences from the latter, improving the quantification accuracy (see Methods for the implementation details and Supplementary Note 3 for a benchmark).
We benchmarked DIA-NN using public datasets that have been specifically created for testing DIA software. First, we evaluated the identification performance using a HeLa whole-proteome tryptic digest recorded on a nanoflow LC-coupled QExactive HF mass spectrometer (Thermo Fisher) with different chromatographic gradient lengths, ranging from 0.5h to 4h12. The same data were processed with OpenSWATH18, Skyline19 and Spectronaut Pulsar5 (Biognosys), that were tuned extensively to achieve the optimal performance (Methods). A key issue in proteomic benchmarks is that each software tool calculates the false discovery rate (FDR) in a different way, so that the reported peptide numbers cannot be directly compared. Indeed, it was demonstrated recently, that even a simple change to a decoy precursor generation algorithm can halve or double the internal FDR estimates reported by an analysis tool12. We therefore estimated the effective FDR using the unbiased two-species spectral library method12, wherein (non-redundant) maize precursors assigned within a human sample are counted as false positives (Methods). DIA-NN achieves substantially better identification performance compared to the other tools tested (Fig. 1B). The biggest differences are observed at strict FDR thresholds (Fig. 1B). Similar improvements were observed when analysing a K562 human cell line whole-cell tryptic digest measured on a microflow LC-coupled TripleTOF 6600 mass spectrometer (Sciex) using a fast 19 min chromatographic gradient. Even with an Orbitrap-based spectral library, DIA-NN consistently identified over 35000 precursors (Supplementary Note 2), more than what was achieved only a few years ago with 2h nanoflow gradients on Orbitrap instruments5.
Importantly, DIA-NN enables confident identification and deep proteome coverage with short chromatographic gradients. DIA-NN identified more precursors from the 0.5h acquisition than either Skyline or OpenSWATH from the same sample analysed using a 1h chromatographic gradient (Fig. 1B), at all FDR thresholds considered, as well as more than either of these tools obtains from the 2h acquisition at 1% FDR and the 4h acquisition at 0.5% FDR.
To validate the identifications DIA-NN achieved at the fast gradient, we compared its analysis to that at longer gradients. Out of the top 50000 precursors reported by DIA-NN at 0.5h, 49694 are confirmed by Spectronaut at either 1h, 2h or 4h (at 1% FDR as calculated by Spectronaut). Furthermore, the quantities produced by DIA-NN at 0.5h are more similar to the quantities generated by Spectronaut from the low-interference 4h acquisition, than the quantities Spectronaut itself extracts at 0.5h, likely due to a higher number of misidentifications for Spectronaut at 0.5h (Fig. 1C). The latter are the probable cause of the clearly visible admixture peak (at ~2.5) in the density plot for differences between the 0.5h and 4h log2-quantities (Spectronaut; Fig. 1C, bottom panel).
We also benchmarked DIA-NN against the Specter software20, but as this tool has a fixed FDR cut-off, we used a different benchmark. DIA-NN also showed better performance (Supplementary Note 4).
We demonstrated the robustness of DIA-NN on large-scale data using 364 yeast acquisitions covering the deletion of non-essential kinases4 (Supplementary Note 5). Protein identification was robust over this large dataset. DIA-NN processed the dataset in just ~3h on a conventional processing PC (Supplementary Note 6).
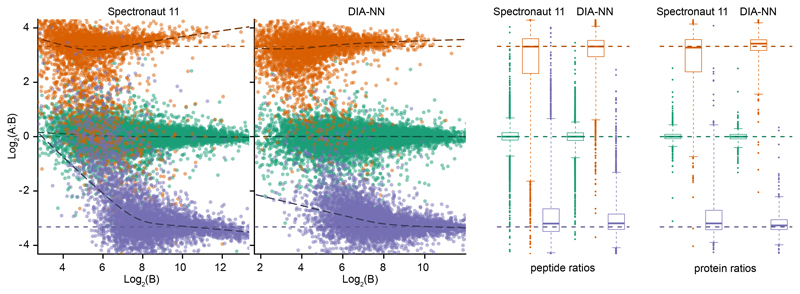
Finally, we used the LFQbench dataset, specifically designed to compare DIA software tools21, to assess quantification precision on the basis of how well known ratios between three species lysates (human, yeast and E.coli) are recovered by the software tool. In comparison to Spectronaut, DIA-NN demonstrated better quantification precision for both yeast and E.coli peptides and proteins (Fig. 2, Supplementary Note 7). In addition, DIA-NN improved median CV values for human peptides and proteins: 5.6% and 3.0%, respectively, compared to 7.0% and 3.8% for Spectronaut (see Supp. Table 2.H, Supp. Figures 7.H and 8.H in the original LFQbench manuscript21 for benchmarks of other software tools on the same dataset). Additionally, we benchmarked DIA-NN against Spectronaut on the LFQbench dataset in library-free mode (Supplementary Note 8). Also here, DIA-NN quantified more human peptides and proteins and with better precision.
Fig. 2. LFQbench test performance of DIA-NN.
Quantification precision was benchmarked using two peptide preparations (yeast and E.coli) that were spiked in two different proportions (A and B, three repeat injections each) into a human peptide preparation21. The data were processed at 1% q-value (reported by the software tools themselves, i.e. the effective FDR for DIA-NN and Spectronaut may be different) using a spectral library generated from a fractionated sample analysis with DDA21. Peptide ratios between the mixtures were visualised using the LFQbench R package (left panel; the dashed lines indicate the expected ratios). Right panel: peptide and protein quantification performance given as box-plots (boxes: interquartile range, whiskers: 1-99 percentile; n = 15442 and 15743 (human), 3403 and 3755 (yeast), 4494 and 4997 (E.coli) for peptide ratios obtained from the reports of Spectronaut and DIA-NN, respectively; n = 1921 and 1950 (human), 529 and 550 (yeast), 566 and 616 (E.coli) for protein ratios obtained from the reports of Spectronaut and DIA-NN, respectively).
In summary, the computational methods introduced in DIA-NN consistently and significantly increase the numbers of identified and precisely quantified precursors and proteins, in the analysis of samples of varied complexity and acquired on different mass spectrometry platforms. Enabling for the first time comprehensive proteome coverage using fast chromatographic gradients, DIA-NN allows for significant reduction of mass spectrometer running times, opening the door for previously inaccessible proteomic experiments that require fast and precise measurements of large numbers of proteomes.
Methods
DIA-NN algorithms
DIA-NN can read mzML files as well as directly import the raw data from Sciex and Thermo acquisitions. To process them, DIA-NN requires either a spectral library or a sequence database to be provided as input. In the latter case, DIA-NN generates a spectral library in silico. For this, DIA-NN can optionally use a fragmentation predictor (based on the approach introduced in MS Simulator22) and a linear retention time predictor. The predictors are trained using any spectral library supplied by the user.
For each target precursor in the spectral library, a decoy precursor is generated, if not provided in the library. By default, this is done by replacing the fragment ion m/z values of the target precursor assuming the amino acids adjacent to the peptide termini were mutated (GAVLIFMPWSCTYHKRQEND to LLLVVLLLLTSSSSLLNDQE mutation pattern is used). Optional pseudo-reverse approach to decoy precursor generation is also supported.
Chromatograms are then extracted for each target and decoy precursor and the respective fragment ions. Potential elution peaks are identified, and for each of these the fragment with the most optimal properties for quantification is selected. This fragment (chosen among the top six based on the reference intensities in the library) maximizes the sum of the Pearson correlations between its elution profile (in the vicinity of the putative peak apex) and the elution profiles of the remaining fragments from the top six list. It is assumed, that this “best” fragment is likely to be the one least affected by interferences, its elution profile thus being representative of the true elution profile of the peptide. A set of 73 scores is calculated for each potential elution peak (Supplementary Note 1). These are used differentially in different processing stages based on algorithmic decision making. The “best” candidate peak is selected per precursor using one of the scores, and a linear classifier is trained to distinguish between target and decoy precursors based on the sets of scores corresponding to the respective best peaks, allowing to calculate a single discriminant score for each peak. These discriminant scores are then used to refine the selection of best peaks, and the procedure is repeated iteratively several times.
Rather than rely on a single linear classifier, DIA-NN can dynamically switch between linear discriminant analysis (LDA; conventionally used in the DIA software that relies on the mProphet algorithm13, e.g. Skyline of Spectronaut) and a custom linear classifier, based on which yields a higher number of identifications. Briefly, the discriminant score is calculated as the weighted sum ∑ wKsK, where s = {s1,…,s73} is the set of scores characterizing the “best” candidate peak corresponding to the precursor. The set of weights w = {w1,…,w73} is obtained as the solution of the equation Rw = b, where b is the vector of average score differences between the target and decoy precursors (i.e. for each target precursor and the respective decoy precursor, the score difference sTARGET – sDECOY is calculated, and b is defined as the average of these differences), and R is either the average of the two covariance matrices that correspond to target and decoy precursor scores (LDA), or the covariance matrix of the score differences between target and decoy precursors (custom linear classifier). Notably, DIA-NN does not restrict the set of precursors used to train the classifier only to those that have been identified with high confidence, as this was found to have a negative effect on the performance.
During the next step, DIA-NN looks for precursors matched to the same retention time which also have interfering fragments. If the degree of interference is deemed significant enough, DIA-NN only reports the precursor with the highest discriminant score as identified. This method effectively allows to combine the advantages of peptide-centric and spectrum-centric approaches to mass-spectrometry data analysis.
An ensemble of deep feed-forward fully connected neural networks (12 by default) is trained (as implemented in the Cranium deep neural network library (available from https://github.com/100/Cranium), supplied with the DIA-NN distribution) via Adam23 (which we integrated in the Cranium code) to distinguish between target and decoy precursors. The Cranium library is written in C and demonstrates very high speed, which led us to choose it among the many alternatives. For each precursor, the set of scores corresponding to the respective best elution peak is provided as input for the networks. Each network comprises a series of tanh hidden layers (5 by default, with i-th hidden layer featuring 5(6 - i) neurons, i = 1…5) and a softmax output layer. Cross-entropy is used as the loss function. The peak scores (73 total) are standardized before training.
Briefly, each neural network is essentially a function, which depends on a number of parameters (connection weights) and outputs a value between 0 and 1 (that is interpreted as the predicted likelihood that the respective precursor is a target precursor, and not a decoy precursor), when provided with a set of peak scores. If the connection weights are altered, the output value also changes, for the same set of peak scores. Thus, the weights can be tuned to make the network a better predictor, i.e. to better distinguish between targets and decoys. This is achieved by sequentially supplying the network with sets of peak scores and adjusting the weights using a gradient descent-type optimization algorithm. Each time, the weights are changed slightly, based on the calculated discrepancy between the prediction produced by the network with the current set of weights and the “ideal’ prediction (the latter being 1 or 0, for a target or decoy precursor, respectively). This change is aimed at reducing the discrepancy. Each training epoch involves looping through all the samples in the dataset. Training using multiple epochs sometimes allows to achieve better prediction accuracy, but can also lead to overfitting.
By default, in DIA-NN training is performed for one epoch only, minimizing the effects of overfitting. The predictions of the neural networks are then averaged for each precursor, resulting in the final set of scores used for q-value calculation. This step, known as ensemble learning, further reduces the effects of potential overfitting. Optionally, DIA-NN can train each network on a part of the dataset, only using it to score precursors it has not been trained on, or use a higher number of training epochs. The use of neural networks allows to effectively utilize all the 73 scores calculated for each elution peak, thus increasing the amount of information extracted from the data in comparison to the use of a linear classifier.
Of note, we experimented with different neural network architectures. e.g. varied the number of hidden layers and the number of neurons in each layer. In general, we observed that changing the complexity of the network in this way had only minimal effect on the performance. We also experimented with adding e.g. dropout or batch normalization layers (using a different neural network library), however this did not result in any significant gains.
For a particular score threshold, DIA-NN calculates a conservative FDR estimate (used to generate the respective q-values24 for precursor identifications), dividing the number of decoys with scores exceeding the threshold by the number of targets with scores exceeding the threshold. Correction based on estimating the prior probability of incorrect identification (π0) is not performed.
DIA-NN uses a conservative protein q-value calculation method, which is applied to individual proteins and not protein groups. To estimate protein-level FDR, only target and decoy precursors specific to the protein of interest are considered. Thus, proteins without any proteotypic precursors identified are automatically assigned a q-value equal to one. The maxima of target and decoy scores are calculated for each protein and the distributions of these are examined. For a given score threshold, FDR is estimated by dividing the number of decoy scores exceeding it by the number of target scores exceeding it.
For each run, DIA-NN quantifies the intensities of all fragment ions associated with each precursor. For this, we have conceived an efficient interference removal algorithm. Importantly, this algorithm does not rely on the accuracy of the reference fragment intensities provided in the spectral library; its performance is thus independent of the quality of the spectral library and its suitability for the specific LC-MS setup. As aforementioned, for each putative elution peak of the precursor, DIA-NN designates one of the fragment ions (among the top six annotated in the library) as the “best”. The interference removal algorithm assumes that the elution profile best(⋅) of this fragment is representative of the true elution profile of the peptide. The elution profile x(⋅) of each fragment is then compared to best(⋅). If x(⋅) has been affected by some interfering peptide (for example, suppose that x(⋅) is the “green” (dashed) elution profile as illustrated in Supplementary Note 3; the best fragment is the one corresponding to the “blue” elution profile), and the elution apex of that peptide (RTIFS) is not the same as the elution apex of the target peptide (RTTARGET) (e.g. RTIFS > RTTARGET in Supplementary Note 3, where RTIFS = 93.1981, RTTARGET = 93.0784), then the ratio x(RTIFS)/best(RTIFS) would be higher than x(RTTARGET)/best(RTTARGET). The idea behind the interference removal in DIA-NN is to detect it when the value of x(RTIFS)/best(RTIFS) is too high, and “correct” the respective values of x(⋅). First, best(⋅) is smoothed to produce the “reference” elution profile ref(⋅). For each fragment, the "weighted" fragment intensity is then calculated as the sum of the fragment elution profile values weighted by the respective squared values of ref(⋅). This emphasizes the contribution of the data points close to the apex of the reference elution profile, thus making the impact of potential interferences manifesting far from the apex negligible. The ratio r of weighted intensities of the fragment under consideration and the best fragment is calculated. If the correlation of x(⋅) and ref(⋅) is below 0.8, however, i.e. the impact of interferences on x(⋅) is likely to be significant, the value of r is replaced by the minimum of x(⋅)/best(⋅) in the vicinity of the peak apex (this completely eliminates the contribution of the data points away from the apex). Interferences are then removed by all values of x(⋅) exceeding 1.5⋅r⋅ref(⋅) being replaced with 1.5⋅r⋅ref(⋅). The area under the resulting profile is then considered to be the intensity of the fragment. Preliminary precursor quantities (before cross-run selection of fragments for quantification, see below) are obtained by summing the intensities of the top six fragments (ranked by their library intensities).
DIA-NN enables cross-run precursor ion quantification. In each acquisition, each fragment is assigned a score which is the correlation score of its elution profile with the respective reference profile, i.e. the smoothed elution profile of the best fragment. For each precursor, three fragments with highest average correlations are selected in a cross-run manner. Only acquisitions where the precursor was identified with a q-value below a given threshold are considered. The intensities of these fragments are then summed in each acquisition to obtain the precursor ion intensity. This approach allows to deal with the situation when in certain acquisitions interferences were not efficiently removed from elution profiles of some fragments, e.g. if interferences manifested close to the apexes.
Protein grouping can be performed either for individual acquisitions or in a cross-run fashion (default). For each precursor, DIA-NN aims to reduce the number of proteins associated with it using the maximum parsimony principle, which is implemented via a greedy set cover algorithm.
After precursor ion quantification, cross-run normalization and protein quantification are performed. All the precursor intensities corresponding to identifications with q-values above a given threshold are replaced with zeros and preliminary cross-run normalization based on the total signal (i.e. the sum of the intensities of all precursors) is performed. Precursors are then ordered by their coefficients of variation. Top pN precursors are selected, where N is the average number of identifications passing the q-value threshold and p is between 0 and 1. Sums of the intensities of these precursors are calculated and are used for normalization, i.e. the levels of all precursors are scaled to make these quantities equal in different acquisitions. A "Top 3" method is eventually used for protein quantification: intensities of protein groups are calculated as sums of the intensities of top 3 most abundant precursors identified with a q-value lower than a given threshold in a particular acquisition.
When generating a spectral library from multiple DIA acquisitions, DIA-NN scores all target and decoy precursors using the minimum q-value across all the acquisitions as the score. These scores are then used to calculate “cross-run” q-values, which allow to filter the output at the specified confidence level. For each target precursor that passes the filtering threshold (set by the user), an acquisition with the lowest q-value (i.e. the minimum one across all the acquisitions) is identified, and chromatograms in the vicinity of the previously determined retention time are extracted. Fragments (y-series and b-series) with charges up to 2 as well as with neutral losses of H2O and NH3 are considered. Correlations with the elution curve of the best fragment are calculated for elution curves of all these fragments. For fragments with the respective correlations exceeding a particular threshold (which depends on the type of the fragment, e.g. it’s higher for b-series fragments than y-series), intensities are determined at the precursor elution peak apex retention time (which has been identified previously) and are saved to the newly generated library.
Benchmark, software versions
DIA-NN (1.6.0) was compared to OpenSWATH18 (part of OpenMS25 2.4.0), Skyline19 (4.1.0.11796) and Spectronaut5 (Pulsar 11.0.15038.17.27438 (Asimov) (Biognosys)).
Previously published mass spectrometry data
Raw analyses of the HeLa cell lysate have been described previously12 and were obtained from ProteomeXchange (data set PXD005573). DIA-NN and Spectronaut accessed these directly; for processing with Skyline and OpenSWATH, .raw files were converted to the mzML format using MSConvertGUI (part of ProteoWizard26 3.0.11537) with MS1 and MS2 vendor peak picking enabled, 32-bit binary precision and all other options unchecked. Raw data files for the LFQbench test were generated by Navarro and colleagues21 and were obtained from ProteomeXchange (data set PXD002952; “HYE110” acquisitions featuring 10:1 yeast and 1:10 E.coli spike-ins A:B ratios, recorded on TripleTOF 6600 with 64-variable windows acquisition). These were directly accessed by DIA-NN and Spectronaut. Acquisitions used for comparison with Specter were obtained from ProteomeXchange (data set PXD006722). The yeast kinase deletions acquisitions are available from ProteomeXchange (data set PXD010529).
Two-species spectral library FDR estimation method
A key problem in proteomic benchmarks is that each software tool calculates the false discovery rate (FDR) in its own way, thus making it impossible to directly compare the identification numbers reported by different tools at a particular FDR threshold. Indeed, it was demonstrated recently, that even a simple change to a decoy precursor generation algorithm can halve or double the internal FDR estimates reported by an analysis tool12. The effective FDR was therefore estimated using an unbiased two-species spectral library method as introduced by Bruderer et al12, a strategy that has recently been adopted also in other benchmarks20. A spectral library created for the target organism (in this case, human) is augmented with spectra from peptides belonging to another organism (in this case, maize), not expected to be found in the sample (extensive filtering against the proteome of the target organism is performed to remove all peptides potentially originating from both organisms). Calls of these extra peptides are then considered false positive, allowing to estimate the effective FDR, and to estimate identification numbers that can be robustly compared irrespective of the differences introduced by the software-integrated FDR estimation methods.
Generation of the two-species spectral library
The human and maize spectral libraries (project specific, obtained via fractionated sample analysis using data-dependent acquisition LC-MS/MS) used to generate the two-species compound library have been described previously12. The maize library was filtered to exclude peptides matched to either the NCBI human redundant database (April 25th, 2018) or the UniProt27 human canonical proteome (3AUP000005640). The human library was filtered to include only peptides matched to the latter. In both cases, filtering was performed with leucine and isoleucine treated as the same amino acid. The libraries were merged, resulting in a library containing only precursor ions matched to either human or maize proteomes, but not both. To enable the use of the library by all of the software tools under consideration, the library was converted to the OpenMS-compatible format with the use of DIA-NN. Following the protocol of Navarro and co-workers21, only precursor ions associated with at least six fragment ions were retained in the library, and all fragments but the top six (ordered by their reference intensities) were discarded. This was done to ensure that there is no bias in terms of the distribution of the number of annotated fragments between human and maize precursors. In addition, although DIA-NN can take advantage of large numbers of fragment ions described in the spectral library, many software tools tend to perform poorly if the number of fragment ions is not restricted, e.g. Spectronaut and Skyline only use the top six fragments by default. Reference retention times (Biognosys iRT scale) below -60.0 were adjusted to -60.0, to enable efficient linear retention time prediction by Skyline and OpenSWATH, as the respective precursors were observed to elute concomitantly. A low number of precursor ions had to be removed from the spectral library, so that the library could be imported error-free into Skyline (Supplementary Note 9). The resulting compound spectral library contained 202310 human precursor ions and 9781 maize precursor ions.
FDR estimation using the two-species spectral library
The HeLa cell lysate proteomic datasets (Fig. 1B) were analysed with each software tool using the human-maize compound spectral library described above. For each identified maize precursor, its score (that was ultimately used to calculate the q-value) was considered. The numbers of human and maize precursors identified with the same or better score were then calculated ([human IDs] and [maize IDs], respectively). A conservative FDR estimate was then obtained:
Here [human total] and [maize total] are the respective numbers of human and maize precursors in the spectral library.
Configuring DIA-NN and Spectronaut
The default settings were used for DIA-NN and Spectronaut, except that protein inference and FDR filtering of the output were turned off to obtain complete reports.
Configuring Skyline and OpenSWATH
The spectral library was directly imported into Skyline using the 0.05 m/z ion match tolerance. Shuffle decoy generation was used. For the use with OpenSWATH, the spectral library was converted (using OpenMS 2.3.0) to the TraML format, decoys were generated using the following options: “-append -exclude_similar -remove_unannotated -enable_detection_specific_losses -enable_detection_unspecific_losses -force”. The spectral library was then converted back to the .tsv format.
We extensively optimised the parameters of OpenSWATH and Skyline, to maximise the number of precursor ion identifications reported by the tools themselves at 1% q-value. We tried different combinations of mass accuracy and retention time window settings, and acted upon the assumption that longer-gradient acquisitions are best processed with stricter mass accuracy settings and a wider retention time window. Mass accuracy settings attempted for Skyline were: 5 ppm, 7 ppm and 10ppm for 0.5h, 7ppm for 1h, 5ppm and 7ppm for 2h, 3ppm, 5ppm and 7ppm for 4h acquisitions. Mass accuracy settings attempted for OpenSWATH were: 15ppm (i.e. 30ppm extraction window) and 20ppm for 1h, 10ppm and 15ppm for 2h, 5ppm, 7ppm, 10ppm for 4h. Eventually, 7ppm for 0.5h, 1h and 2h, 5ppm for 4h were chosen for Skyline, 15ppm for 1h and 2h, 7ppm for 4h – for OpenSWATH. OpenSWATH was not used to analyse the 0.5h acquisition, as it was unable to correctly recognise sufficient number of iRT (Biognosys) or CiRT28 retention time standards in the short gradient. The use of a retention time window speeds up search in Skyline and OpenSWATH, but might potentially impair performance, if too strict a window is chosen. Starting from the recommended values for 2h acquisitions21, namely 20 min for Skyline (i.e. using scans within 10 min from the predicted RT) and 10 min for OpenSWATH, we were able to somewhat improve performance of both of these tools by increasing the retention time window. Eventually, with Skyline we used a 40 minute window for the 2h and 4h acquisitions and 20 minute for 0.5h and 1h, for OpenSWATH we used 80 minute window for the 4h, 60 minute window for the 2h and 20 minute for the 1h acquisition. As the acquisitions in question were obtained using a segmented gradient, rather than a purely linear gradient, we tried different strategies of RT normalisation with OpenSWATH (Skyline supports only linear RT normalisation): linear (with both iRT (in iRTassays.TraML file downloaded from the PeptideAtlas29 repository with the identifier PASS00779) and CiRT peptides), as well as lowess (with CiRT peptides). For the use with CiRT peptides, -RTNormalization:estimateBestPeptides option was enabled, as recommended for OpenSWATH. CiRT peptides proved to be better than iRT peptides for all gradients, with lowess normalisation being beneficial for the 2h and 4h gradients. Skyline was run with the acquisition method set to DIA, product mass analyzer set to centroided and isolation scheme set to “Results (0.5 margin)”. We also attempted running Skyline with product mass analyzer set to Orbitrap (i.e. using profile data, 4h gradient), without converting the .raw files to centroided .mzML, but this resulted in a significantly lower number of identified precursors. Including MS1 isotope peaks in Skyline was also detrimental, so this feature was not used (in line with the default settings of Skyline and its settings for LFQbench21). The retention time calculator was created using the “Biognosys-11” built-in set of retention time standards. The calculation of q-values was performed using the built-in mProphet algorithm and decoy precursors. With OpenSWATH, we tried different background subtraction (Scoring:TransitionGroupPicker:background_subtraction) options (“original” and “none”), with “original” performing better for all the gradients. The following options were further used for OpenSWATH:
“-readOptions cacheWorkingInMemory -batchSize 1000 -Scoring:stop_report_after_feature -1 -Scoring:Scores:use_dia_scores true -ppm -threads 14 -min_upper_edge_dist 1.0 -min_rsq 0.95 -extra_rt_extraction_window 100 -use_ms1_traces”.
OpenSWATH output was further processed using PyProphet30 2.0.4 with the “--level=ms2” option. PyProphet output was further processed to remove decoy precursors and suboptimal peaks. The reports produced by Skyline and OpenSWATH (with all the parameter sets tested) were converted to a common format and have been deposited to the OSF repository among the rest of the supplementary materials: https://doi.org/10.17605/OSF.IO/6G3UX.
Statistics
“Box and whiskers” plots were produced using the LFQbench R package, representing the interquartile ranges as well as the 1-99 percentiles for the A:B peptide and protein ratios21. The respective numbers of peptides and proteins are given in the figure legends.
Sample preparation
The yeast protein extracts (Supplementary Note 10) were prepared from Saccharomyces cerevisiae (BY4743-pHLU31) grown to exponential phase in minimal synthetic nutrient media and processed in a bead beater for 5min at 1500rpm (Spex Geno/Grinder). Plasma samples (Supplementary Note 10) were prepared from commercial plasma (Human Cord Blood Plasma, Stemcell Technologies).
Proteins were denatured in 8M urea/0.1M ammonium bicarbonate pH 8.0 before they were reduced and alkylated in 5mM dithiothreitol and 10mM iodoacetamide, respectively. The sample was diluted to <1.5M urea/0.1M ammonium bicarbonate pH 8.0 before the proteins were digested overnight with trypsin (37°C). Peptides were cleaned-up with 96-well MacroSpin plates (Nest Group) and iRT peptides (Biognosys) were spiked in.
The digested human K562 cell lysate (Supplementary Note 2) was bought commercially (Promega - V6951) and spiked with iRT peptides.
Mass spectrometry
The digested peptides were analysed on a nanoAcquity (Waters) (running as 5µl/min microflow LC) coupled to a TripleTOF 6600 (Sciex). 2 µg of the protein digest was injected and the peptides were separated with a 23-minute (yeast), 21-minute (plasma) or 19-minute (K562) non-linear gradient starting with 4% acetonitrile/0.1 % formic acid and increasing to 36% acetonitrile/0.1% formic acid. A Waters HSS T3 column (150mm x 300µm, 1.8µm particles) was used. The DIA method consisted of an MS1 scan from m/z 400 to m/z 1250 (50ms accumulation time) and 40 MS2 scans (35ms accumulation time) with variable precursor isolation width covering the mass range from m/z 400 to m/z 1250.
The library generation with “gas-phase fractionation” was performed using the same LC-MS/MS setup as mentioned above. The peptides were separated with a 120 minute (plasma samples) and 45 minute (yeast samples) linear gradient (3% acetonitrile/0.1% formic acid to 60% acetonitrile/0.1 formic acid). Repetitive injections were performed to cover the following scan ranges: m/z 400 – 500, m/z 495 – 600, m/z 595 – 700, m/z 695 – 800, m/z 795 – 900, m/z 895 – 1000, m/z 995 – 1100, m/z 1095 – 1250 (yeast) and m/z 400 – 500, m/z 500 – 600, m/z 600 – 700, m/z 700 – 800, m/z 800– 900, m/z 900 – 1000, m/z 1000 – 1250 (plasma). The precursor selection windows were m/z 4 (m/z 1 overlap) for all acquisitions except the yeast m/z 1095 – 1250, for which m/z 5 (m/z 1 overlap) windows were used. For the plasma acquisitions, each acquisition cycle was split into two subcycles with the second subcycle having the isolation windows shifted by m/z 1.5.
Supplementary Material
Acknowledgements
We thank R. Bruderer for providing the spectral libraries. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001134), the UK Medical Research Council (FC001134), and the Wellcome Trust (FC001134), and received specific funding from the BBSRC (BB/N015215/1 and BB/N015282/1), the Wellcome Trust (200829/Z/16/Z) as well as a Crick Idea to Innovation (i2i) initiative (Grant Ref 10658).
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
DIA-NN (1.6.0) is open-source and is freely available at https://github.com/vdemichev/diann under a permissive licence.
Data availability
The newly generated mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE32 partner repository with the dataset identifier PXD014690; previously published data was also used to benchmark the software (repositories with identifiers PXD005573, PXD002952, PXD010529 and PXD006722). All the precursor and protein identification and quantification information has been uploaded to the OSF repository https://doi.org/10.17605/OSF.IO/6G3UX.
Author contributions
V.D., M.R. and K.S.L. designed the study, V.D and M.R wrote the first manuscript draft, V.D. designed and implemented the algorithms, C.B.M., V.D. and S.I.V. performed the experiments, all authors discussed the results and commented on the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Nature. 2016 Sep;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 3.Geyer PE, Holdt LM, Teupser D, Mann M. Mol Syst Biol. 2017 Sep;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zelezniak A, et al. Cell Syst. 2018;7:269–283.e6. doi: 10.1016/j.cels.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruderer R, et al. Mol Cell Proteomics. 2015 May;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meier F, Geyer PE, Virreira Winter S, Cox J, Mann M. Nat Methods. 2018;15:440–448. doi: 10.1038/s41592-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 7.Venable JD, Dong M-Q, Wohlschlegel J, Dillin A, Yates JR. Nat Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 8.Gillet LC, et al. Mol Cell Proteomics. 2012 Jun;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig C, et al. Mol Syst Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins BC, et al. Nat Commun. 2017 Dec;8 [Google Scholar]
- 11.Vowinckel J, et al. Sci Rep. 2018;8 doi: 10.1038/s41598-018-22610-4. 4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruderer R, et al. Mol Cell Proteomics. 2017 Dec;16:2296–2309. doi: 10.1074/mcp.RA117.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiter L, et al. Nat Methods. 2011 May;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 14.Elias JE, Gygi SP. Nat Methods. 2007 Mar;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 15.Ting YS, et al. Nat Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Nat Methods. 2015;12:1106–1108. doi: 10.1038/nmeth.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeCun Y, Bengio Y, Hinton G. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 18.Röst HL, et al. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 19.MacLean B, et al. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peckner R, et al. Nat Methods. 2018;15:371–378. doi: 10.1038/nmeth.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro P, et al. Nat Biotechnol. 2016;34:1130–1136. doi: 10.1038/nbt.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, et al. J Proteome Res. 2012;11:4509–4516. doi: 10.1021/pr300235v. [DOI] [PubMed] [Google Scholar]
- 23.Kingma DP, Ba J. arXiv [cs.LG] 2014 at < http://arxiv.org/abs/1412.6980>. [Google Scholar]
- 24.Storey JD. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. [Google Scholar]
- 25.Röst HL, et al. Nat Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 26.Chambers MC, et al. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The UniProt Consortium. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker SJ, et al. Mol Cell Proteomics. 2015;14:2800–2813. doi: 10.1074/mcp.O114.042267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deutsch EW, Lam H, Aebersold R. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teleman J, et al. Bioinformatics. 2015;31:555–562. doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- 31.Mülleder M, Campbell K, Matsarskaia O, Eckerstorfer F, Ralser M. F1000Res. 2016;5:2351. doi: 10.12688/f1000research.9606.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Riverol Y, et al. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.