Abstract
Avocado (Persea americana Mill.), macadamia (Macadamia integrifolia L.) and mango (Mangifera indica L.) are important subtropical tree species grown for their edible fruits and nuts. Despite their commercial and nutritional importance, the genomic information for these species is largely lacking. Here we report the generation of avocado, macadamia and mango transcriptome assemblies from pooled leaf, stem, bud, root, floral and fruit/nut tissue. Using normalized cDNA libraries, we generated comprehensive RNA-Seq datasets from which we assembled 63420, 78871 and 82198 unigenes of avocado, macadamia and mango, respectively using a combination of de novo transcriptome assembly and redundancy reduction. These unigenes were functionally annotated using Basic Local Alignment Search Tool (BLAST) to query the Universal Protein Resource Knowledgebase (UniProtKB). A workflow encompassing RNA extraction, library preparation, transcriptome assembly, redundancy reduction, assembly validation and annotation is provided. This study provides avocado, macadamia and mango transcriptome and annotation data, which is valuable for gene discovery and gene expression profiling experiments as well as ongoing and future genome annotation and marker development applications.
Subject terms: RNA sequencing, Plant molecular biology, Transcriptomics
| Measurement(s) | transcription profiling assay • sequence_assembly • sequence feature annotation |
| Technology Type(s) | RNA sequencing • sequence assembly process • sequence annotation |
| Factor Type(s) | plant species |
| Sample Characteristic - Organism | Persea americana • Macadamia integrifolia • Mangifera indica |
| Sample Characteristic - Location | Australia |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.11303135
Background & Summary
Fruits and nuts are an important source of vitamins and dietary fibre for consumers and a source of income for farmers. Avocado (Persea americana Mill.), macadamia (Macadamia integrifolia L.) and mango (Mangifera indica L.) are important commercial tree species grown in Australia and other tropical/sub-tropical regions. In 2013, the world production of avocado was about 4.7 million tonnes1. Macadamia is grown commercially for its edible nuts in tropical and subtropical regions, including Australia, Hawaii, China, Thailand, southern and central Africa and Central and South America2. Mangoes are produced commercially in at least 87 countries on an estimated area 5 million hectares with an annual production of over 35 million tonnes3. Despite their commercial and nutritional importance, these tree crops are yet to benefit from a substantial research effort required to generate significant public bioinformatic resources. These resources are essential for functional genomics studies, marker-assisted breeding, cultivar development, and genome annotation efforts. Here, we report on the generation and availing of transcriptomic resources for avocado, macadamia and mango.
Currently a few genomic resources are available for avocado, mango and macadamia. Most of the publicly available de novo transcriptome assemblies of avocado and mango are limited to either leaf or fruit tissue4–7. Only two studies published open-access transcriptome assemblies from several tissues of avocado and mango respectively8–10. However, these assemblies were derived from RNA-Seq libraries that were not normalised and therefore lack some essential yet lowly expressed genes and near-universal single-copy genes (Supplementary Fig. 1). Additionally, the ‘Keitt’ mango transcriptome study9 was designed for SNP discovery and did not produce a reference transcriptome for gene discovery purposes. A reference macadamia genome assembly with its accompanying reference gene set was recently published11. However, this genome assembly comprises 79% of the estimated macadamia genome size11,12. A draft mango genome was published in 2016, although it is not yet be publicly available13. We believe that our de novo transcriptome assemblies derived from normalized RNA-Seq libraries are complimentary to these resources as they accentuate rare/low abundance transcripts. In eukaryotes, the high abundance transcripts (several thousand mRNA copies per cell) from as few as 5–10 genes account for 20% of the cellular mRNA14. The intermediate abundance (several hundred copies per cell) transcripts of 500–2000 genes constitute about 40–60% of the cellular mRNA. The remaining 20–40% of mRNA is represented by rare, low abundance (from one to several dozen mRNA copies per cell) transcripts14. Such an enormous difference in transcript abundance compromises gene discovery, which results in poor detection of genes transcribed at relatively low levels.
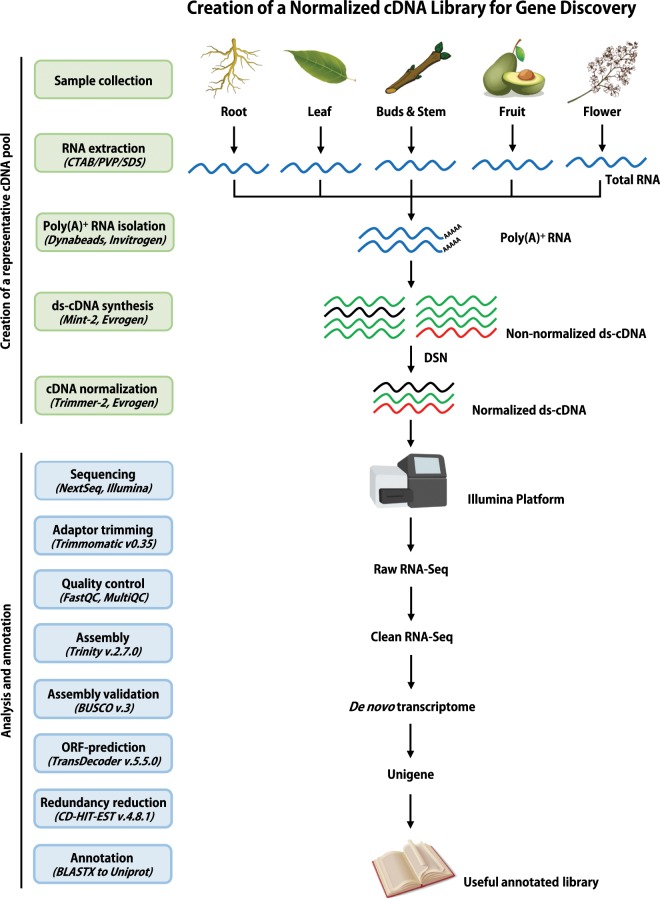
We therefore prepared comprehensive cDNA libraries from RNA pooled from of a wide range of plant tissues (leaf, stem, axillary bud, root and flower and fruit/nut) to maximize the number of transcripts represented in each library. The essential part of the library preparation process was converting the pooled RNA into normalized cDNA using a duplex-specific nuclease (DSN) normalization protocol15. This was done to avoid the dilution of transcripts from lowly expressed genes by those from highly expressed genes (Fig. 1) and therefore to improve gene discovery16. The assemblies generated in this study can be utilized as reference gene sets for a variety of tree genomics studies requiring transcriptome information of Persea americana, Macadamia integrifolia, Mangifera indica and related species. For example, considering that Persea americana, and Mangifera indica, are both prone to alternate/biennial bearing17,18, identification and subsequent manipulation of genes regulating floral induction may greatly contribute to solving this problem. Our transcriptome assemblies will also assist in mRNA-based genome annotation19 for ongoing whole genome sequencing projects of macadamia and mango11,13.
Fig. 1.
Flowchart of the CDNA library preparation, RNA-sequencing setup and de novo transcriptome data analysis steps (created with BioRender.com).
Methods
Sample collection
Tissue samples were collected from mature (7–15 year old) field-grown avocado cv. “Hass”, mango cv. 1243, and macadamia cv. 751 trees in Queensland, Australia. Plant tissue sampled included young and mature leaves, dormant and bursting axillary and terminal buds, mature and elongating stems and roots, a mixture of floral tissues at different stages of development and a mixture of fruit tissue in the case of avocado and mango or nuts in the case of macadamia. Fresh material was flash frozen in liquid nitrogen or dry ice and stored at −80 °C before being homogenized using an automated tissue grinder (Geno/Grinder®, SPEX).
RNA extraction
RNA was extracted from the different samples using a CTAB/PVP/SDS method developed for these types of samples as previously described20. Briefly, frozen powder was lysed using a CTAB/PVP buffer + 1 mM DTT for 10–15 min. One percent SDS was then added to each sample before centrifugation for 15 min at 20,000 g. The liquid phase containing the nucleic acids was up taken and added to an equal volume of isopropanol before centrifugation (20,000 g) for 45–60 min at 4 degrees. The nucleic acid pellet was then washed with 70% ethanol and resuspended in water. DNase treatment was then applied for 25 min and RNA was precipitated in an equal volume of isopropanol to form a nucleic acid pellet. The pellets were washed in 70% ethanol and then resuspended in pure water. RNA concentration was measured using a NanoVue™ Plus Spectrophotometer (GE Healthcare Life Sciences, USA). RNA integrity check was performed by agarose gel electrophoresis.
Normalised cDNA Library preparation
One normalised cDNA library was prepared for each of avocado, macadamia and mango, from equal amounts of mRNA from the different tissue types mentioned above and as described in Fig. 1. Poly(A)-RNA was isolated using oligo(dT) magnetic beads (Invitrogen™ Dynabeads™). 0.5–1 μg of the poly(A)RNA was converted into full-length-enriched double stranded cDNA using the Mint-2 cDNA synthesis kit and following the manufacturer’s instructions (Evrogen, Moscow, Russia). The double stranded cDNA was then normalized using the DSN-based Trimmer-2 cDNA normalization kit and following the manufacturer’s instructions (Evrogen, Moscow, Russia). The normalized cDNA libraries were then sheared into ~300 bp fragments with a sonicator (Bioruptor®, Diagenode) and indexed with adaptors using the NEBNext® DNA Library Prep Master Mix Set for Illumina®. Four technical replicates of each of the three normalized cDNA libraries were sequenced on the Illumina NextSeq. 500 platform (Fig. 1) with the primary objective of enhancing de novo gene discovery.
De novo assembly and dataset annotation
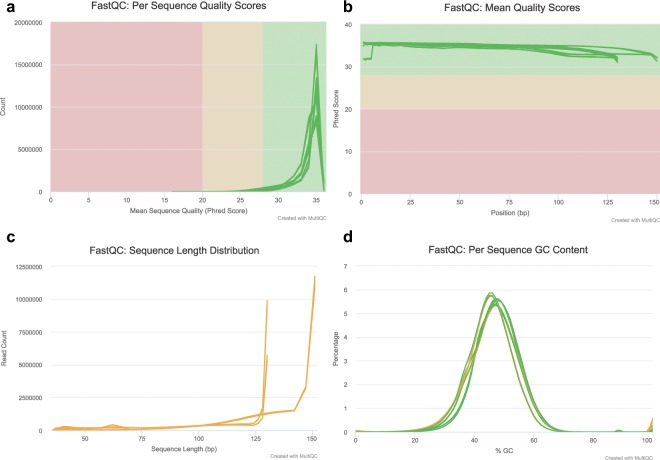
High-quality RNA-Seq reads (sequences) were used in the subsequent de novo transcriptome assembly. Raw RNA-seq reads were pre-processed by removing adapters and low-quality sequences (<Q30) using Trimmomatic (v. 0.35) with default parameters21. Sequencing summary statistics showing the total number of reads before and after trimming and quality filtering is presented in Table 1. RNA-Seq read quality before and after trimming was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and aggregated using MultiQC22, read quality after trimming is presented in Fig. 2. De novo transcriptome assembly was done with Trinity (v. 2.7.0) using default settings23,24. Coding regions of the assembled transcripts were predicted using TransDecoder (v. 5.5.0) with default settings24. We used selected the single best open reading frame (ORF) per transcript longer than 100 peptides. We then used the CD-HIT-EST program (v. 4.8.1) with default parameters (similarity 95%) to reduce transcript redundancy and produce unique genes (“unigenes”)25. We used Basic Local Alignment Search Tool (BLAST) to assign functional annotations to the unigenes26,27.
Table 1.
Read summary statistics and comparative analysis of Avocado and Macadamia RNA-Seq reads and de novo assembled transcripts to publicly available avocado and macadamia genomic resources.
| Avocado | Macadamia | Mango | |
|---|---|---|---|
| NCBI BioSample accession numbers | SRR8926023, SRR8926022, SRR8926017, SRR8926016 | SRR8926019, SRR8926018, SRR8926021, SRR8926020 | SRR8926027, SRR8926026, SRR8926025, SRR8926024 |
| Total number of raw reads | 226341270 | 159438181 | 188997291 |
| Total number of reads after trimming | 209971284 (92.77%) | 150743988 (94.57%) | 167567866 (88.6%) |
| Reference genome size | 912.6 Mbp | 652 Mbp | N/A |
| Number of trimmed reads mapped to reference genome | 166781058 (73.69%) | 127314454 (79.85%) | N/A |
| Average depth of coverage of mapped reads | 29.09 | 20.93 | N/A |
| Reference gene sets (number of sequences) | 24616 | 35337 | N/A |
| Number of unigenes in de novo transcriptome assemblies | 63420 | 78871 | 82198 |
| Unique BLASTN matches to reference gene sets | 22670 (92%) | 27322 (77%) | N/A |
Reference genomes and genesets used for the comparative analysis are Rendón-Anaya et al. (2019) Nock et al. (2016) for avocado and macadamia respectively.
Fig. 2.
Quality assessment metrics for trimmed and filtered RNA-Seq data used to make the de novo transcriptome assembly.
Data Records
Nine datasets were generated in this study. The first datasets consists of RNA-seq raw reads of Persea americana, Macadamia integrifolia and Mangifera indica, which were deposited in the NCBI Sequence Read Archive database under project identification number PRJNA53351828. Datasets containing Persea americana, Macadamia integrifolia and Mangifera indica transcriptome assemblies were deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database under TSA accession numbers GHOF000000029, GHOE0000000030 and GHOG0000000031. Datasets containing Persea americana, Macadamia integrifolia and Mangifera indica raw trinity transcriptome assemblies, unigenes, and functional annotation files were deposited in Figshare32–34.
Technical Validation
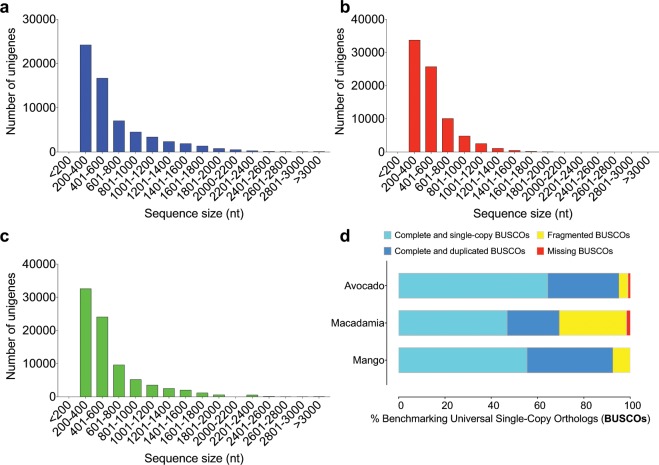
Read quality assessment and by extension, read validation was done using FastQC, quality control (QC) plots were aggregated using MultiQC22 and are presented in Fig. 2. We used HISAT235 to map avocado and macadamia RNA-Seq reads to their respective reference genome assemblies10,11. 73,7 and 79,8% of the avocado and macadamia reads mapped to their respective reference genome assemblies (Table 1). 63420, 78871 and 82198 unigenes of avocado, macadamia and mango were generated from the RNA-Seq data using a combination of de novo transcriptome assembly and redundancy reduction (Fig. 1; Table 2). We used BLASTn (e-value cut-off of 1e-5 and an identity cut-off of 70%) to compare our avocado and macadamia unigenes to the published reference gene sets10,11. 22670 (92%) and 27322 (77%) of the reference avocado and macadamia genes respectively were present in our assemblies (Table 1). The length distribution of “unigenes” was similar across the three species (Fig. 3a–c).
Table 2.
De novo assembly statistics of avocado, macadamia and mango transcriptomes before (Trinity output) and after redundancy reduction (Unigenes).
| Avocado | Macadamia | Mango | ||||
|---|---|---|---|---|---|---|
| Trinity output | Unigenes | Trinity output | Unigenes | Trinity output | Unigenes | |
| # contigs (>=0 bp) | 249765 | 63420 | 225591 | 78871 | 251204 | 82198 |
| # contigs (>=1000 bp) | 42988 | 10981 | 17643 | 4464 | 44854 | 10694 |
| # contigs (>=5000 bp) | 28 | 2 | 0 | 0 | 14 | 1 |
| Total length (>=0 bp) | 154556593 | 41442153 | 106195638 | 40705830 | 156057297 | 49246959 |
| Total length (>=1000 bp) | 69201144 | 16247577 | 23529519 | 5499159 | 72163715 | 15228411 |
| Total length (>=5000 bp) | 153870 | 11058 | 0 | 0 | 76292 | 5547 |
| # contigs | 100110 | 28816 | 68025 | 29090 | 98975 | 34564 |
| Largest contig | 6121 | 5700 | 3594 | 3219 | 6179 | 5547 |
| Total length | 109464144 | 28572369 | 58255825 | 22035423 | 110183488 | 31425165 |
| GC (%) | 43.33 | 46.89 | 45.09 | 48.29 | 41.82 | 45.58 |
| N50 | 1239 | 1104 | 888 | 756 | 1292 | 978 |
| N75 | 817 | 744 | 663 | 606 | 839 | 675 |
| L50 | 29949 | 9111 | 23589 | 10869 | 29822 | 11184 |
| L75 | 57262 | 16985 | 42633 | 19050 | 56299 | 20938 |
| # N’s per 100 kbp | 0 | 0 | 0 | 0 | 0 | 0 |
Fig. 3.
Sequence length distributions and assessment of completeness of the avocado, macadamia and mango unigenes. (a–c) Sequence length distributions, (d) transcriptome completeness as determined by Benchmarking Universal Single-Copy Orthologous (BUSCO). The figure was generated using GraphPad Prism Version 7.0a.
Transcriptome assembly validation was done using Benchmarking Universal Single-Copy Orthologs (BUSCO) v. 336. 70–95% of complete BUSCOs were present in the three de novo transcriptomes indicated high-quality assemblies, particularly for avocado and mango transcriptomes (Fig. 3d). Our normalized avocado assembly lacks only 3 while our normalised mango assembly has all near-universal single-copy genes (Fig. 3d). BUSCO provides a quantitative measure of transcriptome quality and completeness, based on evolutionarily-informed expectations of gene content from the near-universal, ultra-conserved eukaryotic proteins (eukaryota_odb9) database36–38. The BLASTx program (e-value cut-off of 1e-3) was used to annotate the “unigenes” based on UniProtKB/Swiss-Prot, a manually annotated, non-redundant protein sequence database26,27,39. 64–67% of the “unigenes” per species were annotated to the UniProtKB/Swiss-Prot non-redundant protein sequence database. A comprehensive workflow and links to obtain transcriptome data are provided. This dataset adds valuable transcriptome resources for further study of developmental gene expression, transcriptional regulation and functional genomics in avocado, macadamia and mango.
Supplementary information
Acknowledgements
This work is part of the Small Tree – High Productivity Initiative, a research collaboration between the Queensland Department of Agriculture and Fisheries (DAF), NSW Department of Primary Industries and the Queensland Alliance for Agriculture and Food Innovation, and co-funded through Horticulture Innovation Australia Limited (HIA Ltd) using the Hort Innovation Across Horticulture research and development levy (project number AI13004), co-investment from DAF and contributions from the Australian Government. Hort Innovation is the grower owned, not-for-profit research and development corporation for Australian horticulture. This work was financially supported by the Australian Research Council (ARC), the Queensland Government and the Horticulture Innovation Australia Limited. C.A.B. was funded by an ARC Laureate Fellowship FL180100139. We would like to thank Annette Dexter and Rosanna Powell for valuable discussions about the RNA extraction method and Helen Hoffman, John Wilkie, Ian Bally, Siegrid Parfitt, Jarrad Griffin, Hanna Toegel, Natalie Dillon, Paula Ibell and Anahita Mizani for collecting and providing the samples for this work.
Author contributions
T.G.C. processed and analysed data, and wrote the draft manuscript. F.F.B. processed the samples, performed library preparation and assisted in drafting the manuscript. M.T. and C.A.B. designed and supervised the project.
Code availability
Trimmomatic v. 0.35 parameters:
trimmomatic-0.35.jar PE -phred33 in_forward.fq.gz in_reverse.fq.gz out_forward_paired.fq.gz out_forward_unpaired.fq.gz out_reverse_paired.fq.gz out_reverse_unpaired.fq.gz ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
HISAT2 v 2.1.0 parameters:
hisat2-build reference_index_name genome.fa
hisat2 –x reference_index -1 reads_1a.fq,reads_1b.fq, reads_1c.fq,reads_1d.fq -2 reads_2a.fq,reads_2b.fq,reads_2c.fq,reads_2d.fq -S output.sam
SamTools v. 1.9.0 parameters:
samtools view -b -o output.bam samfile_from_hisat2.sam
samtools sort -o sorted.bam output.bam
samtools depth sorted.bam | awk ‘{sum+=$3} END {print “Average = ”,sum/NR}’
Trinity v. 2.7.0 parameters:
Trinity --seqType fq --left reads_1a.fq,reads_1b.fq,reads_1c.fq,reads_1d.fq --right reads_2a.fq,reads_2b.fq,reads_2c.fq,reads_2d.fq --CPU 6 --max_memory 20G
CD-HIT-EST v. 4.8.1 parameters:
cd-hit-est –i trinity_transcripts.fasta –o output file –c 0.9.
TransDecoder v.5.5.0 parameters:
TransDecoder.LongOrfs -t cd-hit-est__0.95_transcripts.fasta
BUSCO v. 3 parameters:
python BUSCO.py -i unigenes -l OrthoDB v9 -o output_name
BLAST v. 2.7.1 parameters:
makeblastdb -in reference_trancriptome assembly.fasta -dbtype “nucl”
blastn -query unigenes.fasta -db reference_trancriptome assembly.fasta -out outputfile.txt -evalue 1e-5 -max_target_seqs. 20 -outfmt 6
makeblastdb -in -in uniprot_sprot.fasta -dbtype “prot”
blastx -query unigenes.fasta -db uniprot_sprot.fasta -out outputfile.txt -evalue 1e-3 -max_target_seqs. 20 -outfmt 6.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Milos Tanurdzic, Email: m.tanurdzic@uq.edu.au.
Christine A. Beveridge, Email: c.beveridge@uq.edu.au
Supplementary information
is available for this paper at 10.1038/s41597-019-0350-9.
References
- 1.Hurtado-Fernández, E., Fernández-Gutiérrez, A. & Carrasco-Pancorbo, A. Avocado fruit— Persea americana. In Exotic Fruits – Reference Guide 37–48 (Academic Press, 2018).
- 2.Stimpson K, Luke H, Lloyd D. Understanding grower demographics, motivations and management practices to improve engagement, extension and industry resilience: a case study of the macadamia industry in the Northern Rivers, Australia. Aust. Geogr. 2019;50:69–90. doi: 10.1080/00049182.2018.1463832. [DOI] [Google Scholar]
- 3.Zaharah SS, Singh Z. Postharvest nitric oxide fumigation alleviates chilling injury, delays fruit ripening and maintains quality in cold-stored ‘Kensington Pride’ mango. Postharvest Biol. Technol. 2011;60:202–210. doi: 10.1016/j.postharvbio.2011.01.011. [DOI] [Google Scholar]
- 4.Azim MK, Khan IA, Zhang Y. Characterization of mango (Mangifera indica L.) transcriptome and chloroplast genome. Plant Mol. Biol. 2014;85:193–208. doi: 10.1007/s11103-014-0179-8. [DOI] [PubMed] [Google Scholar]
- 5.Luria N, et al. De-novo assembly of mango fruit peel transcriptome reveals mechanisms of mango response to hot water treatment. BMC Genomics. 2014;15:957. doi: 10.1186/1471-2164-15-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, et al. Transcriptome and proteomic analysis of mango (Mangifera indica Linn) fruits. J. Proteomics. 2014;105:19–30. doi: 10.1016/j.jprot.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Liqin, L. I. U. et al. Avocado Fruit Pulp Transcriptomes in the after-Ripening Process. Not. Bot. Horti Agrobot. Cluj-Napoca47, 308–319 (2018).
- 8.Ibarra-Laclette E, et al. Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genomics. 2015;16:599–599. doi: 10.1186/s12864-015-1775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman A, et al. Mango (Mangifera indica L.) germplasm diversity based on single nucleotide polymorphisms derived from the transcriptome. BMC Plant Biol. 2015;15:277. doi: 10.1186/s12870-015-0663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rendón-Anaya M, et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA. 2019;116:17081–17089. doi: 10.1073/pnas.1822129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nock CJ, et al. Genome and transcriptome sequencing characterises the gene space of Macadamia integrifolia (Proteaceae) BMC Genomics. 2016;17:937. doi: 10.1186/s12864-016-3272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chagné, D. Chapter One - Whole Genome Sequencing of Fruit Tree Species. In Advances in Botanical Research (eds. Plomion, C. & Adam-Blondon, A.-F.) vol. 74 1–37 (Academic Press, 2015).
- 13.Singh, N. Origin, Diversity and Genome Sequence of Mango (Mangifera indica L.). Indian J. Hist. Sci. 51, 355–368 (2016).
- 14.Vella F. Molecular biology of the cell (third edition): By Alberts, B. et al. Watson. pp 1361. Garland Publishing, New York and London. 1994. Biochem. Educ. 2010;22:164–164. doi: 10.1016/0307-4412(94)90059-0. [DOI] [Google Scholar]
- 15.Bogdanova EA, et al. Normalization of full-length-enriched cDNA. Methods Mol. Biol. 2011;729:85–98. doi: 10.1007/978-1-61779-065-2_6. [DOI] [PubMed] [Google Scholar]
- 16.Ekblom R, Slate J, Horsburgh GJ, Birkhead T, Burke T. Comparison between normalised and unnormalised 454-sequencing libraries for small-scale RNA-Seq studies. Comp. Funct. Genomics. 2012;2012:8. doi: 10.1155/2012/281693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkie JD, Sedgley M, Olesen T. Regulation of floral initiation in horticultural trees. J. Exp. Bot. 2008;59:3215–28. doi: 10.1093/jxb/ern188. [DOI] [PubMed] [Google Scholar]
- 18.Ziv D, Zviran T, Zezak O, Samach A, Irihimovitch V. Expression profiling of FLOWERING LOCUS T-like gene in alternate bearing ‘Hass’ avocado trees suggests a role for PaFT in avocado flower induction. PLoS One. 2014;9:e110613. doi: 10.1371/journal.pone.0110613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L, et al. EAnnot: a genome annotation tool using experimental evidence. Genome Res. 2004;14:2503–9. doi: 10.1101/gr.3152604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbier FF, et al. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods. 2019;15:62. doi: 10.1186/s13007-019-0447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.2019. NCBI Sequence Read Archive. SRP192932
- 29.Chabikwa TG, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Persea americana, transcriptome shotgun assembly. GenBank. GHOF00000000
- 30.Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Macadamia integrifolia, transcriptome shotgun assembly. GenBank. GHOE00000000
- 31.Chabikwa TG, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Mangifera indica, transcriptome shotgun assembly. GenBank. GHOG00000000
- 32.Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. Avocado transcriptome assembly. figshare. [DOI] [PMC free article] [PubMed]
- 33.Chabikwa T, Barbier FF, Tanurdzic M, Beveridge C. 2019. Macadamia Transcriptome Assembly. figshare. [DOI] [PMC free article] [PubMed]
- 34.Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. Mango transcriptome assembly. figshare. [DOI] [PMC free article] [PubMed]
- 35.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse RM, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 38.Zdobnov EM, et al. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2017;45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2019. NCBI Sequence Read Archive. SRP192932
- Chabikwa TG, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Persea americana, transcriptome shotgun assembly. GenBank. GHOF00000000
- Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Macadamia integrifolia, transcriptome shotgun assembly. GenBank. GHOE00000000
- Chabikwa TG, Barbier FF, Tanurdzic M, Beveridge CA. 2019. TSA: Mangifera indica, transcriptome shotgun assembly. GenBank. GHOG00000000
- Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. Avocado transcriptome assembly. figshare. [DOI] [PMC free article] [PubMed]
- Chabikwa T, Barbier FF, Tanurdzic M, Beveridge C. 2019. Macadamia Transcriptome Assembly. figshare. [DOI] [PMC free article] [PubMed]
- Chabikwa T, Barbier FF, Tanurdzic M, Beveridge CA. 2019. Mango transcriptome assembly. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Trimmomatic v. 0.35 parameters:
trimmomatic-0.35.jar PE -phred33 in_forward.fq.gz in_reverse.fq.gz out_forward_paired.fq.gz out_forward_unpaired.fq.gz out_reverse_paired.fq.gz out_reverse_unpaired.fq.gz ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
HISAT2 v 2.1.0 parameters:
hisat2-build reference_index_name genome.fa
hisat2 –x reference_index -1 reads_1a.fq,reads_1b.fq, reads_1c.fq,reads_1d.fq -2 reads_2a.fq,reads_2b.fq,reads_2c.fq,reads_2d.fq -S output.sam
SamTools v. 1.9.0 parameters:
samtools view -b -o output.bam samfile_from_hisat2.sam
samtools sort -o sorted.bam output.bam
samtools depth sorted.bam | awk ‘{sum+=$3} END {print “Average = ”,sum/NR}’
Trinity v. 2.7.0 parameters:
Trinity --seqType fq --left reads_1a.fq,reads_1b.fq,reads_1c.fq,reads_1d.fq --right reads_2a.fq,reads_2b.fq,reads_2c.fq,reads_2d.fq --CPU 6 --max_memory 20G
CD-HIT-EST v. 4.8.1 parameters:
cd-hit-est –i trinity_transcripts.fasta –o output file –c 0.9.
TransDecoder v.5.5.0 parameters:
TransDecoder.LongOrfs -t cd-hit-est__0.95_transcripts.fasta
BUSCO v. 3 parameters:
python BUSCO.py -i unigenes -l OrthoDB v9 -o output_name
BLAST v. 2.7.1 parameters:
makeblastdb -in reference_trancriptome assembly.fasta -dbtype “nucl”
blastn -query unigenes.fasta -db reference_trancriptome assembly.fasta -out outputfile.txt -evalue 1e-5 -max_target_seqs. 20 -outfmt 6
makeblastdb -in -in uniprot_sprot.fasta -dbtype “prot”
blastx -query unigenes.fasta -db uniprot_sprot.fasta -out outputfile.txt -evalue 1e-3 -max_target_seqs. 20 -outfmt 6.