Abstract
Blockading programmed death ligand 1 (PD-L1) shows promising results in patients with some cancers, but not in those with ovarian cancer. V-domain Ig suppressor of T cell activation (VISTA) is a recently discovered immune checkpoint protein that suppresses T cell activation. This study aimed to investigate the expression and clinical significance of VISTA in ovarian cancer as well as its relationship with PD-L1. VISTA and PD-L1 levels in 146 ovarian cancer samples were assessed using immunohistochemistry. We investigated the association between VISTA and other clinicopathological variables, including survival. The associations between the VISTA-encoding C10orf54 gene, other immune checkpoints, and survival were analyzed. VISTA was detected in 51.4% of all samples and 46.6% of PD-L1-negative samples; it was expressed in 28.8%, 35.6%, and 4.1% of tumor cells (TCs), immune cells (ICs), and endothelial cells, respectively. Furthermore, VISTA expression was associated with pathologic type and PD-L1 expression. Moreover, VISTA expression in TCs, but not in ICs, was associated with prolonged progression-free and overall survival in patients with high-grade serous ovarian cancer. The expression of C10orf54 mRNA was associated with prolonged overall survival and immune escape-modulating genes. These results showed that VISTA expression in ovarian tumor cells was associated with a favorable prognosis in patients with high-grade serous ovarian cancer; however, additional studies are required to better understand the expression and role of VISTA in ovarian cancer.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02434-5) contains supplementary material, which is available to authorized users.
Keywords: Ovarian cancer, PD-L1, VISTA, Immune checkpoints, Prognosis
Introduction
Ovarian cancer is the most lethal type of the gynecologic malignancy and accounts for the highest rate of gynecological cancer-related deaths worldwide. Approximately 295,414 patients were newly diagnosed with ovarian cancer in 2018, and 184,799 women died of this disease [1]. More than 75% of patients are diagnosed at an advanced stage (stage III/IV), for which the 5-year overall survival (OS) rate is less than 30%. Although surgery and adjuvant chemotherapy are effective for the majority of patients with ovarian cancer, more than 70% ultimately relapse, and their tumors eventually become chemotherapy resistant.
Immunotherapy with antibodies against negative immune checkpoints such as programmed death 1 (PD-1), its ligand PD-L1, and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) has shown great efficacy in patients with a variety of solid tumors [2]. However, in a study of 26 patients with PD-L1-expressing advanced ovarian cancer, the objective response rate to immunotherapy was 11.5% (only one patient achieved a complete response and two had partial responses) [3]. Therefore, there is an urgent need to improve the outcomes of patients with ovarian cancer by better understanding the immune microenvironment of this tumor type and developing effective immunotherapeutic strategies [4].
V-domain Ig suppressor of T cell activation (VISTA) is a protein encoded by the C10orf54 gene and is also known as PD-1 homolog. VISTA is a recently discovered inhibitory immune checkpoint molecule that belongs to the B7 family and shares sequence homology with both PD-1 and PD-L1 [5–7]. This protein can suppress T cell activation when expressed as a ligand on antigen-presenting cells or when expressed as a receptor on T cells [8, 9]. Liu and colleagues reported a nonredundant, T cell-activating role for VISTA that is distinct from the PD-1/PD-L1 pathway [8]; this provided a rationale for the combined targeting of the VISTA and PD-1 pathways when treating patients with cancer.
Interestingly, Kakavand et al. discovered a significantly higher density of VISTA-positive lymphocytes after treatment with an anti-PD-1 antibody alone or in combination with anti-CTLA4 (ipilimumab); this was associated with adaptive resistance to immune checkpoint blockade [10]. Similarly, Gao et al. found that VISTA expression was elevated after anti-CTLA4 therapy in patients with prostate cancer, suggesting that elevated VISTA expression is a compensatory event in the setting of ipilimumab therapy [11]. When comparing immune cell (IC) infiltrates in melanoma and pancreatic cancer, Blando et al. demonstrated that VISTA is preferentially expressed at higher levels in pancreatic cancer, and highlighted this protein as a potential immunotherapeutic target for patients with this disease [12]. Furthermore, preclinical murine studies have shown that blockading both VISTA and PD-L1 produces a synergistic therapeutic effect in colon cancer models [8]. These studies suggest that VISTA may modulate a novel immune evasion mechanism and is thus a potential target for cancer immunotherapy. VISTA expression in human cancers has been reported in non-small cell lung cancer (NSCLC), hepatocellular carcinoma, colorectal carcinoma, oral squamous cell carcinoma, gastric carcinoma, acute myeloid leukemia, and gestational trophoblastic neoplasia [13–21]. Mulati et al. also found that VISTA was highly expressed in human ovarian and endometrial cancers [20]. However, the relationships between VISTA, PD-L1, clinicopathologic features, and prognoses in patients with ovarian cancer remain unknown.
In the current study, we investigated VISTA and PD-L1 expression using a tumor tissue microarray (TMA) encompassing the major histological subtypes of ovarian cancer. To support our findings, we also performed analyses of The Cancer Genome Atlas (TCGA) ovarian cancer dataset.
Materials and methods
Patient cohort and TMA
The commercialized TMA (panel HOvaC160Su01) and clinical data were provided by Shanghai Outdo Biotech Co. Ltd. (Shanghai, China). The TMA comprised samples from 160 patients with stage I–IV ovarian cancer that were collected between 2009 and 2012. All tumor tissues were obtained at the time of primary surgery from patients who were deemed optimally debulked. A total of 146 samples were included in this analysis after excluding non-epithelial ovarian cancer samples and quality controls. Of these samples, 93 were high-grade serous ovarian carcinoma (HGSOC), 27 were mucinous ovarian carcinoma, 17 were endometrioid ovarian carcinoma, and 9 were ovarian clear cell carcinoma. When categorized by International Federation of Gynecology and Obstetrics (FIGO) stage, eight samples were stage I, 31 were stage II, 75 were stage III, and 32 were stage IV. The mean age of the patients was 51 years (range 23–75 years), and the median follow-up time was 41 months (range 4–109 months).
Immunohistochemistry
Primary antibodies against VISTA (clone D1L2G, 1:200; Cell-Signaling Technology, MA, USA) and PD-L1 (clone E1L3 N, 1:200; Cell-Signaling Technology, MA, USA) were used to detect the expression of VISTA and PD-L1. Human tonsil and placenta tissues that were obtained from the Department of Pathology, Peking Union Medical College Hospital, Beijing, China, were used as positive controls, whereas normal tonsil tissues without primary antibodies were used as negative controls. Immunohistochemistry was performed using our laboratory protocol as described previously [13, 22].
Samples stained for VISTA and PD-L1 were evaluated by two pathologists who were blinded to the clinical outcomes. PD-L1 expression was evaluated with binary positive/negative scoring; PD-L1 positivity was defined as membranous staining on ≥ 1% of the cells using the previously described ‘combined positive score’ [23], which is calculated by summing the number of PD-L1-stained cells (tumor cells [TCs], lymphocytes, and macrophages), dividing the result by the total number of viable TCs, and multiplying the quotient by 100 [23]. VISTA expression was evaluated in both TCs and tumor-infiltrating ICs; TCs were considered VISTA-positive if at least 1% of these cells per histospot had membranous and/or cytoplasmic staining. ICs were defined as VISTA-positive if any staining was present in at least 1% of these cells, which included macrophages and lymphocytes. Endothelial cells were designated positive if any VISTA staining was present within. For purposes of stratification and statistical analysis, VISTA in each sample was defined as positive if any staining was visible in the TCs, ICs, or endothelial cells.
Kaplan–Meier plotter analysis
To analyze the prognostic value of mRNA from the VISTA-encoding gene C10orf54 in HGSOC, we performed survival analysis using the Kaplan–Meier plotter (www.kmplot.com), which contains gene expression data and patient survival information related to 2190 ovarian cancers derived from the Gene Expression Omnibus, European Genome–Phenome Archive, and TCGA. Patients with HGSOC (grades 2–3) who had stage II–IV disease were included in this study for the analysis of OS (n = 405) and progression-free survival (PFS) (n = 402). Patient samples were split into two groups (high vs. low expression) according to the most optimal cutoff of C10orf54 mRNA levels, which was automatically determined by the Kaplan–Meier plotter. The survival rates of patients in the two groups were compared using Kaplan–Meier plots; the hazard ratios, 95% confidence intervals, and log-rank P values were calculated [24].
TCGA data analysis for mRNA expression
We analyzed VISTA mRNA expression in the ovarian serous cystadenocarcinoma samples from the TCGA database using cBioPortal for cancer genomics (http://www.cbioportal.org/) [25, 26]. We included a total of 291 samples with the next-generation sequencing data (RNA Seq V2 RSEM), and evaluated the association between log2-transformed mRNA ‘fragments per kilobase million’ scores of the genes encoding VISTA, CD8A, and other inhibitory immune checkpoints including CTLA4, PD-1, PD-L1, PD-L2, T cell immunoglobulin and mucin domain-3 (TIM-3), lymphocyte activation gene-3 (LAG3), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) according to the cBioPortal instructions [26]. In addition, co-expressed genes with Pearson correlation coefficients > 0.4 were used for gene-ontology enrichment annotation using the cluster Profiler package [27].
Statistical analysis
The Chi-square or Fisher’s exact test was used to analyze the correlations between categorical variables (VISTA, PD-L1, and pathologic subtypes). Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. Statistical analyses were performed using SPSS 20.0 for Windows (IBM Corp., Armonk, NY, USA). Graphs and images were prepared using Prism 5.0 (GraphPad Software, La Jolla, CA, USA) and Photoshop CC 2019 for Windows (Adobe Inc., San Jose, CA, USA). All statistical tests were two-sided, and P values < 0.05 were considered significant.
Results
VISTA and PD-L1 expression in epithelial ovarian cancer
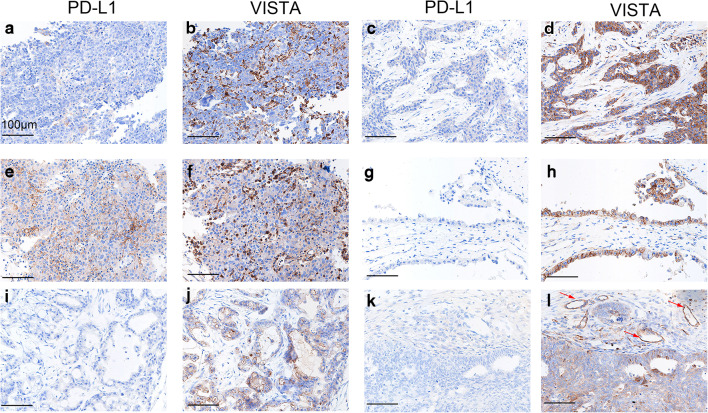
Among the 146 samples overall, VISTA expression was detected in 51.4% (75 samples), while PD-L1 was expressed in 8.9% (13 samples). VISTA protein was detected in TCs, ICs, and endothelial cells, and exhibited a cytoplasmic/membranous staining pattern. Representative stained samples of PD-L1 and VISTA are shown in Fig. 1. Forty-two samples (28.8%) showed VISTA expression in TCs, 52 (35.6%) showed its expression in ICs, and 6 (4.1%) had VISTA-positive endothelial cells. VISTA expression in the four types of ovarian cancer is shown in Table 1. Notably, 46.6% (62/133) of the PD-L1-negative samples expressed VISTA. When comparing non-serous carcinoma with HGSOC, VISTA expression levels in both the TCs and ICs of mucinous carcinomas were lower than those in the TCs and ICs of serous carcinomas; moreover, VISTA expression in the ICs of clear cell carcinomas was lower than that in the ICs of serous carcinomas (Table 1). However, no differences in VISTA expression were observed between serous carcinoma and endometrioid ovarian carcinoma.
Fig. 1.
Representative immunohistochemical staining of V-domain Ig suppressor of T cell activation (VISTA) and PD-L1 in human ovarian cancer. a, b Negative PD-L1 and positive VISTA in immune cells (ICs) in high-grade serous ovarian carcinomas (HGSOC). c, d Negative PD-L1 and positive VISTA in tumor cells (TCs) and ICs in HGSOC. e, f Positive PD-L1 and positive VISTA in ICs in HGSOC. g, h Negative PD-L1 and positive VISTA in TCs in clear cell carcinoma. i, j Negative PD-L1 and positive VISTA in TCs in mucinous ovarian carcinoma. k, l Negative PD-L1 and positive VISTA in TCs, ICs, and endothelial cells (red arrows) in endometrioid carcinoma
Table 1.
VISTA expression in the four pathologic types of ovarian cancer
| VISTA expression | Serous carcinoma | Mucinous carcinoma | Endometrioid carcinoma | Clear cell carcinoma |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | |
| VISTA in all cells | / | P < 0.001 | P = 0.272 | P = 0.751 |
| Positive | 57 (61.3) | 4 (14.8) | 8 (47.1) | 6 (66.7) |
| Negative | 36 (38.7) | 23 (85.2) | 9 (52.9) | 3 (33.3) |
| VISTA in ICs | / | P < 0.001 | P = 0.149 | P = 0.004 |
| Positive | 45 (48.2) | 2 (7.4) | 5 (29.4) | 0 (0) |
| Negative | 48 (51.6) | 25 (92.6) | 12 (70.6) | 9 (100) |
| VISTA in TCs | / | P = 0.016 | P = 0.670 | P = 0.057 |
| Positive | 28 (30.1) | 2 (7.4) | 6 (35.3) | 6 (66.7) |
| Negative | 65 (69.9) | 25 (92.6) | 11 (64.7) | 3 (33.3) |
ICs immune cells, TCs tumor cells, VISTA V-domain Ig suppressor of T cell activation
VISTA expression is associated with favorable prognoses in patients with HGSOC
We investigated the associations between VISTA expression, FIGO stage, PD-L1 expression, and survival in patients with HGSOC. The expression of VISTA was positively correlated with that of PD-L1. VISTA expression in ICs as well as in all cells (TCs, ICs, and endothelial cells) combined was significantly more frequent in PD-L1-positive cells. However, VISTA expression in TCs alone was not associated with PD-L1 expression (Table 2).
Table 2.
Association between VISTA expression, stage, PD-L1, and survival in patients with high-grade serous ovarian cancer
| VISTA in all cells | VISTA in ICs | VISTA in TCs | |||||
|---|---|---|---|---|---|---|---|
| Total | Positive | Negative | Positive | Negative | Positive | Negative | |
| n (%) | n (%) | i (%) | n (%) | n (%) | n (%) | n (%) | |
| Stage | 93 | P = 0.289 | P = 0.794 | P = 0.146 | |||
| I | 3 (3.2) | 1 | 2 | 1 | 2 | 0 | 3 |
| (33.3) | (66.7) | (33.3) | (66.7) | 0 | (100) | ||
| II | 18 (19.4) | 14 | 4 | 10 | 8 | 8 | 10 |
| (77.8) | (22.2) | (55.6) | (44.4) | (44.4) | (55.6) | ||
| III | 44 (47.3) | 27 | 17 | 22 | 22 | 15 | 29 |
| (61.4) | (38.6) | (50) | (50) | (34.1) | (65.9) | ||
| IV | 28 (30.1) | 15 | 13 | 12 | 16 | 5 | 23 |
| (53.6) | (46.4) | (42.9) | (57.1) | (17.9) | (82.1) | ||
| PD-L1 | P = 0.006 | P = 0.018 | P = 0.297 | ||||
| Positive | 11 (11.8) | 11 | 0 | 9 | 2 | 5 | 6 |
| (100) | (0) | (81.8) | (18.2) | (45.5) | (54.5) | ||
| Negative | 82 (88.2) | 46 | 36 | 36 | 46 | 23 | 59 |
| (56.1) | (43.9) | (43.9) | (56.1) | (28) | (72) | ||
| PFS | P = 0.151 | P = 0.201 | P = 0.043 | ||||
| Mean (months) | 38.4 | 42 | 32.8 | 42.9 | 34.5 | 46.4 | 33.8 |
| 95% CI | 31.6–45.2 | 33.1–50.9 | 22.8–42.9 | 32.7–53.2 | 25.6–43.3 | 35.0–57.8 | 26.2–41.3 |
| OS | P = 0.225 | P = 0.427 | P = 0.040 | ||||
| Mean (months) | 57.2 | 60.8 | 51.2 | 60.3 | 54.5 | 71.1 | 51.2 |
| 95% CI | 49.2–65.2 | 50.8–70.9 | 43.1–68.1 | 38.5–64.0 | 43.8–65.3 | 58.5–83.6 | 41.5–60.9 |
“All cells” include ICs, TCs, and endothelial cells
PD-L1 programmed cell death ligand 1, PFS progression-free survival, CI confidence interval, OS overall survival, ICs immune cells, TCs tumor cells, VISTA V-domain Ig suppressor of T cell activation
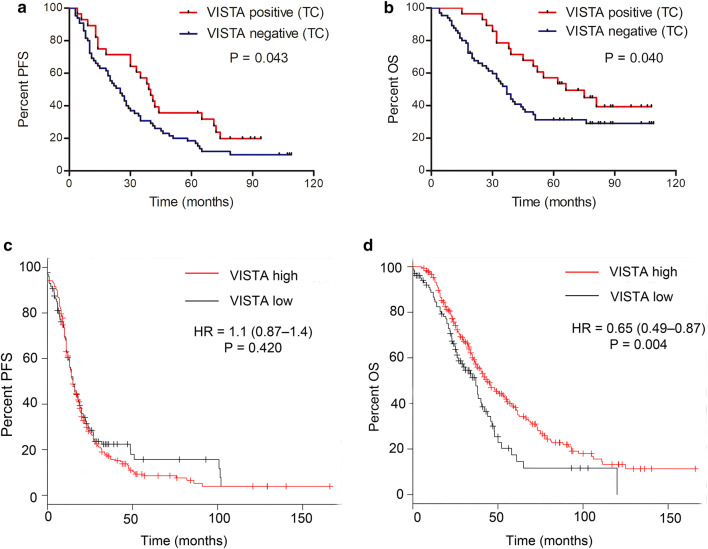
Survival analyses showed that VISTA-positive staining in TCs was significantly associated with prolonged PFS (P = 0.043, Fig. 2a) and OS (P = 0.040, Fig. 2b); however, VISTA expression in ICs or in all cells combined was not associated with survival (Supplementary Fig. S1). Moreover, no association between PD-L1 expression and survival was observed in these patients (Supplementary Fig. S2). We also performed survival analyses to determine the prognostic value of VISTA-encoding gene C10orf54 expression in patients with HGSOC. The Kaplan–Meier plots revealed that high C10orf54 mRNA expression did not influence PFS (P = 0.42, Fig. 2c), but was significantly associated with longer OS (P = 0.004, Fig. 2d). These results indicate that VISTA is associated with a favorable prognosis in patients with HGSOC.
Fig. 2.
Association between V-domain Ig suppressor of T cell activation (VISTA) expression and survival in patients with high-grade serous ovarian carcinomas (HGSOC). VISTA-positive staining in tumor cells (TCs) was significantly associated with prolonged PFS (a) and OS (b). Using the Kaplan–Meier plotter database, patients with HGSOC (grades 2–3) with stage II–IV disease were subjected to analysis of OS (n = 405) and PFS (n = 402). High expression of VISTA mRNA (encoded by C10orf54) was not associated with PFS (c), but was significantly associated with prolonged OS (d). HR, hazard ratio
Biological processes modulated by genes co-expressed with C10orf54
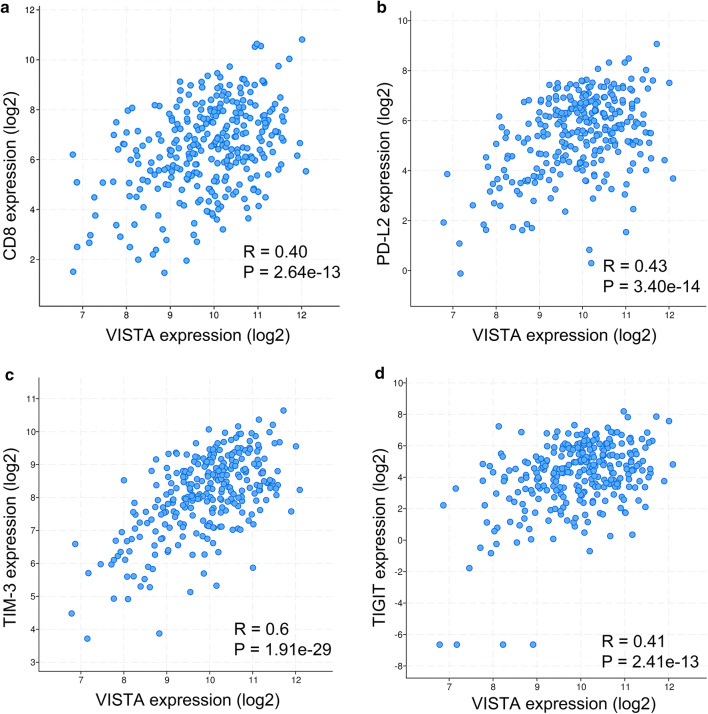
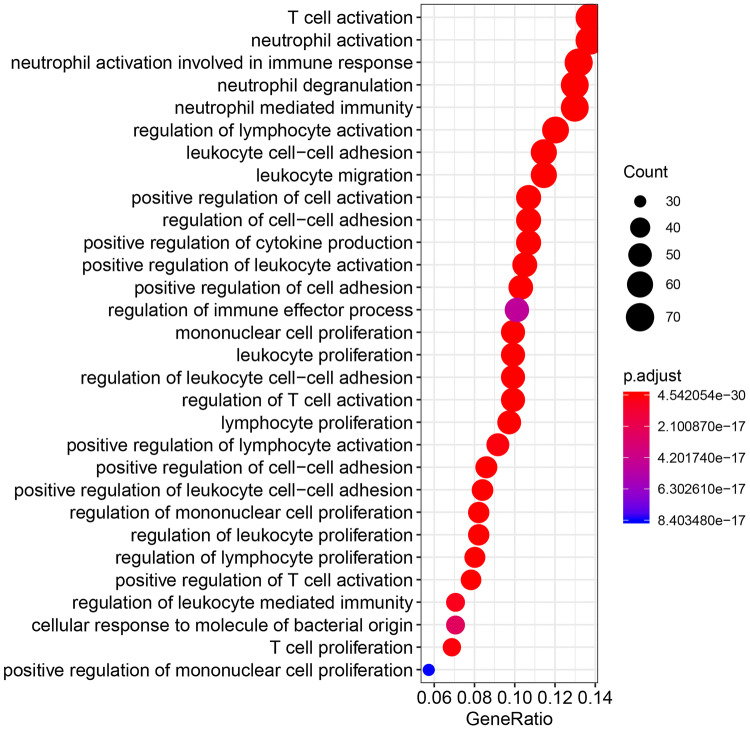
To assess the biological relevance of VISTA expression in serous ovarian cancer, we evaluated the association between the C10orf54 gene (encoding VISTA), CD8A (encoding CD8), and other inhibitory immune checkpoints including PDCD1 (encoding PD-1), CD274 (encoding PD-L1), PDCD1LG2 (encoding PD-L2), HAVCR2 (encoding TIM-3), LAG3 (encoding LAG3), and TIGIT (encoding TIGIT) based on TCGA mRNA expression profiles. Expression of the gene encoding VISTA was positively correlated with the expression of genes encoding CD8 (R = 0.40, Fig. 3a), PD-L2 (R = 0.43, Fig. 3b), TIM-3 (R = 0.6, Fig. 3c), and TIGIT (R = 0.41, Fig. 3d). Although VISTA was positively associated with CTLA4 (R = 0.38, P < 0.01), PD-1 (R = 0.38, P < 0.01), PD-L1 (R = 0.34, P < 0.01), and LAG3 (R = 0.32, P < 0.01), these correlations were weak (R < 0.4). Using a Pearson correlation coefficient of > 0.4 as a cutoff, we analyzed 588 genes co-expressed with VISTA for gene-ontology enrichment annotation. As shown in Fig. 4, these genes were mainly involved in modulating T cell activation, neutrophil activation, neutrophil-mediated immunity, and lymphocyte activation.
Fig. 3.
V-domain Ig suppressor of T cell activation (VISTA)-encoding gene is correlated with expression of genes that encode CD8 (a), programmed death ligand 2 (PD-L2) (b), T cell immunoglobulin and mucin domain-3 (TIM-3) (c) and T cell immunoreceptor with Ig and ITIM domains (TIGIT) (d) in ovarian serous cystadenocarcinoma samples from The Cancer Genome Atlas database
Fig. 4.
Gene-ontology biological process terms associated with V-domain Ig suppressor of T cell activation (VISTA) co-expressed genes that have Pearson correlation coefficients > 0.4
Discussion
VISTA has attracted wide interest as a novel immune checkpoint and a potential target of cancer immunotherapy. However, little is known about its expression in ovarian cancer, especially in PD-L1-negative tumors. In the present study, we profiled VISTA and PD-L1 expression in epithelial ovarian cancer and found that VISTA protein was expressed not only in tumor-infiltrating ICs but also in TCs themselves as well as endothelial cells. We discovered that VISTA is frequently expressed in PD-L1-negative ovarian cancer and that high VISTA expression is associated with a favorable prognosis in patients with HGSOC. Moreover, our study investigated the expression of VISTA and PD-L1 in ovarian cancer using a relatively large sample size.
Using real-time polymerase chain reaction, Lines et al. found that VISTA was predominantly expressed in human placental and hematopoietic tissues, as well as in tissues that exhibit markedly high leukocyte infiltration [28]. We also found that normal human organs other than lymphoid tissues and placental trophoblastic cells did not express VISTA at the protein level [13]. Although VISTA protein has been detected in gastric cancer, oral squamous cell carcinoma, NSCLC, colorectal carcinoma, hepatocellular carcinoma, endometrial cancer, ovarian cancer, melanoma, and gestational trophoblastic neoplasia, its expression and relationship with patient survival vary according to the type of cancer [13–21]. While VISTA was mainly expressed by tumor-infiltrating lymphocytes, it was only present in 8.8% of gastric cancer cells and in 19.4–22.8% of NSCLC lesions [15, 16]. We previously reported that VISTA protein was widely overexpressed in 98.2% of all the gestational trophoblastic neoplasia [13]. Mulati et al. reported that VISTA was expressed in 84 (91.3%) of the 92 ovarian cancer tissues they investigated, and that there was no difference in survival as a function of VISTA expression [20]. Contrary to their results, we found that 75 (51.4%) of the 146 ovarian cancer tissues we investigated expressed VISTA protein in TCs and/or tumor-infiltrating ICs. Furthermore, we found that VISTA expression in TCs (but not in ICs) was significantly associated with prolonged PFS and OS in patients with HGSOC. The discrepancy between these studies may be attributed to the use of different VISTA antibodies and variable sample sizes with distinct stages and pathological distributions. Recent studies demonstrated that VISTA expression in TCs (but not ICs) was associated with significantly longer OS in patients with hepatocellular carcinoma and NSCLC [16, 21], which correspond to our findings in patients with HGSOC. Contrary to these results, Kuklinski et al. reported that VISTA expression in tumor-infiltrating inflammatory cells was correlated with worse disease-specific survival in patients with primary cutaneous melanoma [19]. These data suggest that VISTA expressed in TCs and ICs may exert different functions. However, the exact role of VISTA expressed on TCs remains unknown, and there is an urgent need to further clarify the significance and possible function of VISTA in the tumor immune environment.
Although the immune checkpoint molecules PD-L1 and VISTA belong to the B7 family, they play nonredundant roles in suppressing T cell activation and immune regulation [8]. In the present study, we found that VISTA was expressed in 46.6% of PD-L1-negative ovarian cancers, indicating that this protein might serve as an alternative immune checkpoint that suppresses T cell activation. VISTA expression was associated with that of PD-L1, and corresponded to a favorable prognosis in patients with HGSOC. However, no association between PD-L1 expression and survival was observed in our study. The prognostic significance of PD-L1 in ovarian cancer remains controversial. Hamanishi et al. reported that PD-L1 was associated with poor prognosis in patients with this disease [29], whereas Webb et al. found that PD-L1 expression was correlated with a favorable prognosis in patients with HGSOC [30]. Moreover, no association was observed between PD-L1 expression and the survival of patients with HGSOC in Mills et al.’s study [31]. Consistent with our results, a recent meta-analysis showed that PD-L1 protein expression was not associated with the survival of patients with ovarian cancer. In contrast, VISTA appears be an indicator of favorable prognosis in patients with HGSOC, although this needs to be verified in larger cohorts.
In our study, we found a significant positive correlation between C10orf54 and genes such as TIM-3, TIGIT, and PDCD1LG2, which are responsible for tumor immune escape and immune suppression in human cancers. Similarly, Xie et al. found a significantly positive correlation between the expression of C10orf54 and that of TIGIT, HAVCR2, BTLA, CD274, and PDCD1 in colorectal carcinoma [17]. Furthermore, Villarroel-Espindola et al. found a consistent and prominent association between VISTA, PD-1, and PD-L1 in NSCLC at both the protein and mRNA levels [16]. Taken together, these data suggest that VISTA might contribute to immune escape in some other tumor types and could represent an effective target for immunotherapy in patients with other cancers. However, our findings suggest that this may not be the case for patients with ovarian cancer. Abiko et al. found that interferon-γ secreted by CD8-positive lymphocytes upregulated PD-L1 on ovarian cancer cells and promoted tumor growth. However, the mechanisms through which VISTA expression is regulated in the ovarian cancer microenvironment remain unknown. Therefore, additional studies are warranted to further elucidate the molecular regulation and mechanism of VISTA in ovarian cancer.
Despite our novel findings, our study had some limitations including those inherent to a retrospective study. Given the heterogeneity of tumor markers, TMA sections may not be representative of the entire tumor insofar as PD-L1 and VISTA expression. Moreover, we had a relatively small number of non-serous ovarian cancer samples; therefore, we did not analyze the relationship between VISTA and survival in patients with this tumor subtype. Furthermore, it was difficult to assess the expression of VISTA in the different subgroups of tumor-infiltrating ICs and to analyze the co-expression of VISTA and PD-L1 using immunohistochemistry. Future studies using multiplexed quantitative immunofluorescence are warranted to elucidate the expression and function of VISTA within the tumor immune microenvironment.
In conclusion, ours is the first investigation of VISTA and PD-L1 expression in a single large-size cohort of patients with ovarian cancer. Our study showed that VISTA was frequently expressed in PD-L1-negative ovarian cancer specimens and that VISTA expression in TCs, but not in ICs, was associated with the expression of genes that modulate tumor immune escape on one hand, but was also associated with prolonged survival in patients with HGSOC on the other. Further studies are urgently needed to explore the regulation and functions of VISTA in ovarian cancer to identify patients eligible for intensive treatments based on their expected prognoses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- CTLA4
Cytotoxic T-lymphocyte-associated protein 4
- FIGO
International federation of gynecology and obstetrics
- HGSOC
High-grade serous ovarian carcinoma
- IC
Immune cell
- LAG3
Lymphocyte activation gene-3
- TCGA
The cancer genome atlas
- TC
Tumor cell
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- TIM-3
T cell immunoglobulin and mucin domain-3
- TMA
Tumor tissue microarray
- VISTA
V-domain Ig suppressor of T cell activation
Author contribution
JC and YX conceived and designed the study. LZ and MZ performed experiments and evaluated the results of immunohistochemical staining. LZ and YZ analyzed VISTA mRNA from TCGA database and performed statistical analyses. LZ prepared the first draft of the manuscript. All authors made substantial contributions to the review and revision of the manuscript, and approved the final manuscript.
Funding
This work was supported by Grants from the National Natural Science Foundation of China (Nos. 81772783 and 81971475) and the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (No. CAMS-2017-I2 M-1-002) to Yang Xiang.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Institutional Review Board of Peking Union Medical College Hospital (approval number S-K888).
Informed consent
For this type of study, informed consent was not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Song W, Rubinstein M, Liu D. Recent updates in cancer immunotherapy: a comprehensive review and perspective of the 2018 China Cancer Immunotherapy Workshop in Beijing. J Hematol Oncol. 2018;11(1):142. doi: 10.1186/s13045-018-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga A, Piha-Paul S, Ott PA, Mehnert JM, Berton-Rigaud D, Morosky A, Yang P, Ruman J, Matei D. Pembrolizumab in patients with programmed death ligand 1-positive advanced ovarian cancer: analysis of KEYNOTE-028. Gynecol Oncol. 2019;152(2):243–250. doi: 10.1016/j.ygyno.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Pakish JB, Jazaeri AA. Immunotherapy in gynecologic cancers: are we there yet? Curr Treat Options Oncol. 2017;18(10):59. doi: 10.1007/s11864-017-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, Lu LF, Gondek D, Wang Y, Fava RA, Fiser A, Almo S, Noelle RJ. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208(3):577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flies DB, Han X, Higuchi T, Zheng L, Sun J, Ye JJ, Chen L. Coinhibitory receptor PD-1H preferentially suppresses CD4(+) T cell-mediated immunity. J Clin Invest. 2014;124(5):1966–1975. doi: 10.1172/JCI74589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowak EC, Lines JL, Varn FS, Deng J, Sarde A, Mabaera R, Kuta A, Le Mercier I, Cheng C, Noelle RJ. Immunoregulatory functions of VISTA. Immunol Rev. 2017;276(1):66–79. doi: 10.1111/imr.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Yuan Y, Chen W, Putra J, Suriawinata AA, Schenk AD, Miller HE, Guleria I, Barth RJ, Huang YH, Wang L. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T cell responses. Proc Natl Acad Sci USA. 2015;112(21):6682–6687. doi: 10.1073/pnas.1420370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wu G, Manick B, Hernandez V, Renelt M, Erickson C, Guan J, Singh R, Rollins S, Solorz A, Bi M, Li J, Grabowski D, Dirkx J, Tracy C, Stuart T, Ellinghuysen C, Desmond D, Foster C, Kalabokis V. VSIG-3 as a ligand of VISTA inhibits human T cell function. Immunology. 2019;156(1):74–85. doi: 10.1111/imm.13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakavand H, Jackett LA, Menzies AM, Gide TN, Carlino MS, Saw RPM, Thompson JF, Wilmott JS, Long GV, Scolyer RA. Negative immune checkpoint regulation by VISTA: a mechanism of acquired resistance to anti-PD-1 therapy in metastatic melanoma patients. Mod Pathol. 2017;30(12):1666–1676. doi: 10.1038/modpathol.2017.89. [DOI] [PubMed] [Google Scholar]
- 11.Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, Zhao H, Chen J, Chen H, Efstathiou E, Troncoso P, Allison JP, Logothetis CJ, Wistuba II, Sepulveda MA, Sun J, Wargo J, Blando J, Sharma P. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blando J, Sharma A, Higa MG, Zhao H, Vence L, Yadav SS, Kim J, Sepulveda AM, Sharp M, Maitra A, Wargo J, Tetzlaff M, Broaddus R, Katz MHG, Varadhachary GR, Overman M, Wang H, Yee C, Bernatchez C, Iacobuzio-Donahue C, Basu S, Allison JP, Sharma P. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. Proc Natl Acad Sci USA. 2019 doi: 10.1073/pnas.1811067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zong L, Zhang M, Wang W, Wan X, Yang J, Xiang Y. PD-L1, B7-H3 and VISTA are highly expressed in gestational trophoblastic neoplasia. Histopathology. 2019;75(3):421–430. doi: 10.1111/his.13882. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Jia B, Claxton DF, Ehmann WC, Rybka WB, Mineishi S, Naik S, Khawaja MR, Sivik J, Han J, Hohl RJ, Zheng H. VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML. Oncoimmunology. 2018;7(9):e1469594. doi: 10.1080/2162402x.2018.1469594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boger C, Behrens HM, Kruger S, Rocken C. The novel negative checkpoint regulator VISTA is expressed in gastric carcinoma and associated with PD-L1/PD-1: a future perspective for a combined gastric cancer therapy? Oncoimmunology. 2017;6(4):e1293215. doi: 10.1080/2162402X.2017.1293215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villarroel-Espindola F, Yu X, Datar I, Mani N, Sanmamed M, Velcheti V, Syrigos K, Toki M, Zhao H, Chen L, Herbst RS, Schalper KA. spatially resolved and quantitative analysis of VISTA/PD-1H as a novel immunotherapy target in human non-small cell lung cancer. Clin Cancer Res. 2018;24(7):1562–1573. doi: 10.1158/1078-0432.ccr-17-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie S, Huang J, Qiao Q, Zang W, Hong S, Tan H, Dong C, Yang Z, Ni L. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol Immunother. 2018;67(11):1685–1694. doi: 10.1007/s00262-018-2227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Deng WW, Huang CF, Bu LL, Yu GT, Mao L, Zhang WF, Liu B, Sun ZJ. Expression of VISTA correlated with immunosuppression and synergized with CD8 to predict survival in human oral squamous cell carcinoma. Cancer Immunol Immunother. 2017;66(5):627–636. doi: 10.1007/s00262-017-1968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuklinski LF, Yan S, Li Z, Fisher JL, Cheng C, Noelle RJ, Angeles CV, Turk MJ, Ernstoff MS. VISTA expression on tumor-infiltrating inflammatory cells in primary cutaneous melanoma correlates with poor disease-specific survival. Cancer Immunol Immunother. 2018;67(7):1113–1121. doi: 10.1007/s00262-018-2169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulati K, Hamanishi J, Matsumura N, Chamoto K, Mise N, Abiko K, Baba T, Yamaguchi K, Horikawa N, Murakami R, Taki M, Budiman K, Zeng X, Hosoe Y, Azuma M, Konishi I, Mandai M. VISTA expressed in tumour cells regulates T cell function. Br J Cancer. 2019;120(1):115–127. doi: 10.1038/s41416-018-0313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Pang HJ, Zhao W, Li YF, Yan LX, Dong ZY, He XF. VISTA expression associated with CD8 confers a favorable immune microenvironment and better overall survival in hepatocellular carcinoma. BMC Cancer. 2018;18(1):511. doi: 10.1186/s12885-018-4435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Zong L, Wang J, Wan X, Feng F. Epithelioid trophoblastic tumors: treatments, outcomes, and potential therapeutic targets. J Cancer. 2019;10(1):11–19. doi: 10.7150/jca.28134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulangara K, Zhang N, Corigliano E, Guerrero L, Waldroup S, Jaiswal D, Ms MJ, Shah S, Hanks D, Wang J, Lunceford J, Savage MJ, Juco J, Emancipator K. Clinical utility of the combined positive score for programmed death ligand-1 expression and the approval of pembrolizumab for treatment of gastric cancer. Arch Pathol Lab Med. 2019;143(3):330–337. doi: 10.5858/arpa.2018-0043-OA. [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, Ceeraz S, Suriawinata AA, Yan S, Ernstoff MS, Noelle R. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74(7):1924–1932. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, Honjo T, Fujii S. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci USA. 2007;104(9):3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb JR, Milne K, Kroeger DR, Nelson BH. PD-L1 expression is associated with tumor-infiltrating T cells and favorable prognosis in high-grade serous ovarian cancer. Gynecol Oncol. 2016;141(2):293–302. doi: 10.1016/j.ygyno.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Mills AM, Peres LC, Meiss A, Ring KL, Modesitt SC, Abbott SE, Alberg AJ, Bandera EV, Barnholtz-Sloan J, Bondy ML, Cote ML, Funkhouser E, Moorman PG, Peters ES, Schwartz AG, Terry PD, Wallace K, Schildkraut JM. Targetable immune regulatory molecule expression in high-grade serous ovarian carcinomas in African American Women: a study of PD-L1 and IDO in 112 cases from the African American Cancer Epidemiology Study (AACES) Int J Gynecol Pathol. 2019;38(2):157–170. doi: 10.1097/pgp.0000000000000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.