Abstract
All viruses require strategies to inhibit or evade the immunity pathways of cells they infect. The viruses that infect bacteria, bacteriophages (phages), must avoid nucleic-acid targeting immune pathways such as CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes) and restriction-modification (R-M) systems to replicate efficiently1. Here, we show that jumbo phage ΦKZ, infecting Pseudomonas aeruginosa, segregates its DNA from immunity nucleases by constructing a proteinaceous nucleus-like compartment. ΦKZ resists many DNA-targeting immune systems in vivo, including two CRISPR-Cas3 subtypes, Cas9, Cas12a, and the restriction enzymes HsdRMS and EcoRI. Cas and restriction enzymes are unable to access the phage DNA throughout the infection, but engineered re-localization of EcoRI inside the compartment enables phage targeting and cell protection. Moreover, ΦKZ is sensitive to the RNA targeting CRISPR-Cas enzyme, Cas13a, likely due to phage mRNA localizing to the cytoplasm. Collectively, we propose that Pseudomonas jumbo phages evade a broad spectrum of DNA-targeting nucleases through the assembly of a protein barrier around their genome.
Jumbo phage ΦKZ Resists CRISPR-Cas targeting
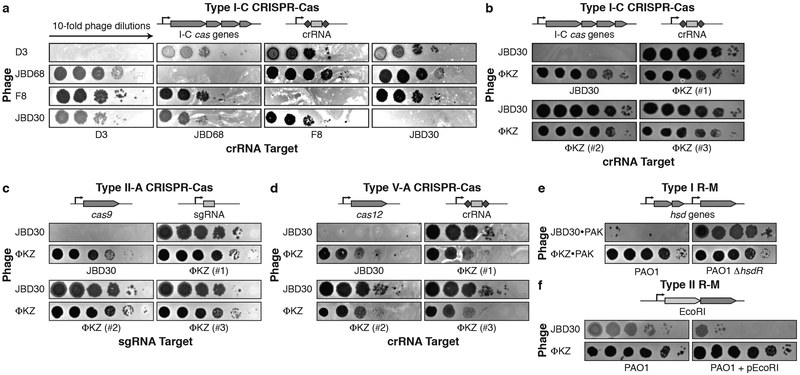
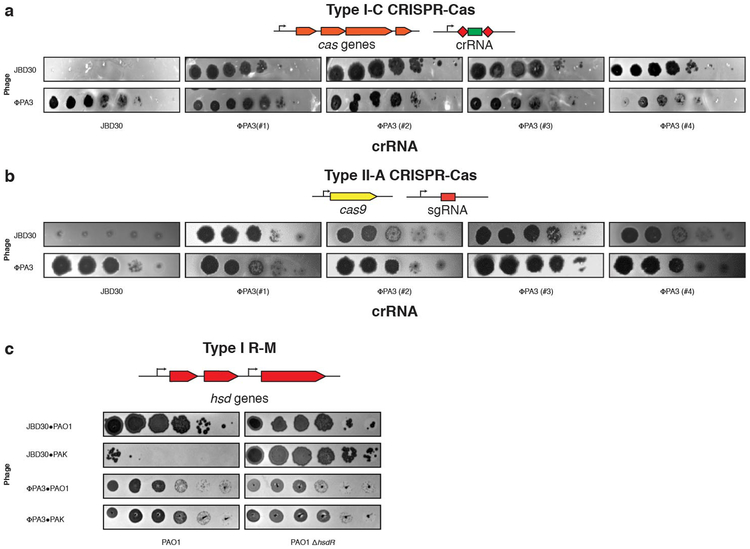
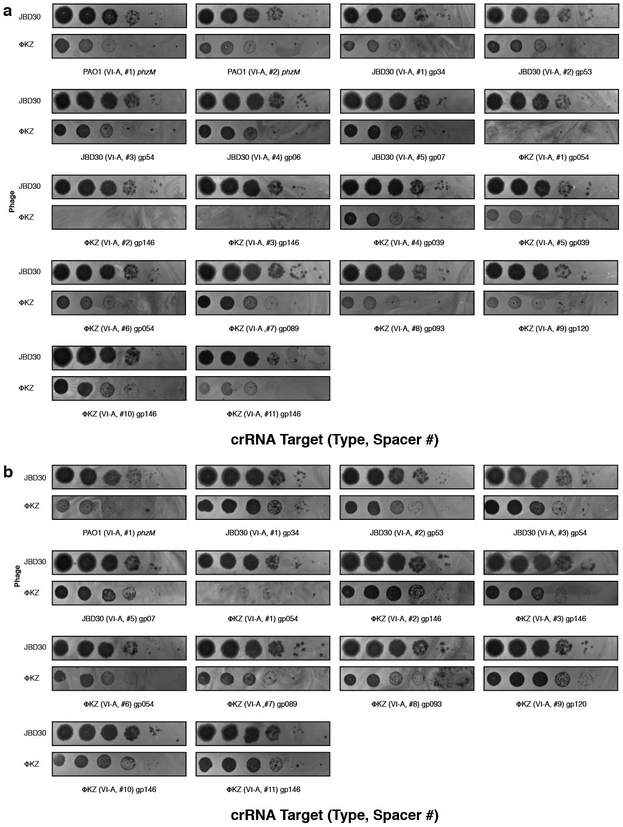
Phages that infect Pseudomonas aeruginosa can avoid CRISPR-mediated destruction by encoding “anti-CRISPR' (Acr) proteins that inhibit the Type I-E and I-F CRISPR-Cas systems2-4. To determine whether any P. aeruginosa phages are resistant to the P. aeruginosa Type I-C CRISPR-Cas system5, a common and understudied variant6, a strain engineered to express Type I-C cas3, cas5, cas8, cas7 7 was provided with a panel of crRNAs targetin phages from five taxonomic groups: JBD30, D3, and JBD68 (distinct temperate siphophages), F8 and ΦKZ (distinct lytic myophages). All phages succumbed to targeting (Fig. 1a), except ΦKZ (Fig. 1b) and a related phage ΦPA3 (Extended Data Fig. 1a). ΦKZ titer did not decrease when challenged using 11 different Type I-C crRNAs (Fig. 1b, Extended Data Fig. 2a) nor when exposed to the type I-F CRISPR-Cas system of P. aeruginosa8 (Extended Data Fig. 2b).
Figure 1: Identification of a phage that resists targeting by diverse CRISPR-Cas and restriction-modification systems.
Plaque assay with the indicated phage spotted in ten-fold serial diluations on a lawn of P. aeruginosa, dark clearings in the lawn represent phage replication. Strain PAO1 expressing: a, Type I-C cas genes (cas3-5-8-7) and crRNAs targeting the indicated phages. b, Type I-C cas genes and crRNAs targeting phage JBD30 or distinct phage ΦKZ-targeting crRNAs (#1-#3). c, Type II-A cas9 and distinct single guide RNAs (sgRNAs) targeting the indicated phage. d, Type V-A cas12a and distinct crRNAs against the indicated phage. e, Endogenous Type I R-M system (hsdRSM) in strain PAO1 was assayed using phages propagated on PAK (e.g. JBD30•PAK was first propagated on strain PAK). Together with an isogenic PAO1∆hsdR knockout. f, Type II EcoRI R-M system. Restriction activity was assayed using phages JBD30 and ΦKZ. All plaque assays replicated ≥ 2 times with similar results.
The ΦKZ genome possesses no homologs of known acr2-4,9,10 or anti-CRISPR associated (aca) genes that have previously enabled identification of acr genes4,9,10. Moreover, gene knockout approaches have not been previously established for ΦKZ. To determine the mechanism for CRISPR evasion, we attempted to utilize the Type II-A CRISPR-Cas9 (SpyCas9) as a genetic tool. SpyCas9 and sgRNAs robustly targeted control phage JBD30 but ΦKZ replication and associated cell lysis was unaffected (Fig. 1c). An additional six sgRNA sequences also failed to target ΦKZ (Extended Data Fig. 3a), as did four against ΦPA3 (Extended Data Fig. 1b). Given the ability of this phage to evade unrelated CRISPR systems (Type I and II), including one from a microbe that this phage does not infect (Streptococcus pyogenes), we considered that ΦKZ may be generally resistant to CRISPR-Cas immunity, as opposed to relying on specific inhibitor proteins. Type V-A Cas12a (Cpf1) CRISPR-Cas system from Moraxella bovoculi was expressed in P. aeruginosa and sucessfully targeted the control phage, but not ΦKZ with any of the nine crRNAs tested (Fig. 1d, Extended Data Fig. 3b). The ability of this phage to resist CRISPR systems found in its natural host, Pseudomonas (Type I-C and I-F), and those not naturally present (Type II-A and V-A) suggests that this phage possesses a mechanism enabling “pan-CRISPR” resistance.
Restriction-modification systems are the most common form of bacterial immunity in nature and pose a significant impediment to phage replication1. Type I R-M (HsdRMS from P. aeruginosa) and Type II R-M (EcoRI from E. coli) were next tested, possessing 24 and 184 cut sites in the ΦKZ genome, respectively. ΦKZ was propagated on strain PAK, an isolate that generates phages susceptible to PAO1 HsdRMS restriction. When phage JBD30 (5 Type I R-M sites) was assayed in this manner, its titer was reduced by ~5 orders of magnitude, dependent on hsdR (Fig. 1e). In contrast, no restriction was observed for ΦKZ (Fig. 1e) or ΦPA3 (Extended Data Fig. 1c). Similarly, the expression of EcoRI reduced JBD30 titer by 3 orders of magnitude (12 EcoRI sites) but had no impact on ΦKZ (184 EcoRI sites) (Fig. 1f). Together, these experiments demonstrate that ΦKZ is refractory to the six selected CRISPR-Cas and restriction endonucleases in vivo.
Phage nucleus-like structure excludes immune enzymes
ΦKZ and ΦKZ-like phages infecting P. aeruginosa and P. chlororaphis were recently shown to construct an elaborate proteinaceous nucleus-like compartment where phage DNA replicates11,12. Additionally, a phage-encoded tubulin homologue, PhuZ, centers the compartment within the host cell11-15. Proteins involved in DNA replication, transcription, and recombination localize inside the shell, while mRNA and proteins mediating translation localize in the cytoplasmic space11, akin to the eukaryotic nucleus. Given the apparent exclusion of select proteins, we considered whether this structure was responsible for the pan-resistance of ΦKZ to such distinct immune processes.
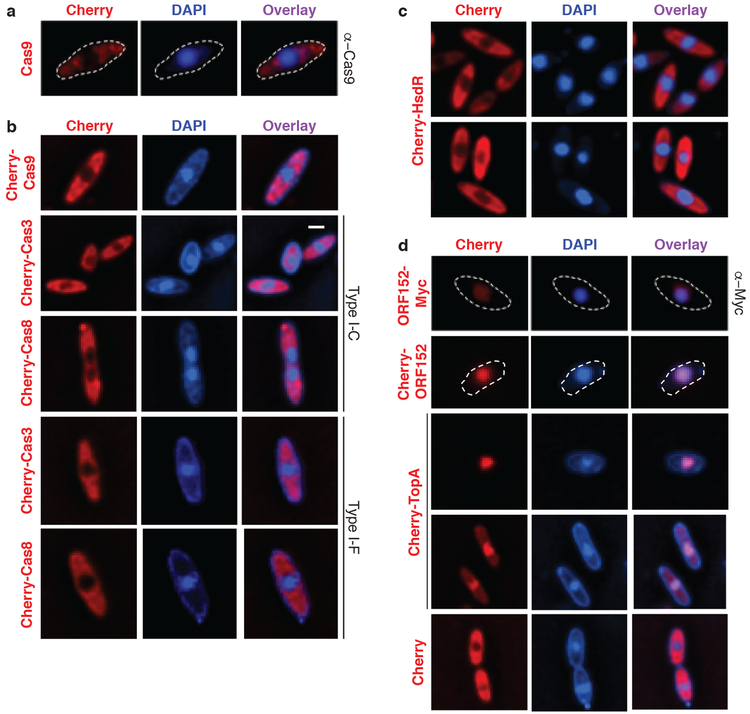
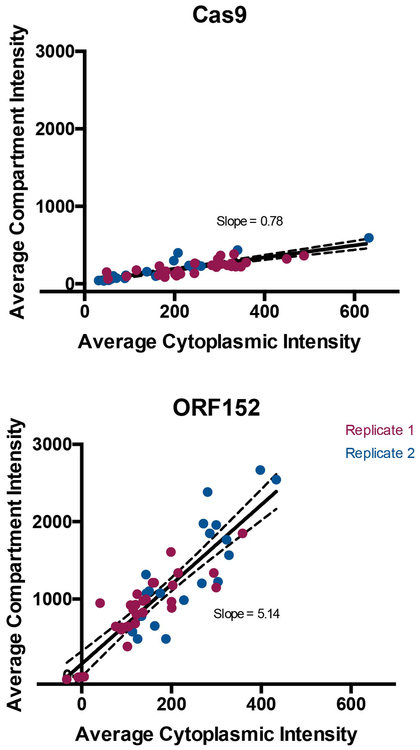
P. aeruginosa cells infected with ΦKZ were imaged with immunofluorescence to detect Cas9 (Fig. 2a, Extended Data Fig. 4); likewise, Cherry fusions with Cas9, two signature proteins from the Type I-C and I-F CRISPR-Cas systems (Cas8 and Cas3, Fig. 2b), as well as the restriction enzyme HsdR (Fig. 2c) were imaged using live cell imaging. These experiments revealed that the immune enzymes are excluded from the shell during phage infection. DAPI staining revealed the phage DNA inside the shell, while the host genome was degraded14. Proteins previously shown to be internalized in the shell, phage ORF152 (imaged with anti-myc immunofluorescence and Cherry fusion) and host Topoisomerase I (Cherry fusion) co-localized with the DAPI signal, while Cherry was excluded (Fig. 2d). Although the rules for protein internalization in the shell are currently unknown, each protein of known function that localizes inside of the shell interacts with DNA11,12 (i.e. DNA replication and transcription machinery), suggesting that the exclusion of DNA-binding Cas and restriction proteins is an adaptive function of the shell.
Figure 2: CRISPR-Cas and restriction proteins are excluded from ΦKZ’s nucleus-like structure.
a, Fluorescence microscopy of P. aeruginosa immunostained for Cas9, DAPI stain shows the phage DNA within the nucleus-like structure. Live fluorescence microscopy of P. aeruginosa strains engineered to express b, II-A Cas9 or I-C or I-F Cas8 or Cas3 proteins fused to Cherry, c, a Cherry-HsdR fusion, d, Immunostained for Myc-ORF152 (top panels), or live imaging of ORF152 and TopA proteins fused to Cherry, or Cherry alone. All experiments were replicated ≥ 2 times with similar results.
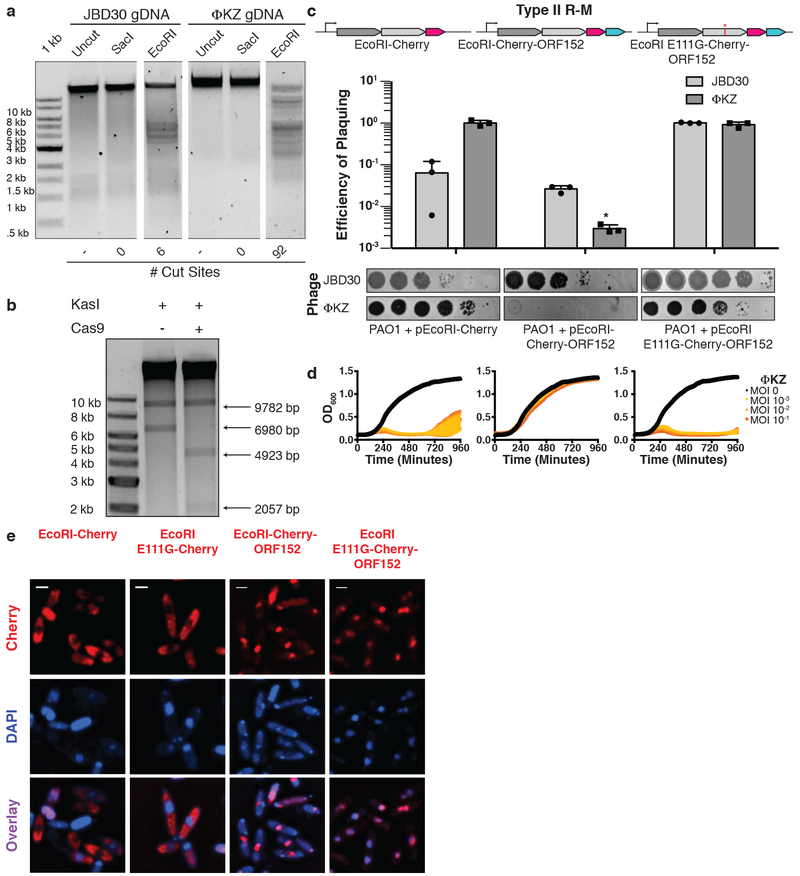
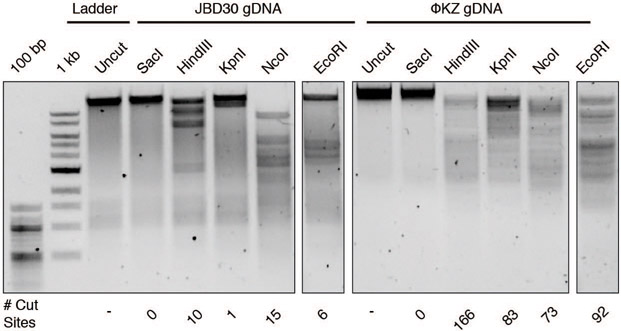
To confirm that the ΦKZ phage genome could be a substrate for DNA cleavage, if accessed, two enzymes that failed in vivo, EcoRI and Cas9, were assayed in vitro. ΦKZ DNA was extracted from virions and subjected to restriction digestion reactions with a panel of restriction enzymes. EcoRI, HindIII, KpnI, and NcoI each cleaved the DNA, while SacI lacks a sequence recognition motif in the ΦKZ genome and did not (Fig. 3a, Extended Data Fig. 5a). Cas9’s ability to cleave ΦKZ gDNA in vitro was next assessed. Due to the large size of the ΦKZ genome (280 kbp), we first subjected purified phage DNA to the restriction enzyme KasI to liberate a 6.98 kbp product and then cleaved that species with a dual crRNA:tracrRNA-loaded SpyCas9 nuclease in vitro. This reaction depleted the substrate, liberating the expected 4.9 kb and 2.1 kb fragments, confirming that the phage gDNA is sensitive to Cas9 cleavage (Fig. 3b). Together these results demonstrate that immune enzymes are capable of cleaving ΦKZ gDNA when accessed and that immune evasion is likely not due to an intrinsic feature of the phage DNA, such as base modifcations that can impede Cascade-Cas3, Cas9, and EcoRI16-19.
Figure 3: ΦKZ genomic DNA can be cleaved by immune enzymes.
a, ΦKZ and JBD30 genomic DNA digested with the indicated restriction enzymes in vitro. The first lane contains a 1 kb DNA ladder. The number of cut sites for each enzyme is shown at the bottom of the gels. Products were visualized on a 0.7% agarose gel, visualized with SYBR Safe nucleic acid stain. b, ΦKZ phage genomic DNA digested in vitro using KasI, and incubated with and without Cas9 loaded with crRNA:tracrRNA targeting the fragment liberated by KasI. Products were visualized on a 0.7% agarose gel, visualized with SYBR Safe nucleic acid stain. c, Strain PAO1 expressing EcoRI-Cherry, EcoRI-Cherry-ORF152, or EcoRI E111G-Cherry-ORF152 fusion protein. Plaque assays were conducted as in Fig. 1a and quantified (n=3). Mean values are plotted as bars with error bars representing standard deviation. A t-test comparing ΦKZ EOP on PAO1 pEcoRI-Cherry to PAO1 pEcoRI-Cherry-ORF152 yielded a p-value of 2.88×10−4. d, Growth curves monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). e, Live fluorescence imaging of infected P. aeruginosa strains engineered to express EcoRI-Cherry, EcoRI E111G-Cherry, EcoRI-Cherry-ORF152 or EcoRI E111G-Cherry-ORF152. DAPI stain shows the phage DNA. Cherry shows EcoRI fusion protein. In vitro digestion experiments a and b replicated twice with similar results. Plaque assays (c), growth curves (d), and microscopy (e) were replicated ≥ 3 times.
Immune enzyme re-localization enables phage targeting
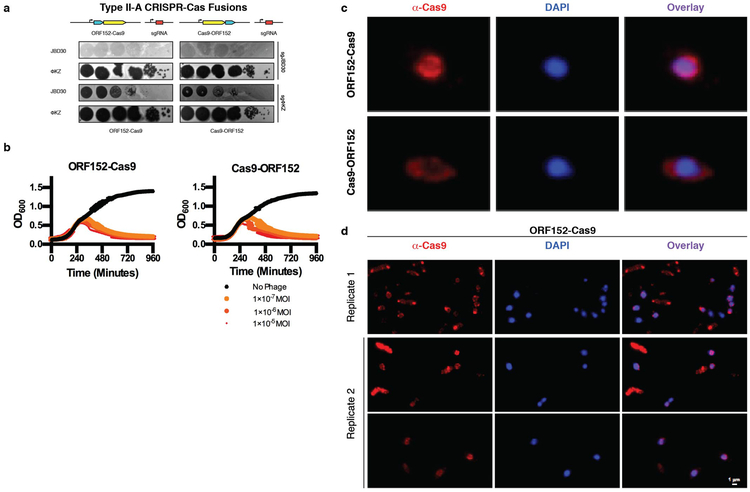
Due to a current inability to mutate, weaken, or knockout the shell structure, we next sought to enable an immunity enzyme to bypass the shell to access phage DNA in vivo. The single effector enzyme Cas9 was fused to ORF152, a phage-encoded RecA-like protein that is internalized within the shell11,12. Independent fusions to the N- or C- terminus did not affect ΦKZ replication, however (Extended Data Fig. 6a, 6b). Imaging of one fusion orientation (ORF152-Cas9) revealed peri-shell localization (Extended Data Fig. 6c, 6d), suggesting that the fusion redirected Cas9 from its previously diffuse state, but perhaps the large Cas9 protein (1,368 amino acids, 158 kDa) was unable to traverse the shell border.
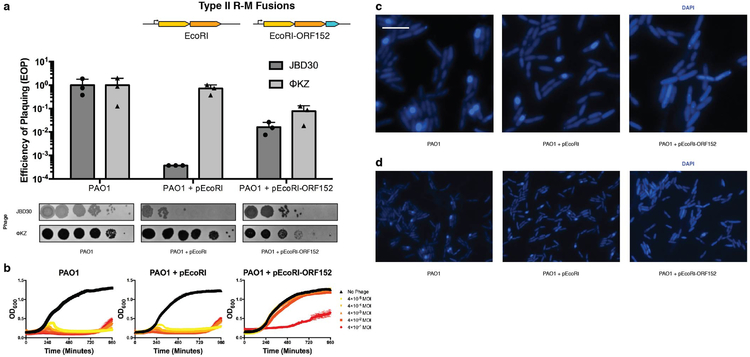
Given the large size (e.g. Cas12a) and complexity (e.g. Cascade, HsdRMS) of each of the other immune systems, we next fused the small, single effector protein EcoRI (278 amino acids, 31.5 kDa) to Cherry-ORF152. This fusion resulted in a notable >2.5 order of magnitude reduction in ΦKZ phage titer and markedly reduced plaque sizes (Fig. 3c), the first successful in vivo DNA targeting observed in this study. Liquid infections revealed that cells expressing EcoRI-Cherry-ORF152 were extremely well protected, gaining >5 orders of magnitude resistance to phage-induced lysis (Fig. 3d). A catalytic mutant EcoRI(E111G) fused to Cherry-ORF152 displayed no immune activity against ΦKZ in either assay, nor did active EcoRI-Cherry without the ORF152 fusion (Fig. 3c, 3d). Similar results were observed in the absence of the Cherry tag (i.e. EcoRI-ORF152), however ΦKZ targeting was more modest in that case, reducing titers by ~10-fold and protecting cells by a factor of ~104 (Extended Data Fig. 7). Imaging of infected cells expressing EcoRI-Cherry or EcoRI(E111G)-Cherry demonstrated that these proteins are excluded from the shell, while EcoRI(E111G)-Cherry-ORF152 is successfully localized inside (Fig. 3e, Supplementary Video 1 for time lapse images). EcoRI-Cherry-ORF152 impaired the ability of infected cells to form full shells and proceed through the infection process, and the EcoRI fusion protein was often localized within or adjacent to the DAPI-stained puncta (Fig. 3e, Supplementary Video 2). Some host DNA can be seen in these cells and infection does not proceed (Fig. 3e, Extended Data Fig. 7c, 7d). By bypassing the physical barrier of the shell using rational engineering, we conclude that the shell is the cause of resistance to immunity displayed by jumbo phage ΦKZ.
Phage mRNA is sensitive to Cas13a
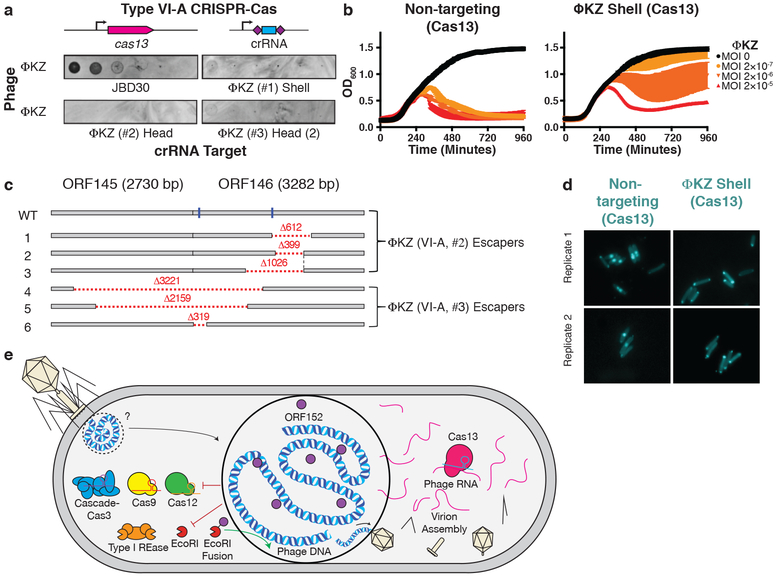
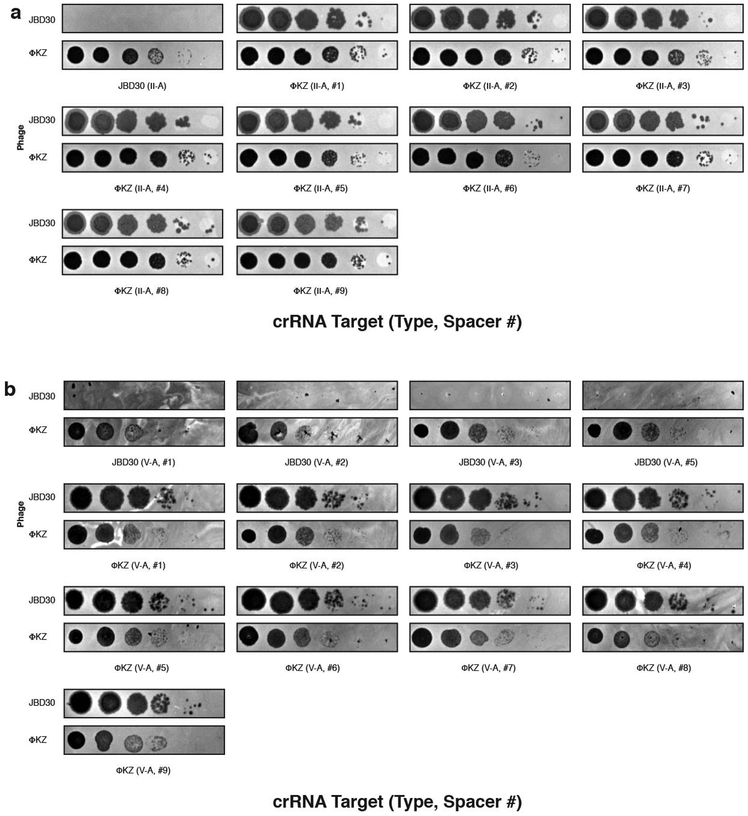
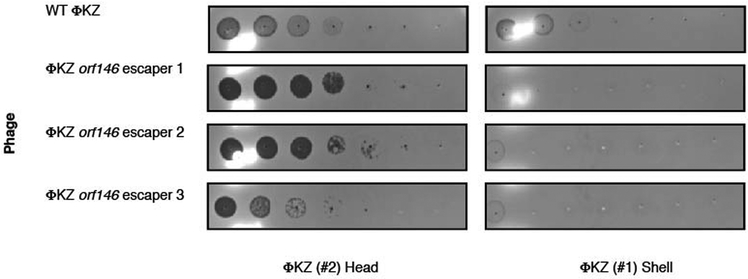
The nucleus-like structure produced by ΦKZ provides robust resistance to DNA-targeting immune systems, but natural immune systems may exist that can evade this mechanism. We envisaged that the mRNA exported for translation in the cytoplasm is likely susceptible to targeting and that a ribonuclease could provide anti-ΦKZ immunity. To test this, we adapted the Type VI-A CRISPR RNA-guided RNA nuclease LseCas13a20,21 for phage targeting in P. aeruginosa. Three LseCas13a spacers (two targeting head gene gp146 and one targeting the shell gene gp054) decreased ΦKZ plaquing efficiency by >106-fold (Fig. 4a, Extended Data Fig. 8). Corroborating the plaquing results, LseCas13a was also effective at protecting P. aeruginosa in liquid cultures, with 1–2 orders of magnitude resistance to phage induced lysis (Fig. 4b). We suspect that the previously demonstrated collateral damage caused by Cas13a22 is detrimental to cell fitness, leading to a modest enhancement to the growth curves compared to EcoRI targeting. At a frequency of ~10-6–10-7, Cas13 escaper phages were identified (Extended Data Fig. 9) and surprisingly contained genomic deletions at the site of targeting (Fig. 4c). Notably, all deletions were in-frame and 5/6 escapers had clear microhomology regions ranging from 7–21 bp flanking the deletion. One guide targeting the 5ʹ end of orf146 selected for deletions where the upstream gene (orf145) became fused, in frame, to the targeted gene (Fig. 4c). These data support the conclusion that sequence-specific CRISPR-Cas RNA targeting can inhibit this phage.
Figure 4: Phage ΦKZ DNA is sensitive to RNA-targeting Cas13.
a, PAO1 expressing LseCas13a and a crRNA targeting phage ΦKZ. Plaque assays were conducted as in Fig. 1a. b, Growth curves measuring the OD600 of PAO1 infected with the indicated ΦKZ MOI. c, Cas13 escaper ΦKZ phage mutant deletions (red dashed lines) with target sites indicated (blue line). d, Live fluorescence imaging of P. aeruginosa strains engineered to express LseCas13a and crRNAs targeting ΦKZ. Cyan stain shows the phage DNA. e, Model summarizing the ΦKZ nucleus-like structure excluding (flat arrow) Cas9, Cas12, Cascade-Cas3 (Type I-C, Type I-F) and Type I restriction endonucleases (REase) and Type II REase, while the mRNA (red) is exported and can be targeted by Cas13. The nucleus-like structure is resistant to the indicated nucleases, but EcoRI fusion (to internal protein ORF152) enables targeting. Cas13 plaque assays a were replicated >3 times with similar results. Growth curve experiments b were replicated twice with similar results. Escaper mutants c were isolated once and verified by PCR, sequencing, and plaque assays. Microscopy d was replicated twice with similar results.
Prior to shell construction, it is unknown what the state of the phage DNA is. During Cas13a targeting of the shell mRNA, imaging revealed that infections arrested before the phage DNA proceeded from its injection site at the poles. Most cells had two polar puncta, even one hour after infection (Fig. 4d). The absence of phage DNA diffusion or clearance (e.g. by the endogenous Type I R-M system) suggests that the injected phage genome may be protected by a yet unknown mechanism involving injected, rapidly synthesized, or pre-existing host factors, prior to shell assembly. This is additionally corroborated by observations in the absence of Cas13a arrest, where we have observed DAPI-stained phage DNA adjacent to: i) EcoRI-Cherry-ORF152 puncta (Fig. 3e) or ii) to a nascent shell localizing TopA (Extended Data Fig. 10). The molecular nature and mechanisms underlying this early protection event remain to be elucidated.
Discussion
The assembly of a proteinaceous compartment to house the replicating phage DNA creates a physical protective barrier resulting in the resistance of phage ΦKZ to DNA-cleaving enzymes (Fig. 4e). Evasion of endogenous P. aeruginosa Type I CRISPR-Cas systems by ΦKZ suggests that these jumbo phages are likely Type I CRISPR-Cas resistant in nature. Supporting this hypothesis, our analysis of >4,000 non-redundant P. aeruginosa spacers (Type I-C, I-E, and I-F) reported by van Belkum et al. (2015) found no spacers against ΦKZ or its jumbo phage relatives ΦPA3, PaBG, KTN4, and PA7 (Table 1). This is in contrast to the many spacer matches from each system matching diverse P. aeruginosa phages, such as those assayed in our screen (Fig. 1) and those encoding anti-CRISPR proteins. Additionally, given the efficacy of the RNA-targeting CRISPR-Cas13 system, we propose that Type VI CRISPR systems are well-suited to target the mRNA of DNA phages when the DNA is inaccessible (i.e. due to base modifications or physical segregation).
Table 1:
ΦKZ and ΦKZ-like phages have no natural spacers matching their genomes from a natural collection of >4000 P. aeruginosa spacers.
| Phage | # spacers | Type I CRISPR sensitivity | Reference |
|---|---|---|---|
| DMS3m | 75 | I-C: sensitive I-E: resistant (AcrIE3) I-F: sensitive |
This study, ref.3,8 |
| JBD30 | 51 | I-C: sensitive I-E: sensitive I-F: resistant (AcrIF1) |
This study, ref. 2,3 |
| JBD18 | 51 | I-F: sensitive | Ref. 8 |
| JBD25 | 46 | I-F: sensitive | Ref. 8 |
| JBD68 | 28 | I-C: sensitive | This study |
| D3 | 49 | I-C: sensitive | This study |
| F8 | 3* | I-C: sensitive | This study |
| ΦKZ | 0 | I-C: resistant I-F: resistant |
This study |
| phiPA3 | 0 | I-C: resistant | This study |
| PaBG | 0 | not assayed | |
| KTN4 | 0 | not assayed | |
| PA7 | 0 | not assayed |
The total number of Type I CRISPR spacers with a perfect match to the indicated P. aeruginosa phages assayed in this study and previous CRISPR-Cas studies. The experimental sensitivity of each phage to the indicated subtypes are shown. AcrIE3 and AcrIF1 are I-E and I-F anti-CRISPR proteins, respectively.
indicates that all spacers have mismatches (≤4) to the F8 genome.
This phage compartment has only been documented among the jumbo phages of Pseudomonas11,12, however, we consider that physical occlusion of phage genomic DNA through this and other mechanisms may comprise a novel route to immune system evasion by phages. Indeed, recently discovered “mega phages” were reported to encode homologues of phage tubulin, which centers this compartment during infection23. The pan-resistance of ΦKZ to DNA-targeting enzymes provides an explanation for the elaborate and impressive nucleus-like structure. Furthermore, the polar localization of the injected phage DNA during mRNA targeting suggests a poorly understood early protective mechanism. Considering the pronounced resistance to overexpressed immune enzymes and the previously observed docking of capsids at the shell periphery11,24, we propose that the phage DNA is never exposed to the cytoplasm (Fig. 4e). Other hypotheses to explain the importance of the shell remain to be addressed, including protection from phage-derived nucleases that degrade the bacterial genome and spatial organization of the large phage genome during replication and packaging. Regardless, we conclude that the phage-assembled nucleus-like structure provides a strong protective barrier to DNA-targeting immune pathways.
Methods
Bacterial growth and genetic manipulation
Strains, plasmids, phages, and spacer sequences used in this study are listed in Supplementary Information. Pseudomonas aeruginosa strain PAO1 was grown in LB at 37 °C with aeration at 225 RPM. When necessary, plating was performed on LB agar with carbenicillin (250 μg/ml) or gentamicin (50 μg/ml). Gene expression was induced by the addition of L-arabinose (0.1% final) and/or isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.5 mM or 1 mM final). For chromosomal insertions at the attTn7 locus, P. aeruginosa cells were electroporated with the integrating vector pUC18T-lac and the transposase expressing helper plasmid pTNS3, and selected on gentamicin. Potential integrants were screened by colony PCR with primers PTn7R and PglmS-down. Electrocompetent cell preparations, transformations, integrations, selections, plasmid curing, and FLP recombinase mediated marker excision with pFLP were performed as described previously25.
Phage growth and DNA extraction
Phage growth was conducted in LB at 37 °C with PAO1 as a host. Growth curves were conducted in a Biotek Synergy plate reader at 37 °C with orbital shaking set to maximum speed. Phage stocks were diluted and stored in SM buffer8 and used for routine plaquing assays. Plaque assays were conducted at 37 °C with 20 mL of bottom agar containing 10 mM MgSO4 and 0.35% or 0.7% top agar (often both concentrations were used in parallel) also containing 10 mM MgSO4 and any inducer molecules. 3 μL spots were applied to the top agar after it had been poured and solidified. For high titer lysates to generate phage DNA, plates with a high number of plaques were flooded with SM buffer and collected8. The lysates were subsequently DNase treated. Phage DNA was extracted with the Wizard Genomic DNA Purification Kit (Promega). DNA restriction assays were performed according to standard NEB protocols and restriction fragments were assessed by agarose gel electrophoresis.
Type I-C CRISPR-Cas system assay and expression in P. aeruginosa PAO1
Type I-C CRISPR-Cas function was tested by electroporating a strain that naturally contains I-C cas genes (strain: F11) with pHERD30T plasmids encoding crRNAs that target phages. To express this system heterologously in PAO1, the four effector cas genes (cas3–5-8–7) were cloned into pUC18T-lac and inserted in the PAO1 chromosome as described above. After removal of the gentamicin marker, this strain was electroporated with the same pHERD30T crRNA-encoding plasmids to confirm function upon IPTG/arabinose induction.
Type I-F CRISPR-Cas system expression in P. aeruginosa PAO1
To express the Type I-F system heterologously in PAO1, all I-F cas genes (cas1, cas3, csy1–4) were cloned into pMMBHE plasmid and transformed into PAO1. Subsequently, this strain was electroporated with the pHERD30T crRNA-encoding plasmids to confirm function upon IPTG/arabinose induction. To maintain pHERD30T and pMMBHE in the same strain of P. aeruginosa, double selection of 30 μg/mL gentamicin and 100 μg/mL carbenicillin was employed.
crRNA cloning and expression
All crRNAs used here, were cloned into established entry vectors in the pHERD30T background. After removing a pre-existing BsaI cut site in the vector by mutagenesis, a pseudo-CRISPR array (i.e. repeat-spacer-repeat for Type I, V, VI, or a sgRNA scaffold for Type II) was then cloned into the vector, where the spacing sequence possessed two inverted BsaI digest sites, to facilitate scarless cloning of new spacers. Desired spacer sequences were chosen randomly across the phage genome (with the correct PAM sequence for the cognate CRISPR-Cas system) and ordered as two complementary oligonucleotides that generate sticky ends when annealed, to be cloned into the entry vector, which was BsaI digested. Spacer oligonucleotides were PNK treated, annealed, and ligated into the entry vector.
Streptococcus pyogenes (Spy) Cas9 and sgRNA expression in P. aeruginosa
The S. pyogenes Cas9 gene was cloned into a pUC18T-Ara integration vector and then inserted into the attTn7 locus of PAO1. A single guide RNA scaffold was constructed based on a previous design26 with internal BsaI cut sites to enable insertion of pre-annealed oligos for scarless sgRNA design. This sgRNA scaffold was amplified with primers p30T-gRNA_BsaI and p30T-gRNA_BsaI_rev. The resulting product was inserted into the pHERD30T vector via Gibson assembly following backbone (pJW1) amplification by inverse PCR with primers gRNA_BsaI-p30T and gRNA_BsaI-p30T-rev. The sgRNA scaffold was positioned into pJW1 so that following BsaI cleavage the spacer insert +1 position would conincide with the pBAD TSS +1 position. The resulting plasmid, pJB1, was BsaI digested (NEB) followed by ligation of indicated pre-annealed oligos. Supplemental Table 3 contains a complete list of all target sequences. The sequence of the sgRNA construct with BsaI site locations is shown in Supplemental Table 3.
Cas9 in vitro cleavage
Cas9-based phage genome cleavage in vitro was conducted with purified Cas9 protein (NEB #M0386S), and the Cas9-gRNA-tracrRNA based cleavage reaction was then performed according to the manufacturer’s (NEB) instructions. Cas9 crRNAs (Supplemental Table 3) were ordered as Alt-R CRISPR-Cas9 crRNAs from IDT and utilized without further modification. The tracrRNA was amplified using primers tracrRNA-FOR and tracrRNA-REV from a plasmid (pBR62). The tracrRNA was produced through a T7 RNAP reactions using dsDNA encoding the tracrRNA downstream of a T7 RNAP promoter. Cas9 protein (NEB) was combined with pre-annealed crRNA and tracRNA complex at a 1:1 molar ratio. For targeting KasI-liberated ΦKZ genomic DNA, 500 ng of ΦKZ DNA was co-incubated with Cas9 RNP, which would cleave at pos. 183,270, and KasI in NEB buffer 3.1 for 1 hour. After stopping the reaction by proteinase K treatment, products were assessed by agarose gel electrophoresis. For Cas9 digestion of whole DNA, the reaction was performed at 37 °C for 4 hrs with 300 ng of ΦKZ or DMS3 genomic DNA and the products were assessed by agarose gel electrophoresis. Two Cas9 guides were selected that would cleave at pos. 158,649 and 168,715 of the ΦKZ genome to liberate an ~10 kb fragment.
Cas12a and crRNA design for PA expression
The humanized allele of the cpf1 gene of Moraxella bovoculli (MBO_03467, KDN25524.1) was sub-cloned from pTE4495 (Addgene) into pUC18T-lac using primers pUC_cpf1_F and pUC_cpf1_R and inserted in the PAO1 chromosome as described above. A Cpf1 repeat-spacer-repeat pseudo-CRISPR array was synthesized as oligonucleotides, annealed, and ligated into a pHERD30T vector, digested with NcoI and HindIII. Spacer sequences were cloned into the resulting vector (pJB2) following BsaI digestion and ligation of pre-annealed spacer oligonucleotide pairs.
Cas13a and crRNA design for PA expression
The wild type allele of the cas13 gene of Listeria seeligeri and Leptotrichia shahii were sub-cloned from p2CT-His-MBP-Lse_C2c2_WT and p2CT-His-MBP-Lsh_C2c2_WT (Addgene) into pUC18T-lac. LseCas13 and Lsh Cas13 were inserted in the PAO1 chromosome as described above. An Lse and an Lsh Cas13a repeat-spacer-repeat pseudo-CRISPR array were synthesized as oligonucleotides, annealed, and ligated into a pHERD30T vector, digested with NcoI and EcoRI. Spacer sequences were cloned into the resulting vectors (pSDM057 and pSDM070, respectively) following BsaI digestion and ligation of pre-annealed spacer oligonucleotide pairs. crRNA expression vectors were introduced into PAO1 tn7::cas13Lse and PAO1 tn7::cas13Lsh. The resulting strains were were assayed for phage sensitivity under standard phage plaing conditions, with induction of both Cas13 and the gRNAs (50 μg/mL gentamicin, 0.1% (L)-arabinose, 1 mM IPTG).
EcoRI expression in P. aeruginosa
The wild type allele of the M.EcoRI (Methyltransferase) and R.EcoRI (Endonuclease) genes were sub-cloned from pSB1A3 EcoRI Methylase-AmilCP and pSB1A3 EcoRI-RTX with EcoRI Methylase-AmilCP (Addgene plasmid # 85166, Addgene plasmid # 85165)p2CT-His-MBP-Lse_C2c2_WT and p2CT-His-MBP-Lsh_C2c2_WT (Addgene) into pHERD30T using Gibson assembly. The resulting plasmids, pSDM160 and pSDM161, were electroporated into PAOI ΔhsdR (SDM020). The resulting strains were were assayed for phage sensitivity under standard phage plaing conditions, with induction of both M.EcoRI (for genome protection) and R.EcoRI (50 μg/mL gentamicin, 0.1% (L)-arabinose).
Restriction-Modification Assay
PAO1, PAK, and PAO1 ΔhsdR were grown to saturation in LB at 37 °C. 4 mL of 0.7% agar, 10 mM MgSO4 molten top agar were seeded with 100 μL saturated culture and spread on 20 mL 10 mM MgSO4 LB agar plates. 2.5 μL 10-fold serial dilutions of bacteriophage JBD30 and ΦKZ propagated on strain PAO1 and PAK were spotted on plates. Plates were incubated at 37°C overnight and were imaged the following day.
Chromosomal Knockout of hsdR
To delete hsdR (NP_251422.1) from the PAO1 genome, a PAO1 strain expressing Cas9 from S. pyogenes was programmed to express a sgRNA against hsdR: GCCCTCATCGAAGAAACCAG. Additionally, the pHERD30T plasmid expressing this sgRNA was engineered to carry a repair template. This repair template consisted of the 500 bp upstream of hsdR and the 500 bp downstream of hsdR directly enjoined to one another. Induction of Cas9 and sgRNA led to cellular toxicity, as measured by OD600 (data not shown). Survivors of Cas9-mediated targeting were isolated. The hsdR locus of survivors was then amplified by colony PCR with primers binding outside of the region encompassed by the repair template. Products were resolved by gel electrophoresis in a 1.0% agarose TAE gel at 100 V and visualized using SYBR safe DNA stain. Amplicons of a reduced size were then sequenced, confirming the chromosomal deletion of hsdR. A clone with the correct deletion, SDM020, was chosen for downstream experiments.
Construction of fusion proteins
Plasmids expressing cherry alone or cherry tagged with cas3, cas8 (of Type I-C and Type I-F system) and TopA were constructed by Gibson assembly in pHERD30T plasmid digested with SacI and PstI. These fusions have ggaggcggtggagcc (G-G-G-G-A) linker sequence in between them. Cherry was amplified from SF-pSFFV-sfCherryFL1M3_TagBFP (kindly provide by Bo Huang lab, UCSF). cas3, cas8 of the Type I-C and Type I-F systems were amplified from LL77 and PA14 respectively. topA was amplified from gDNA of PAO1. All the primers used for the construction of the plasmids are listed in Supplemental Table 3.
Plasmids expressing cherry-Cas9 (pESN28) (primers: prESN74, prESN75, prESN76, prESN77) and Cherry-Cas9-ORF152 (pESN29) (prESN80, prESN81, prESN82, prESN83) were constructed using Gibson Assembly in the pHERD30T plasmid. Cherry-ORF152 (pESN32) (prESN91, prESN92) was constructed by PCR amplification to omit Cas9 and ligated using a KLD reaction. An sgaaaaggsqk linker connects Cherry to the other proteins. A ggggs linker connects Cas9 and ORF152. The plasmid expressing cMyc-ORF152 (prESN153, prESN154) was constructed. All plasmids were designed using SnapGene.
To make translational fusions of proteins, desired gene fragments including Cas9, M.EcoRI, R.EcoRI, ORF152 (NP_803718.1), and sfCherry2 were amplified by PCR from templates pHERD30T::Cas9, pSB1A3::M.EcoRI-AmilCP, pSB1A3::M.EcoRI-AmilCP;R.EcoRI-RTX, ΦKZ genomic DNA, and pHERD30T::sfCherry2, respectively. pHERD30T was linearized by restriction digest with SacI-HF and PstI-HF or by PCR. PCR primers included 15–30 bp overhangs for Gibson assembly. Overhangs enjoining genes included linkers GGGGS or GGSGGS. PCRs were treated with DpnI to eliminate template DNA. All PCRs and restriction digests were purified using a PCR clean up kit (Zymo Research DNA Clean and Concentrator Kits). Linearized vector and gene fragments treated with Gibson assembly. Gibson assembly reactions were transformed into competent E. coli. Plasmid products were isolated by miniprep and submitted for sequencing. Correct assemblies were electroporated into appropriate P. aeruginosa strains.
Cas13 escaper isolation
For identifying escapers of Cas13a RNA-targeting, 3 μl of high concentration ϕKZ lysates were mixed with 150 μl of overnight cultures of SDM078, SDM109 or SDM107 (strains expressing Cas13a and a gRNA against ϕKZ) . After incubating at 37 °C for 10 minutes, samples were mixed with 4 mL of 0.7% agar, 10 mM MgSO4, 1 mM IPTG and 0.1% arabinose and plated in LB agar plates with Gentamicin (50 μg/ml). After overnight incubation, plates were examined for the presence of escaper plaques. Escapers were formed in SDM078 and SDM109 but not in SDM107. 10 phages that escaped targeting from SDM078 and SDM109 were purified and the protospacer locus was amplified using PCR and subsequently sequenced. Six unique outcomes were identified and shown in Figure 4.
Growth curve experiments:
Growth curve experiments were carried out in a plate reader as in Borges AL et al 2018 Cell. Briefly, cells were diluted 1:100 from a saturated overnight culture with 10 mM MgSO4 and antibiotics and inducers, as appropriate. 140 μL of diluted culture was added, together with 10 μL of phage to wells in a 96-well plate. This plate was cultured with maximum double orbital rotation at 37 °C for 24 hours with OD600 measurements every 5 minutes
Immunofluorescence
Sample Growth
5 mL overnight cultures of a strain expressing Cas9 and an sgRNA targeting ΦKZ (SDM065) and a strain expressing cMyc-ORF152 (bESN27) were grown at 30 °C in LB media with gentamicin. A 1:30 back-dilution of the overnight culture into LB was grown at 30 °C for 1 h. Protein and guide expression was induced with 0.1% arabinose and 0.5 mM IPTG, respectively. After 1 h of expression, an aliquot of uninfected cells was fixed while the remaining cultures were infected with ΦKZ using MOI 1.5. Infected cell aliquots were collected and fixed at 60 mpi.
Fixation
This protocol was adapted from ref. 27. Samples were fixed with 5X Fix Solution (12.5% paraformaldehyde, 150 mM KPO4, pH 7.2) and incubating for 15 minutes at room temperature followed by 20 minutes on ice. Samples were then washed in PBS 3 times and finally resuspended in GTE (50 mM glucose, 10 mM EDTA, pH 8.0, 20 mM Tris-HCl, pH 7.65) with 10 ug/mL lysozyme. Resuspended cells were transferred to polylysinated coverslips and dried. Once dry, coverslips were incubated in cold methanol for 5 minutes followed by cold acetone for 5 minutes. Cells were rehydrated by a rinse in PBS followed by a 3-minute incubation in PBS + 2% BSA blocking solution. Cells were incubated with a 1:50 dilution of primary antibody (Cas9 (7A9–3A3): sc-517386 or cMyc (9E10): sc-40) in PBS + 2% BSA for 1 hr followed by 3, 7 minute washes in fresh PBS + 2% BSA. Coverslips were then incubated in the dark for 1 hr with secondary antibody (goat anti-mouse Alexa Fluor 555, Life Technologies A-21424) diluted 1:500 in PBS + 2% BSA. DAPI was added for the final 10 minutes of the incubation. Cells were washed in PBS 3 times for 7 minutes. Coverslips were then placed on slides using mounting media (v/v 90% glycerol, v/v 10%Tris pH 8.0 and w/v 0.5% propyl-gallate) and sealed with clear nail polish.
Microscopy and Analysis
Images were collected using a Zeiss Axiovert 200M microscope. Images were later processed using NIS Analysis software at the UCSF NIC. Compartments and cells were manually selected using the Simple ROI Editor. Background subtractions were conducted for each cell and compartment separately. The corrected intensities for the cytoplasmic and compartment regions were averaged over the area of each cell and compartment using Matlab and plotted using Prism 6. Data from two pooled replicates was fitted with a line, showing 95% confidence intervals with dashed lines. The slope is as reported in the plots.
Live cell imaging
For live cell imaging of ɸKZ infection, freshly grown cells from LB plates were picked and resuspended in 100 μl of LB media. 5–10 μL of samples are spotted on 0.85% agarose pads with 1:5 diluted LB. Arabinose (0.01% to 0.05%) and DAPI (2 μg/ml). Samples were then incubated in a humidified chamber and allowed to grow for 3 hours at 30 °C. 5 μL ɸKZ lysate was spotted on top of the agar pad and the samples were grown for an additional 1 hour. Cells were visualized in the microscope after covering them with a cover slip. Nikon Ti2-E inverted microscope equipped with Perfect Focus System (PFS) and Photometrics Prime 95B-25mm camera were used for live cell imaging. Images were processed using NIS Elements-AR software.
Extended Data
Extended Data Figure 1: Jumbo phage ΦPA3 resists targeting by CRISPR Cas and a restriction-modification system.
a, Strain PAO1 was engineered to express the I-C cas genes and distinct crRNAs targeting the indicated phages, and plaque assays were conducted as in Fig. 1a. b, Strain PAO1 was engineered to express the Type II-A Cas9 protein and distinct single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Figure 1a. c, The endogenous Type I R-M system (hsdRSM) in strain PAO1 was assayed using phages propagated on PAO1 or PAK as indicated (e.g. JBD30•PAO1 was first propagated on strain PAO1). Together with an isogenic PAO1∆hsdR knockout, all strains were subjected to a plaque assay as in Figure 1a. All plaque assays were replicated twice with similar results.
Extended Data Figure 2: Phage ΦKZ resists P. aeruginosa Type I-C and Type I-F CRISPR-Cas immunity.
a, Strain PAO1 was engineered to express the I-C cas genes and distinct crRNAs targeting phage JBD30 and phage ΦKZ, and plaque assays were conducted as in Fig. 1a. b, Strain PAO1 was engineered to express the I-F cas genes and distinct crRNAs targeting phage JBD30 and phage ΦKZ, and plaque assays were conducted as in Fig. 1a. All plaque assays replicated ≥ 2 times with similar results.
Extended Data Figure 3: Phage ΦKZ resists targeting by heterologous Type II-A and V-A CRISPR-Cas systems.
a, Strain PAO1 was engineered to express the Type II-A Cas9 protein and distinct single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Figure 1a. b, Strain PAO1 was engineered to express the Type V-A Cas12a protein and distinct crRNAs against the indicated phage. Plaque assays were conducted as in Figure 1a. All plaque assays replicated ≥ 2 times with similar results.
Extended Data Figure 4: Quantification of Cas9 localization during phage ΦKZ infection of P. aeruginosa.
Localization of Cas9 and ORF152 in the cytoplasm and shell during ΦKZ infection were quantified. Data points (individual cells) from two pooled replicate experiments were fitted with a line, showing 95% confidence intervals with dashed lines. The slope is as reported in the plots.
Extended Data Figure 5: Phage ΦKZ genomic DNA is susceptible in vitro to cleavage by Restriction Endonucleases.
Genomic DNA was isolated from phages ΦKZ and JBD30 and was subjected to digestion with the indicated restriction enzymes in vitro. The number of cut sites for each enzyme is shown at the bottom of the gels. Products were visualized on a 0.7 % agarose gel, visualized with SYBR Safe nucleic acid stain. In vitro digestion experiment was replicated twice with similar results.
Extended Data Figure 6: Cas9 fusion to ORF152 localizes to periphery of shell, but does not enable immune activity against ΦKZ.
a, Strain PAO1 was engineered to express Cas9 fused to ORF152 at either the N- or C-terminus, with single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Fig. 1a, b, growth curves were conducted, monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). c, Fluorescence microscopy of PAO1 fusion strains, immunostained for Cas9, in cells expressing ORF152-Cas9 or Cas9-ORF152. DAPI stain shows the phage DNA within the shell. d, Fluorescence microscopy of P. aeruginosa, immunostained for Cas9, in cells expressing ORF152-Cas9. DAPI stain shows the phage DNA within the shell. All plaque assays were replicated 4 times with similar results. Growth curve experiments were replicated three times with similar results. Microscopy was replicated twice with similar results.
Extended Data Figure 7: Fusion of EcoRI restriction enzymes to ORF152 enables immune activity.
a, Strain PAO1 was engineered to express EcoRI or EcoRI-ORF152 fusion protein. Plaque assays were conducted as in Fig. 1a and quantified (n=3). b, Growth curves were conducted, monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). c, Live fluorescence imaging of P. aeruginosa strains engineered to express EcoRI or EcoRI-ORF152. DAPI stain shows the phage DNA. d, Live fluorescence imaging of P. aeruginosa strains engineered to express EcoRI, or EcoRI-ORF152. DAPI stain shows the phage DNA within the shell. Wide field of view. All plaque assays were replicated 3 times with similar results. Growth curve experiments were replicated twice with similar results. Microscopy was replicated twice with similar results.
Extended Data Figure 8: Phage ΦKZ DNA is sensitive to RNA-targeting Cas13.
a, Strain PAO1 expressing LseCas13a and distinct crRNAs targeting the indicated gene. Plaque assays conducted as in Figure 1a. b, Strain PAO1 expressing LshCas13a and distinct crRNAs targeting the indicated gene. Plaque assays conducted as in Figure 1a. All plaque assays were replicated twice with similar results.
Extended Data Figure 9: Phage ΦKZ escaper mutants are selected by Cas13-mediated RNA targeting.
Strain PAO1 expressing LseCas13a and a crRNA targeting the indicated gene. Plaque assays conducted as in Figure 1a using wild type and escaper mutant ΦKZ. WT ΦKZ is targeted by both strains and the faint clearings observed here are not observed as plaques in a full-plate assay. All plaque assays were replicated twice with similar results.
Extended Data Figure 10: Observation of DAPI-stained phage DNA adjacent to a Cherry-TopA nascent shell.
Live fluorescence imaging of P. aeruginosa strains engineered to express Cherry-TopA infected with ΦKZ. DAPI stain labels DNA. Microscopy was replicated three times with similar results.
Supplementary Material
Acknowledgements:
Research in the Bondy-Denomy lab was supported by the University of California San Francisco Program for Breakthrough in Biomedical Research, funded in part by the Sandler Foundation, and an NIH Office of the Director Early Independence Award DP5-OD021344 (JBD) and R01-GM127489 (JBD). This work was also supported by HHMI (DAA) and NIH grants R35GM118099 (DAA), and GM104556 (DAA, JP). ΦKZ, JBD30, JBD68, D3, and F8 were provided by Alan Davidson’s lab and ΦPA3 from David Agard’s lab. Phage DMS3m was a gift from the O’Toole lab and the S. pyogenes cas9 expression plasmid for integration in the PAO1 chromosome is from Jason M. Peters and Carol A. Gross. pTE4495 (MbCpf1/MbCas12a) was a gift from Ervin Welker (Addgene plasmid # 80339), and LseCas13a (Addgene plasmid #83486) is a gift from Jennifer Doudna. pSB1A3 EcoRI Methylase-AmilCP and pSB1A3 EcoRI-RTX with EcoRI Methylase-AmilCP were gifts from Robin Dowell (Addgene plasmid # 85166, Addgene plasmid # 85165). The type I-F crRNA expression plasmid pAB04 was contributed by Adair Borges (Bondy-Denomy lab).
Footnotes
Competing Interests: J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences.
Data Availability Statement: All data generated or analysed during this study are included in this published article (and its supplementary information files).
REFERENCES
- 1.Koonin EV, Makarova KS & Wolf YI Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu Rev Microbiol 71, annurev–micro–090816–093830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondy-Denomy J, Pawluk A, Maxwell KL & Davidson AR Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL & Davidson AR A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5, e00896–e00896–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawluk A et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology 1, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 5.van Belkum A et al. Phylogenetic Distribution of CRISPR-Cas Systems in Antibiotic-Resistant Pseudomonas aeruginosa. mBio 6, e01796–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Micro 13, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marino ND et al. Discovery of widespread Type I and Type V CRISPR-Cas inhibitors. Science 362, eaau5174–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR & O'Toole GA The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. 194, 5728–5738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawluk A et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauch BJ et al. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168, 150–158.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaikeeratisak V et al. Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaikeeratisak V et al. The Phage Nucleus and Tubulin Spindle Are Conserved among Large Pseudomonas Phages. Cell Rep 20, 1563–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer JA et al. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149, 1488–1499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erb ML et al. A bacteriophage tubulin harnesses dynamic instability to center DNA in infected cells. Elife 3, e03197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zehr EA et al. The structure and assembly mechanism of a novel three-stranded tubulin filament that centers phage DNA. Structure 22, 539–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryson AL et al. Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9. mBio 6, e00648–15–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strotskaya A et al. The action of Escherichia coli CRISPR-Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Research 45, 1946–1957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlot M et al. Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Research 46, 873–885 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang LH, Farnet CM, Ehrlich KC & Ehrlich M Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Research 10, 1579–1591 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gootenberg JS et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, eaam9321–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meeske AJ & Marraffini LA RNA Guide Complementarity Prevents Self-Targeting in Type VI CRISPR Systems. Mol Cell 71, 791–801.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Shayeb B et al. Clades of huge phage from across Earth’s ecosystems. bioRxiv 3, 572362 (2019). [Google Scholar]
- 24.Chaikeeratisak V et al. Viral Capsid Trafficking along Treadmilling Tubulin Filaments in Bacteria. Cell 177, 1771–1780.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 25.Choi K-H & Schweizer HP mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1, 153–161 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Jinek M et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowles KN et al. The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Molecular Microbiology 90, 923–938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mesyanzhinov VV et al. The genome of bacteriophage phi KZ of Pseudomonas aeruginosa. Journal of Molecular Biology 317, 1–19 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Bondy-Denomy J et al. Prophages mediate defense against phage infection through diverse mechanisms. The ISME Journal 10, 2854–2866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kropinski AM Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. 182, 6066–6074 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu D, Damron FH, Mima T, Schweizer HP & Yu HD PBAD-Based Shuttle Vectors for Functional Analysis of Toxic and Highly Regulated Genes in Pseudomonas and Burkholderia spp. and Other Bacteria. Applied and Environmental Microbiology 74, 7422–7426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi K-H & Schweizer HP mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1, 153–161 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Choi K-H et al. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Applied and Environmental Microbiology 74, 1064–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.