Abstract
Purpose:
We previously determined that cancer-stromal interaction was a direct route to tumor cell heterogeneity progression, since cancer-stromal cell fusion in co-culture resulted in the creation of heterogeneous clones of fusion hybrid progeny. In this report, we modified the cancer-stromal co-culture system to establish optimal experimental conditions for investigating cell fusion machinery and the mechanism of heterogeneity progression.
Experimental design:
Red fluorescence protein-tagged LNCaP cells were co-cultured with green fluorescence protein-labeled prostate stromal cells for cancer-stromal cell fusion, which was tracked as dual fluorescent cells by fluorescence microscopy.
Results:
We identified the most efficient strategy to isolate clones of fusion hybrid progenies. From the co-culture, mixed cells including fusion hybrids were subjected to low-density re-plating for colony formation by fusion hybrid progeny. These colonies could propagate into derivative cell populations. Compared to the parental LNCaP cells, clones of the fusion hybrid progeny displayed divergent behaviors and exhibited permanent genomic hybridization.
Conclusions:
Cancer-stromal cell fusion leads to cancer cell heterogeneity. The cancer-stromal co-culture system characterized in this study can be used as a model for molecular characterization of cancer cell fusion as the mechanism behind the progression of heterogeneity observed in clinical prostate cancers.
Keywords: prostate cancer, mesenchymal stromal cells, tumor microenvironment, cancer-stromal interaction, cell fusion, tumor cell heterogeneity, fluorescence protein
Introduction
A salient feature of prostate cancer (PCa) is its progressive acquisition of tumor cell heterogeneity, which underlies every aspect of PCa clinical histopathology and malignant behavior 1–6. At an early stage, the primary tumor remains dependent on male hormones for survival and is sensitive to hormonal-deprivation therapy. Upon disease progression, tumors in recurrent and metastatic cases become composed of highly heterogeneous cancer cells, with individual cells displaying varied aneuploidy and genomic alterations 2–6. Tumor cells at advanced stages are also heterogeneous in malignant behavior. Individual cells display different levels of androgen independence, and different degrees of migration, invasion, and therapeutic resistance 7–9. Cellular heterogeneity is the ultimate strategy and culminating event in cancer progression, because it ensures that some tumor cells will survive and adapt to any adverse conditions or therapeutic insults. The elucidation of the underlying mechanism may provide a basis for rational treatment of clinical disease.
Progressive acquisition of heterogeneity is a common feature in many human cancer types 10. The underlying mechanism remains to be defined. Cancer cells may diverge from each other due to successive genetic mutations as activation of oncogenes and/or loss of tumor suppressor functions drives cancer progression 11,12. Mutations result in genomic instability 13,14, thus explaining intra-tumoral heterogeneity. Cancer cells perpetuated by self-renewal and may contribute to heterogeneity progression, as such cells carry stem cell properties 12,15,16. Tumor cells may also be reprogrammed into cancer stem cells by lineage plasticity 17–19. These cells may then transdifferentiate into cells of other lineages through epigenetic regulation. We demonstrated that cancer cells also interacted with varieties of resident bystander cells in the tumor microenvironment, and such interaction resulted in cancer cell heterogeneity. We showed that 3-dimensional (3-D) co-culture with bone marrow stromal cells caused permanent genomic and behavioral changes in PCa cells 20, while co-inoculation with stromal cells made human LNCaP PCa cells heterogeneous, yielding individual clones including the C4, C4–2, and C4–2B derivative sublines with distinct levels of androgen independence, tumorigenicity and metastatic capability 21–23. In another study, we investigated heterogeneity progression in ARCaPE, a defined subclone of the human ARCaP PCa cells. Through in vivo xenograft tumor formation, epithelial ARCaPE cells became highly heterogeneous, as derivative subclones acquired variable mesenchymal stromal characteristics, which were permanent and irreversible 24. Our studies thus defined cellular interaction as a direct route to tumor cell heterogeneity progression.
How does cellular interaction result in cancer cell heterogeneity? The interaction between apposed cells may involve dynamic reciprocal communication via plural factors 25. Conventional models of cellular interaction mostly involve paracrine communication through the extracellular matrix, exosomes, soluble factors, or other macromolecules 26,27. We realized that heterogeneity progression could be a result of fusion between PCa cells and bystander cells in the tumor microenvironment. Using fluorescence protein tracking technology, we determined that human PCa cells are inherently fusogenic, capable of fusing with apposing mesenchymal stromal cells in co-culture 28. Cell fusion is critical to heterogeneity progression, because it can lead to the creation of hybrid progeny with divergent genomic makeup and phenotypic behavior unlike either parental cell 29–31. Isolation and characterization of the hybrid progeny could validate the role of cell fusion in PCa progression.
In this report, we assessed the use of fluorescently labeled mesenchymal stromal cells in cancer-stromal interaction to facilitate isolation of fusion hybrids. Accumulating evidence show that cancer cell fusion with various bystander cells is a frequent and dynamic event highly consequential for cancer progression and metastasis 32–35, while all the commonly used PCa cell lines can fuse with prostate or bone marrow stromal cells 28. Application of this protocol to the study of cellular interaction will expedite the mechanistic elucidation of PCa progression.
Materials and Methods
Cell lines and cell culture.
The LNCaP human PCa cell line was originally provided by late Dr. Gary Miller (University of Colorado, Denver, CO). We reported the isolation of the RL-1 clone from LNCaP cells expressing an AsRed2 red fluorescence protein (RFP) by G418 (300 μg/ml) selection and limiting dilution cloning 36. Isolation and characterization of the HPS-15 human prostate stromal cell line have been reported 28,36. For green fluorescence protein (GFP) tagging, HPS-15 cells were infected with TurboGFP lentiviral particles (SHC003V, Sigma-Aldrich, St. Louis, MO) using the manufacturer’s recommended protocol. After selection with puromycin (0.5 μg/ml) for 2 weeks, clones with GFP expression were isolated by limiting dilution. A representative clone, GHPS-15, was used in this study. Both the PCa cells and stromal cells were maintained in T-medium (Formula LS0020056DJ, Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA), penicillin (100 U/ml) and streptomycin (100 μg/ml) in a humidified incubator at 37°C in atmospheric air supplemented with 5% CO2. G418 and puromycin were purchased from Life Technologies and the antibiotic stock solutions were prepared in phosphate buffered saline (PBS).
Cancer-stromal cell co-culture.
The cancer-stromal co-culture protocol has been reported previously 28,36. Briefly, 2.5 × 104 PCa cells in single-cell suspension were overlaid on a monolayer of stromal cells in each well of a 6-well plate, so the well contains a co-culture of equal numbers of cancer and stromal cells in 4 ml of culture medium. The number of stromal cells in a monolayer was deduced by counting cells in 12 random viewfields, 6 mm in diameter under low magnification (40×). Depending on the experimental design, co-cultures were maintained for 4 or 8 weeks with weekly changes of culture medium.
Assessing for dual antibiotic resistance.
Clones displaying dual red and green fluorescence were isolated from co-culture with disk cloning method. The clones were first cultured in the presence of G418 (600 μg/ml) and puromycin (2 μg/ml) for 2 weeks to remove any contaminating cells. The clones were then cultured for 30 passages with a 1:5 re-plating ratio without G418 or puromycin. To assess stability of the clones, cells (5 × 104/ml) of the 31st passage were seeded to 96-well plate (100 μl/well) in the presence of G418 (600 μg/ml) and puromycin (2 μg/ml). RL-1 and GHPS-15 cells were used as control. After 7 days, cell survival and proliferation were assayed with the crystal violet staining method as we previously reported 24,36.
Determining androgen-induced production of prostate specific antigen (PSA).
Cell cultures were subjected to enzyme-linked immunosorbent assay (ELISA) for PSA concentration by our reported protocol 28,37. Briefly, cells in 6-well plates at 70% confluence were kept in androgen-starvation medium (phenol red-free RPMI 1640, Life Technologies) for 48 hours, and were treated with regular culture medium (Control group), androgen-starvation medium containing 1% dextran/charcoal absorbed FBS (Androgen-deprivation group), and 5 nM synthetic androgen methyltrienolone (R1881, Perkin Elmer, Waltham, MA) in androgen-starvation medium containing 1% dextran/charcoal absorbed FBS (R1881 group) for 24 hours, after which the culture medium was sampled for PSA.
Genotyping analysis for mixed genomes.
Short tandem repeat (STR) genotyping was used to demonstrate mixed genomes, using the commercial Human STR Profiling Cell Authentication Service (ATCC, Manassas, VA). Cloned cells after continued culture for 30 passages were collected by trypsin detachment. After washing in PBS, 1×106 cells in 100 μl PBS were spotted onto the provided sample collection card. After air drying for 2 hours, the card was sent for STR profiling. Authenticity of the RL-1 cells as a clone of the LNCaP cells, uniqueness of HPS-15 and the identity of GHPS-15 cells, and mixed genomes in cloned cells were confirmed by searching through the ATCC STR profile database.
Fluorescence microscopy.
The protocol for fluorescence imaging was previously reported 36. In this study, for comparison purposes all the fluorescent images were taken with fixed settings: 3 seconds for RFP imaging and 12 seconds for GFP imaging at 40× magnification; 1 second for RFP imaging and 4 seconds for GFP imaging at 100× magnification; and 0.5 second for RFP imaging and 2 seconds for GFP imaging at 200× magnification. Photoshop CS4 (Adobe Systems, San Jose, CA) was used to overlap images and Layer Style Blending Option software was used to demonstrate localization of green or red fluorescence in cultured cells.
Results
We previously determined that human PCa cells are fusogenic. Upon direct contact, RFP-labeled LNCaP cells of the RL-1 clone fused with prostate stromal cells 28. A comprehensive examination of the fate of hybrid progenies should unveil the impact of cancer-stromal cell fusion on PCa progression. Optimizing a strategy for isolating the clones of fusion hybrid progeny, we tested whether the fate of the fusion hybrids could be conveniently studied with dual-fluorescence tracking, and whether hybrid progeny could be isolated based on dual fluorescence or dual antibiotic selection.
1. Fluorescence protein-tagged cells for cancer-stromal interaction in co-culture.
We previously tagged human PCa LNCaP cells with RFP and used a representative clone, RL-1, for studying cancer-stromal interaction in co-culture 28. All RL-1 cells remained fluorescently red after being cultured continuously for 30 passages in the absence G418, confirming the stability of RFP expression (Figure 1A).
Figure 1. Cell fusion in cancer-stromal co-culture as revealed by fluorescence tracking.
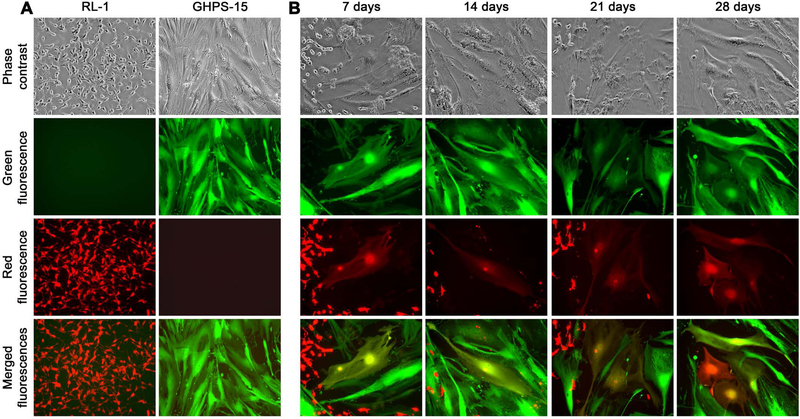
A, fluorescence profiles of participant RL-1 and GHPS-15 cells in the co-culture. B, representative cancer-stromal cell fusion events in the co-culture were documented weekly for 4 weeks (100×). Marked differences in cellular morphology, cell size, and fluorescence color between RL-1 and GHPS-15 cells are shown.
We reported the isolation and characterization of pair-matched human prostate stromal cell lines 36. By continuous passaging, we determined that the HPS-15 stromal cells established from the tumor zone of a prostatectomy specimen could be grown continuously, albeit at a slower rate than LNCaP cells 36, for more than 30 passages without showing signs of replication quiescence. HPS-15 can thus be used as a cancer-associated prostate stromal cell line for studying cancer-stromal interaction. In such a study, we found that HPS-15 cells could fuse with PCa cells 28. Almost 25% of the HPS-15 cells in a monolayer, for instance, were involved in fusion with RL-1 cells in a 4-week co-culture 28. The HPS-15 cell line, therefore, was used in the current study to evaluate an optimal protocol for isolating cancer-stromal fusion hybrids.
We tagged HPS-15 prostate stromal cells with the TurboGFP reporter carrying a puromycin selection marker gene. Clones of green fluorescent cells were isolated after antibiotic selection and limiting dilution. A representative clone, GHPS-15, was cultured continuously for 15 passages to confirm stability of the GFP expression (Figure 1A). The sharp contrast between large green fluorescent stromal cells and small red fluorescent RL-1 PCa cells is a distinguishing feature facilitating the study of cancer-stromal interaction.
2. Cancer-stromal cell fusion in co-culture.
Using fluorescence protein-tagged cells, we assembled cancer-stromal co-cultures to recapitulate the fusion between PCa cells and prostate stromal cells 28. GHPS-15 cells were cultured to form a stromal cell monolayer. As the growth of these cells was controlled by contact inhibition 36, the stromal monolayer could remain at confluence for more than 8 weeks without noticeable overgrowth or cell loss. Cancer-stromal cell co-culture was initiated by overlaying an equal number of RL-1 cells in single cell suspension on the stromal monolayer. The co-culture was maintained for 4 weeks with daily microscopic examination.
In co-culture, RL-1 PCa cells had direct contact with the stromal layer. Growth of RL-1 cells slowed but was constant, occasionally forming red fluorescent cell clusters. Relative to the stromal cells of the monolayer, RL-1 cells were much weaker in adhesion. Many of the RL-1 cell clusters were removed during weekly medium changing, rarely affecting observation of cancer-stromal interaction.
Identification and observation of cancer-stromal cell fusion became simple and straightforward once the two cell types in co-culture were labeled with distinct fluorescence proteins. In this study, cancer-stromal cell fusion was defined by the appearance of dual fluorescent cells showing both fluorescent red and fluorescent green 38,39. In combination with the marked differences in cell size and shape, cell fusion could be conveniently identified as cells that were fluorescent red and fluorescent green at the same time, most of which had a stromal cell shape (Figure 1B). With RL-1 and GHPS-15 stromal cells, individual fusion events started appearing 7 days into the co-culture and accumulated gradually to become frequent at two weeks (Figure 1B). The incidence of fusion kept increasing over the next two weeks, reaching a plateau at which 20% to 25% of the stromal cells in the monolayer were involved. These results were in agreement with our previous findings 28.
3. Isolation of clones of hybrid progeny based on differential growth rates.
GHPS-15 cells have slower proliferation rates than RL-1 cells, and when plated at low density GHPS-15 cells have poorer colony formation potential. These features were exploited to isolate clones of hybrid progeny. Cells in co-culture for 4 weeks were collected in single-cell suspension and re-cultured at low density (5 × 104 cells to a 15-cm dish, 3 dishes/re-culture) for another 4 weeks. Under these culture conditions, individual red fluorescent PCa cells from the co-culture started to grow into colonies, as did the fluorescently green stromal cells. Though most fusion hybrids were either remained in a state of growth arrest or perished by mitotic catastrophe 28, there was always a small fraction displaying proliferative activity. This fraction of hybrids would shrink gradually in size to adopt a smaller but thicker shape, entering the cell cycle to form multi-cell clusters, some of which had enough proliferative activity for colony formation (Figure 2A).
Figure 2. Colony formation by fusion hybrid progeny.
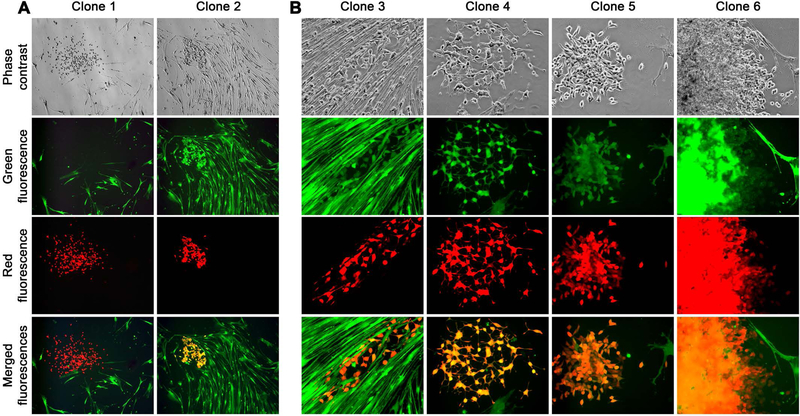
Mixed cells from a RL-1 and GHPS-15 co-culture were re-cultured at low density for colony formation. Images were taken 2 weeks into the re-culture. A, Low magnification views (40×) show the relationship of different colonies in the re-culture. A red RL-1 colony (Clone 1) in re-culture is shown, together with a representative dual fluorescent colony of fusion hybrid progeny (Clone 2). B, another 4 colonies are shown in larger magnification (100×) to show heterogeneous features. As images for all 6 colonies were taken on the same day and from the same re-culture, differences in colony sizes may reflect different growth rates in individual colonies.
The most prominent feature among colonies of hybrid progeny were their heterogeneous growth rates and morphology relative to the parental RL-1 cells (Figure 2B). Cloned from LNCaP cells, subclones of RL-1 displayed similar growth rates and homogenous morphology in general (Figure 1A). In contrast, clones of hybrid progeny derived from RL-1 and GHPS-15 co-culture had markedly different growth rates, and cells in different clones showed either scattered, clustered, or overlapping growth (Figure 2B). These observations suggested that clones of hybrid progeny derived from cancer-stromal fusion were divergent from parental PCa cells.
The formation of hybrid colonies is highly consequential to the study of cancer-stromal interaction, because these colonies could represent the creation of new cell types with mixed genotypic and behavioral heterogeneity. We used triplicate re-cultures of three separate co-cultures to estimate the rate of hybrid colony formation. In three separate re-cultures of 5 × 104 cells from 4-week co-culture of RL-1 with GHPS-15 cells, estimated rates of hybrid colony formation were 46 ± 11, 95 ± 24, and 61 ± 7, respectively. Considering the chronic nature of cancer-stromal interaction in clinical PCa progression, hybrid colony formation may well be a contributing mechanism of heterogeneity progression.
4. Stability of the dual fluorescent clones.
To determine whether the observed features of hybrid progeny clones were stable, we isolated the first 43 dual fluorescent clones, named RLGH15-clones, from the first re-culture of RL-1 and GHPS-15 co-culture. Many slow-growing clones in the same re-culture were not picked. After recovering the picked clones for 2 weeks through dual antibiotic selection with G418 (600 μg/ml) and puromycin (2 μg/ml), we randomly selected 8 of the clones (RLGH15–5, 6, 9, 14, 24, 27, 30, and 32) for further analyses. By continuously culturing them for 30 passages, we found that their dual fluorescence feature was stable, as none of the clones became mono-fluorescent or null-fluorescent (Figure 3A), although some cells in individual clones could be seen with a much brighter fluorescence for one color than the other. Occurrence of differential fluorescence intensities, however, was transient and haphazard throughout the 30 passages, excluding the possibility that it was caused by loss of fluorescence gene expression. This observation was supported by assays of dual antibiotic selection, as all the 8 clones at 31st passage remained resistant to dual G418 and puromycin treatment (Figure 3B). Based on these results, we concluded that clones of hybrid progeny are quite stable, carrying permanent and irreversible features acquired from the process of PCa-stromal cell fusion.
Figure 3. Stability of the clones of fusion hybrid progeny.
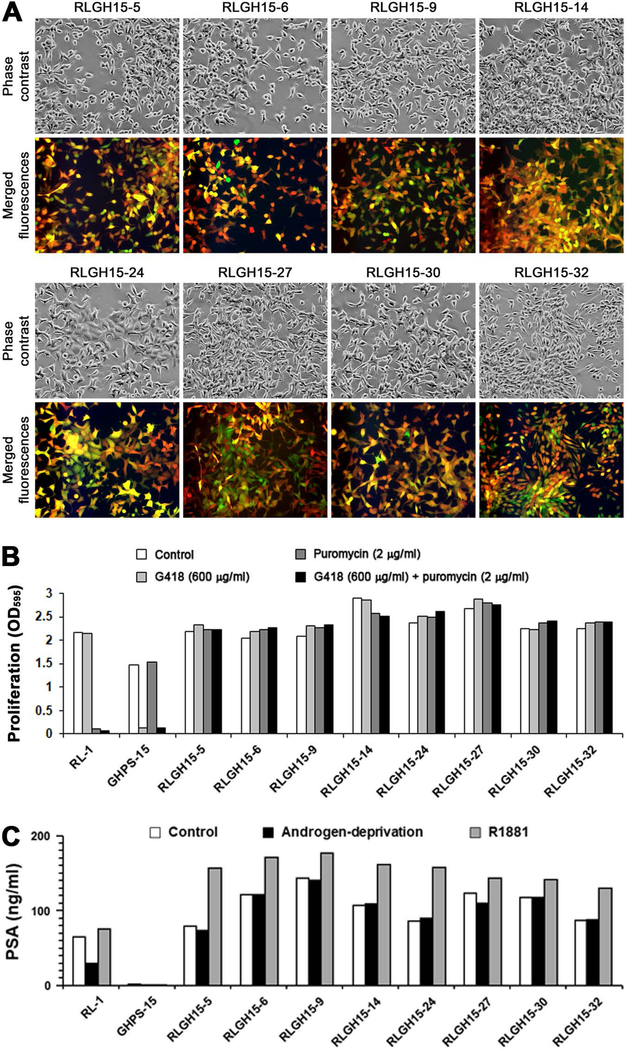
A, varied morphologies of the 8 RLGH clones after 30 continued passages (100×). These clones are dual fluorescent and much large than RL-1 cells in size as seen in Figure 1A. B, stability of the 8 RLGH clones at 31st passage was assayed for their resistance to dual G418 and puromycin selection. After 7 days of dual antibiotic treatment, survival and proliferation of the cells were determined by the method of crystal violet staining. For each data point, the mean of triplicate assays is shown; and standard deviation is less than 5% of the mean and is not shown. C, heterogeneous PSA production in the 8 RLGH clones was determined with ELISA. The data are shown with the mean of triplicate assays. For all the data points, standard deviations are less than 5% of the mean and are not shown.
5. Behavioral changes and genomic heterogeneity confirming cancer-stromal cell fusion.
LNCaP cells produce PSA in an androgen-dependent manner, which can be assayed as a marker for heterogeneity progression as derivative cells often acquire androgen-independent PSA expression 28. We used ELISA to assess the 8 RLGH15 clones (Figure 3C) at passage 30 for PSA production in 24-hour culture medium. Compared to an equal number of parental RL-1 cells, since HPS-15 cells did not express detectable PSA 36, these derivative clones produced more PSA in general. The salient feature of these clones was their insensitivity to androgen depletion, because similar amounts of PSA were produced when the cells were grown in regular culture medium (Control group) or under androgen-starvation conditions (Androgen-deprivation group). On the other hand, these clones remained responsive to androgen stimulation, since addition of R1881 induced marked PSA production (R1881 group). Similar to our previous report 28, the expression of androgen receptor in these clones were not significantly changed as detected by western blotting (data not shown). These results were in good agreement with our findings in the clones of fusion hybrid progeny in a previous study 28.
The clones of fusion hybrid progeny contain mixed genomes of the parental cells. We tested whether their genomic hybridization could be detected by STR genotyping. Unlike the genome of diploid cells that contain 2 alleles, LNCaP cells are aneuploid and polyallelic in many genomic loci. From RLGH15–5 and RLGH15–6, the first 2 of the 8 RLGH clones at passage 30, STR analyses identified 3 loci that were informative of the genomic mix between RL-1 and GHPS-15 cells (Figure 4A). Both clones contained 3 CSF1PO alleles, 2 from the RL-1 PCa cell and 1 from the GHPS-15 stromal cell. The same was true for the D19S433 locus. A unique fourth allele was present in the D19S433 locus in RLGH-6 cells, probably a result of mutation during the 30 continuous passaging. In addition, cells in RLGH15–5 clone carried 1 extra allele from the GHPS-15 genome. Informative results from the STR analyses are outlined in Figure 4B, together with 3 non-informative STR loci (FGA, Penta_D, and D2S1338). Based on these results, we concluded that the dual GFP- and RFP-positive RLGH15 clones recovered from co-culture are indeed fusion hybrid progenies.
Figure 4. Genomic hybridization detected from RLGH clones.

A, partial electrophotograph of the STR analyses for RLGH-5 and RLGH-6 clones are shown together with the parental RL-1 and GHP-15 cells. STR analyses of the original LNCaP and HPS-15 are included. Genomic mixes of the parental cells can be seen at the CSF1PO, D8S1179 and D19S433 loci. B, a summary of the genomic hybridization. STR alleles from red fluorescent RL-1 cells are indicated in red. STR alleles from GHPA-15 cells are indicated in green. STR counts non-informative for this analysis is indicated in black.
Discussions
Following our initial discovery of PCa cancer-stromal fusion, we conducted additional studies to conclude that cancer cell fusion with bystander cells in the tumor microenvironment is a direct route to progression of tumor cell heterogeneity. A reproducible and reliable experimental system can facilitate study of PCa cancer cell fusion relevant to clinical PCa progression and metastasis. In this study, we examined every step of our cancer-stromal co-culture system with the intention of establishing an optimal experimental system.
We used an RFP-tagged LNCaP clone, RL-1, in co-culture with GFP-tagged GHPS-15 prostate stromal cells (Figure 1A). The use of fluorescence protein tagging greatly assisted long-term tracking of fusion hybrids in real-time under an inverted fluorescence microscope (Figure 1B). We simplified the isolation of clones of fusion hybrid progenies based on differential growth in re-culture of the mixed cells from co-culture (Figures 2A and 2B). All the isolated clones were composed of dual RFP- and GFP-expressing cells (Figure 3A) with dual G418 and puromycin resistance (Figure 3B). Remarkably, these clones were heterogeneous in their growth rate and cell shape, many with enhanced PSA gene activity in the absence of androgen stimulation (Figure 3C). We used STR genotyping to confirm that cells with dual red and green fluorescence were bona fide fusion hybrids (Figures 4A and 4B). Dual red and green fluorescence could therefore be used as a surrogate marker of cancer cell fusion in our experimental design. This greatly reduces the labor and cost and simplifies the detection of cell-cell fusion compared to STR analysis, providing an easy assay for studying the molecular mechanism of cancer cell fusion.
The role of cell fusion in cancer progression and its underlying mechanisms are the least understood topics in cancer research, even though somatic cell fusion is one of the original theories of cancer etiology. Because of the isogenic relationship between cancer and normal cells in a given patient, there is no easy method for detecting cell fusion in clinical specimens. In the study of female cancer patients who previously received male tissue or organ transplants, large numbers of tumor cells, as well as circulating tumor cells, were found to contain a Y chromosome 40,41, the concrete evidence for the involvement of cell fusion in cancer progression and metastasis. In myeloma patients, more than 30% of osteoclasts were detected to be osteoclast-myeloma fusion hybrids 42. On the other hand, frequent cancer cell fusion has been observed in xenograft tumors 43 and between cultured cells 35. Because of its spontaneity, cancer cell fusion should be considered a progressive and dynamic process, leading to endless genotypic and phenotypic changes during disease progression 35. Based on these results, it was estimated that a large fraction of tumor cells in advanced cancers are fusion hybrid progeny 40,42,44,45, an estimate in agreement with our findings from cancer-stromal co-culture.
How does a cancer cell fuse with a bystander cell? Much of our knowledge about cell fusion is derived from research on gamete fusion in plants, enveloped virus fusion to eukaryotic cells, and membrane fusion of subcellular organelles in the context of endocytosis and exosome release 46,47. These studies identified fusogens, specialized surface proteins functioning to directly fuse membranes. The cell-cell fusion machinery is proposed to be a fusogen-centered protein complex executing membrane fusion by pulling apposing lipid bilayers together and opening membrane pores between the two cells. In humans, fusogens in cell-cell fusion remain elusive and unidentified 48. A list of surface proteins is considered fusion-related because their loss of expression affected the process of cell-cell fusion 47,48. Though cell fusion is an essential mechanism for fertilization, embryotic development, and tissue and organ maturation and homeostasis 29,30,49, the molecular mechanism of cell fusion is poorly elucidated in humans. Studying the molecular mechanism of cell-cell fusion in humans remains difficult, partly due to the isogenic nature of each person’s cells, and partly due to the lack of suitable bioethical, reproducible and reliable experimental models for cell-cell fusion in real time. In this sense, our well-characterized PCa cell-stromal fusion in co-culture will be a valuable experimental model for the characterization of cell fusion machinery and the investigation of the molecular mechanisms of cancer cell fusion.
LNCaP cells are well known for androgen-dependent PSA production and poor xenograft tumor formation in athymic mice. We used LNCaP cells in our co-culture because gain of cellular heterogeneity could be assessed by androgen-independent PSA production and increased tumorigenic potential in clones of fusion hybrid progenies. On the other hand, LNCaP cells have inherently low fusogenic activity, with only about 20% of the stromal cells in the co-culture affected by fusion. Low fusogenic activity makes it difficult to investigate the molecular mechanism of cancer cell fusion. Recently, we isolated and characterized HPE-15, a new human PCa cell line 50. Though these cells display low PSA production and weak tumorigenic potential, HPE-15 cells recovered from co-culture with PCa or bone marrow stromal cells became more malignant, producing much more PSA and displaying drastically increased tumorigenicity in xenograft tumor formation. As HPE-15 cells share with LNCaP cells the susceptibility of becoming more malignant through co-culture with stromal cells, further work is warranted to examine the extent to which HPE-15 cells are more suitable for studying PCa cell fusion.
Conclusions
Cancer-stromal cell fusion leads to cancer cell heterogeneity. The cancer-stromal co-culture system characterized in this study can be used as a model for molecular characterization of cancer cell fusion as the mechanism behind the progression of heterogeneity observed in clinical prostate cancers.
Acknowledgements
This work was supported in part by NIH grants CA098912 (LWKC) and CA112330 (RXW), and by DoD grants CA170974 (LWKC) and PC040578 (RXW).
Footnotes
Conflicts of interest statement: The authors declare no potential conflicts of interest.
References
- 1.Kucuk O, Demirer T, Gilman-Sachs A, et al. Intratumor heterogeneity of DNA ploidy and correlations with clinical stage and histologic grade in prostate cancer. J Surg Oncol. 1993;54(3):171–174. [DOI] [PubMed] [Google Scholar]
- 2.Liu AY, Roudier MP, True LD. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. Am J Pathol. 2004;165(5):1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyahira AK, Den RB, Carlo MI, et al. Tumor cell heterogeneity and resistance; report from the 2018 Coffey-Holden Prostate Cancer Academy Meeting. Prostate. 2019;79(3):244–258. [DOI] [PubMed] [Google Scholar]
- 4.Sheng S, Margarida Bernardo M, Dzinic SH, Chen K, Heath EI, Sakr WA. Tackling tumor heterogeneity and phenotypic plasticity in cancer precision medicine: our experience and a literature review. Cancer Metastasis Rev. 2018;37(4):655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolkach Y, Kristiansen G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology. 2018;85(1–2):108–116. [DOI] [PubMed] [Google Scholar]
- 6.Yadav SS, Stockert JA, Hackert V, Yadav KK, Tewari AK. Intratumor heterogeneity in prostate cancer. Urol Oncol. 2018;36(8):349–360. [DOI] [PubMed] [Google Scholar]
- 7.Honn KV, Aref A, Chen YQ, et al. Prostate Cancer - Old Problems and New Approaches. (Part II. Diagnostic and Prognostic Markers, Pathology and Biological Aspects). Pathol Oncol Res. 1996;2(3):191–211. [DOI] [PubMed] [Google Scholar]
- 8.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12(6):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rezania S, Amirmozaffari N, Rashidi N, et al. The same and not the same: heterogeneous functional activation of prostate tumor cells by TLR ligation. Cancer Cell Int. 2014;14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunewald TG, Herbst SM, Heinze J, Burdach S. Understanding tumor heterogeneity as functional compartments--superorganisms revisited. J Transl Med. 2011;9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8):3230–3239. [PubMed] [Google Scholar]
- 12.Paduch R. Theories of cancer origin. Eur J Cancer Prev. 2015;24(1):57–67. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Rahman WM. Genomic instability and carcinogenesis: an update. Curr Genomics. 2008;9(8):535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charames GS, Bapat B. Genomic instability and cancer. Curr Mol Med. 2003;3(7):589–596. [DOI] [PubMed] [Google Scholar]
- 15.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. [DOI] [PubMed] [Google Scholar]
- 16.Romanska-Knight H, Abel P. Prostate cancer stem cells. Cent European J Urol. 2011;64(4):196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol. 2018;15(5):271–286. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien-Ball C, Biddle A. Reprogramming to developmental plasticity in cancer stem cells. Dev Biol. 2017;430(2):266–274. [DOI] [PubMed] [Google Scholar]
- 19.Rapp UR, Ceteci F, Schreck R. Oncogene-induced plasticity and cancer stem cells. Cell Cycle. 2008;7(1):45–51. [DOI] [PubMed] [Google Scholar]
- 20.Rhee HW, Zhau HE, Pathak S, et al. Permanent phenotypic and genotypic changes of prostate cancer cells cultured in a three-dimensional rotating-wall vessel. In Vitro Cell Dev Biol Anim. 2001;37(3):127–140. [DOI] [PubMed] [Google Scholar]
- 21.Thalmann GN, Anezinis PE, Chang SM, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54(10):2577–2581. [PubMed] [Google Scholar]
- 22.Thalmann GN, Sikes RA, Wu TT, et al. LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate. 2000;44(2):91–103 Jul 101;144(102). [DOI] [PubMed] [Google Scholar]
- 23.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994;57(3):406–412. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Wang R, Xie ZH, et al. Prostate cancer metastasis: role of the host microenvironment in promoting epithelial to mesenchymal transition and increased bone and adrenal gland metastasis. Prostate. 2006;66(15):1664–1673. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Chu GCY, Mrdenovic S, et al. Cultured circulating tumor cells and their derived xenografts for personalized oncology. Asian J Urol. 2016;3(4):240–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josson S, Matsuoka Y, Chung LW, Zhau HE, Wang R. Tumor-stroma co-evolution in prostate cancer progression and metastasis. Semin Cell Dev Biol. 2010;21(1):26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrissey C, Vessella RL. The role of tumor microenvironment in prostate cancer bone metastasis. J Cell Biochem. 2007;101(4):873–886. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Sun X, Wang CY, et al. Spontaneous cancer-stromal cell fusion as a mechanism of prostate cancer androgen-independent progression. PLoS One. 2012;7(8):e42653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson LI, Bjerregaard B, Talts JF. Cell fusions in mammals. Histochem Cell Biol. 2008;129(5):551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podbilewicz B. Cell fusion. WormBook. 2006:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Primakoff P, Myles DG. Cell-cell membrane fusion during mammalian fertilization. FEBS Lett. 2007;581(11):2174–2180. [DOI] [PubMed] [Google Scholar]
- 32.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3(5):445–448. [DOI] [PubMed] [Google Scholar]
- 33.He X, Tsang TC, Pipes BL, Ablin RJ, Harris DT. A stem cell fusion model of carcinogenesis. J Exp Ther Oncol. 2005;5(2):101–109. [PubMed] [Google Scholar]
- 34.Heppner GH, Miller FR. The cellular basis of tumor progression. Int Rev Cytol. 1998;177:1–56. [DOI] [PubMed] [Google Scholar]
- 35.Weiler J, Dittmar T. Cell Fusion in Human Cancer: The Dark Matter Hypothesis. Cells. 2019;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X, He H, Xie Z, et al. Matched pairs of human prostate stromal cells display differential tropic effects on LNCaP prostate cancer cells. In Vitro Cell Dev Biol Anim. 2010;46(6):538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R, Xu J, Saramaki O, et al. PrLZ, a novel prostate-specific and androgen-responsive gene of the TPD52 family, amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res. 2004;64(5):1589–1594. [DOI] [PubMed] [Google Scholar]
- 38.Suetsugu A, Matsumoto T, Hasegawa K, et al. Color-coded Live Imaging of Heterokaryon Formation and Nuclear Fusion of Hybridizing Cancer Cells. Anticancer Res. 2016;36(8):3827–3831. [PubMed] [Google Scholar]
- 39.Sun C, Zhao D, Dai X, et al. Fusion of cancer stem cells and mesenchymal stem cells contributes to glioma neovascularization. Oncol Rep. 2015;34(4):2022–2030. [DOI] [PubMed] [Google Scholar]
- 40.Yilmaz Y, Lazova R, Qumsiyeh M, Cooper D, Pawelek J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005;35(10):1021–1024. [DOI] [PubMed] [Google Scholar]
- 41.Gast CE, Silk AD, Zarour L, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4(9):eaat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen TL, Boissy P, Sondergaard TE, et al. Osteoclast nuclei of myeloma patients show chromosome translocations specific for the myeloma cell clone: a new type of cancer-host partnership? J Pathol. 2007;211(1):10–17. [DOI] [PubMed] [Google Scholar]
- 43.Hoffman RM, Bouvet M. Imaging the microenvironment of pancreatic cancer patient-derived orthotopic xenografts (PDOX) growing in transgenic nude mice expressing GFP, RFP, or CFP. Cancer Lett. 2016;380(1):349–355. [DOI] [PubMed] [Google Scholar]
- 44.Ramakrishnan M, Mathur SR, Mukhopadhyay A. Fusion-derived epithelial cancer cells express hematopoietic markers and contribute to stem cell and migratory phenotype in ovarian carcinoma. Cancer Res. 2013;73(17):5360–5370. [DOI] [PubMed] [Google Scholar]
- 45.Shabo I, Olsson H, Sun XF, Svanvik J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int J Cancer. 2009;125(8):1826–1831. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez JM, Podbilewicz B. The hallmarks of cell-cell fusion. Development. 2017;144(24):4481–4495. [DOI] [PubMed] [Google Scholar]
- 47.Brukman NG, Uygur B, Podbilewicz B, Chernomordik LV. How cells fuse. J Cell Biol. 2019;218(5):1436–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308(5720):369–373. [DOI] [PubMed] [Google Scholar]
- 49.Oren-Suissa M, Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17(11):537–546. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Chu GC, Wang X, et al. Establishment and characterization of a prostate cancer cell line from a prostatectomy specimen for the study of cellular interaction. Int J Cancer. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


