Mass spectrometry is well-established as one of the primary methods used to characterize peptides and proteins. New advances in instrumentation and innovative analytical workflows have continued to expand the range of applications of mass spectrometry for proteomics. The ability to determine sequences, pinpoint modifications, and quantify vast numbers of peptides or proteins remains the key mission of mass spectrometry-based proteomics. There also remains great fundamental interest in understanding the intrinsic properties of peptides and proteins in the gas phase, such as mechanisms of charging, conformations, and reactivity. One of the cornerstones of the comprehensive analysis of peptides and proteins by mass spectrometry is the utilization of ion activation methods, primarily in the context of tandem mass spectrometry (MS/MS), to cause informative dissociation of the ions of interest. An array of activation methods has been developed to energize ions and promote reproducible fragmentation in a way that generates a molecular fingerprint. These fingerprints can be used to assign a sequence of a peptide or protein, localize its modifications, and/or allow high throughput quantitation based on specific fragmentation pathways. The critical role of ion activation in mass spectrometry-based proteomics strategies has had a tremendous impact in fields ranging from cancer biology to structural biology to the development of biotherapeutics. The broad utility of MS/MS explains why there continues to be new developments and applications of ion activation for peptides and proteins despite the maturity of the field. This Review will provide an overview of some of the latest advances of ion activation methods over the past four years (2016 through 2019) as a sequel to a comprehensive review that was published in January 2016.1 Owing to the widespread use of mass spectrometry for proteomics, there have been an array of outstanding reviews over the past few years, including a number of technique-based reviews of MS/MS that provide key insight and foundational knowledge to which readers are directed.2–22 After introducing the fundamental concepts and nomenclature of MS/MS, the next sections of this review discuss the most common activation methods used for peptides and proteins. This is followed by an overview of new mass spectrometry applications in which ion activation plays a key role.
TANDEM MASS SPECTROMETRY AND PROTEOMICS
The core of most tandem mass spectrometry approaches for analysis of peptides and proteins is the ion activation method used to energize ions and promote a reaction, most often fragmentation. The resulting fragmentation patterns hold the key information to assign a sequence, localize modifications, and in some cases, differentiate isomers. Traditionally, ions have been isolated on the basis of mass-to-charge (m/z) ratio prior to activation, thus generating a specific fragmentation pattern for a selected precursor ion. This requirement for ion isolation is no longer the universal norm, owing to the development of elegant multiplex methods in which broader ranges of precursor ions are simultaneously activated, generating composite fragmentation patterns that are deciphered using postprocessing approaches (as described in a later section). The majority of ion activation methods for peptides and proteins focus on the addition of energy to induce fragmentation, while not promoting other types of reactions. Thus, these dissociation methods are emphasized in this review. The general mode of energizing ions occurs in a stepwise manner, such as via a series of low-energy collisions with a target gas (collision induced dissociation (CID) or collisional activated dissociation (CAD)) or via absorption of low-energy infrared (IR) photons (infrared multiphoton dissociation (IRMPD)). Alternatively, ion activation may involve faster, higher energy deposition via collisions with surfaces (surface induced dissociation (SID)) or absorption of ultraviolet photons (ultraviolet photodissociation (UVPD)). Activation via electron transfer has led to a number of methods, including electron capture dissociation (ECD), electron transfer dissociation (ETD), and a host of related offshoots (electron induced dissociation (EID), electronic excitation dissociation (EED), electron detachment dissociation (EDD)), many of which have been applied for the analysis of peptides and proteins. The types, abundances, and distribution of fragment ions are modulated by the mechanism and net energy deposition of activation and are not always predictable. In addition, specific structures of the resulting fragment ions are frequently not fully elucidated. Despite some of the yet unresolved details of ion activation mechanisms and structures of the product ions, what has become clear is that utilizing multiple activation methods, either in parallel or combined in hybridized strategies, often provides the most comprehensive insight for analysis of peptides and proteins. The quest for even more information-rich MS/MS spectra that reveal previously uncharted structural details continues to motivate the development of new ion activation strategies and their application toward increasingly challenging problems in proteomics and structural biology.
For applications of ion activation methods to proteomics, the approaches are categorized as bottom-up, middle-down, and top-down, depending on whether proteins are digested into peptides (bottom-up and middle-down) or analyzed as intact molecules (top-down). Analysis of small peptides, done in bottom-up methods, is arguably the easiest of the three categories, both in terms of separation of complex mixtures, using chromatographic or electrophoretic methods, and production of informative fragmentation patterns. It is the robust and reproducible nature of bottom-up methods, in addition to the advanced state of high proficiency data interpretation algorithms (database search and de novo methods), that have made the popularity of bottom-up methods unsurpassed by any other proteomics method. However, one of the recognized drawbacks of peptide-based analysis is the potential loss of contextual patterns of post-translational modifications when reconstructing proteins based on limited subsets of small peptides. The “middle-down” approach is aimed at generating and analyzing significantly longer peptides on the basis of restricted proteolysis of proteins, thus focusing on peptides that cover longer sequence stretches while bypassing the challenges raised by analyzing intact proteins. The top-down approach directly interrogates intact proteins, offering the potential to maximize sequence information, but encompasses more significant hurdles with separations of mixtures of proteins and data analysis. With respect to the role of ion activation in these different proteomic workflows, the greatest challenges are encountered with the analysis of intact proteins owing to their large sizes and complexity. All of the aforementioned methods have been supported and advanced by the development of sophisticated algorithms and search engines for comprehensive peptide and protein identification. Readers are directed to numerous reviews and references that cover mass spectrometry workflows for proteomics and software tools.2–9
FRAGMENTATION NOMENCLATURE
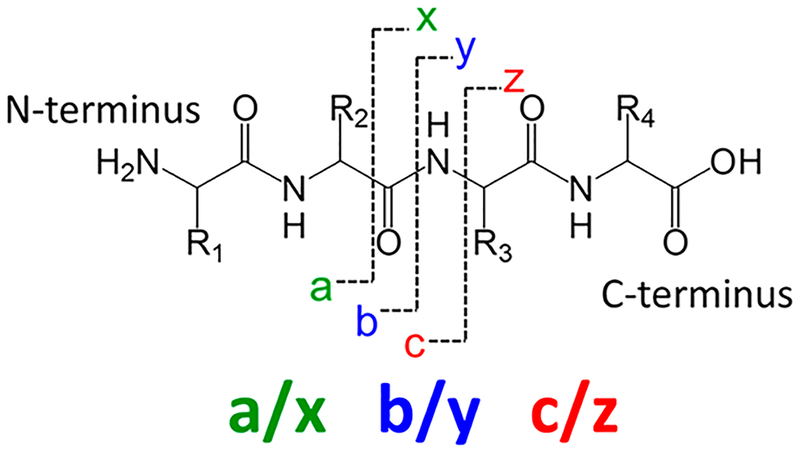
The nomenclature developed for classifying the types of fragment ions produced by peptides and proteins is a well-established code-based system. The fragment ions are categorized alphabetically on the basis of the backbone bond or side chain that is cleaved, with subscript numbers designating the number of amino acids contained in the fragment ion relative to the N- or C-terminus. The nomenclature system is summarized in Figure 1.23 Cleavages of the labile C–N amide bonds of the peptide or protein backbone are prominent upon low-energy activation methods, such as CID and IRMPD, resulting in formation of b- and y-type ions. The loss of NH3 or H2O from b/y ions, as well as the loss of CO from b-type ions to produce a-type ions, is common. Electron-based activation methods, including ECD and ETD, promote the cleavage of N–Cα bonds, which results in c/z-type fragment ions. Activation methods that result in higher energy deposition allow one to access a more diverse array of fragmentation processes. For example, UVPD generates a/x ions via cleavage of Cα–C bonds as well as b/y and c/z ions. In general, the a/x, b/y, and c/z ions are the ones most useful for identifying the primary sequence of a peptide or protein. The production of internal ions, ones arising from the cleavage of multiple backbone bonds, is also possible, and they are typically more difficult to assign because they are not anchored by the N- or C-terminus. Two other types of informative but less frequently generated ions include immonium ions, which are low mass ions composed of an individual amino acid, and side-chain loss products classified as d, v, and w ions. The latter are particularly useful for differentiating isomeric leucine/isoleucine and aspartic acid/isoaspartic acid residues in peptides or proteins.
Figure 1.
Nomenclature for peptide and protein fragmentation.
COLLISIONAL ACTIVATION
Collisional activation continues to be the most popular method for characterization of peptides and proteins.24,25 The activation process originates from collisions between ions and inert gas molecules, thus promoting the conversion of kinetic energy to internal energy and causing fragmentation. The simplicity and high fragmentation efficiency of collisional activation methods has ensured their widespread implementation on all types of mass spectrometers. CID has been utilized in countless investigations of peptides and proteins, and its exceptionally robust performance has been showcased in numerous high throughput proteomics applications, resulting in the identification of thousands of proteins. CID for the characterization of peptides or intact proteins has several well-known limitations, largely related to the stepwise mechanism of energy deposition, which leads to preferential cleavage of the lowest energy bonds. This shortcoming hampers the localization of post-translational modifications and limits the effective dissociation of larger proteins due to the limited energy input. In addition, certain residue-specific backbone cleavages may dominate the fragmentation patterns of peptides and proteins, depending on the charge states selected for analysis, an outcome related to the number of mobile protons. The role and impact of mobile protons was recognized as a key concept that influenced peptide fragmentation, ultimately leading to the formulation of the landmark mobile proton model. The mobile proton model rationalizes the fragmentation pathways of protonated peptides on the basis of the number of ionizing protons and their ability to migrate along the peptide backbone to catalyze charge-site initiated backbone cleavages. For example, the propensity for backbone cleavages adjacent to specific amino acids, particularly proline and aspartic acid, may be enhanced, depending on the charge state of the peptide or protein ion, thus suppressing fragmentation at other backbone positions. Despite the impressive metrics of collisional activation, its limitations have motivated the exploration of other ion activation methods, as well as hybrid (combination) methods, many of which are now used as complementary strategies in addition to CID for analysis of peptides and proteins.
IN-SOURCE DISSOCIATION
Conventional CID is typically performed in a collision cell after isolation of a specific precursor ion or set of precursor ions. Alternatively, ions can be activated as they exit the source region during transfer to the ion focusing or mass analyzer regions in a process named in-source dissociation (ISD). ISD is controlled by the voltages applied during the ion transfer process, leading to acceleration of the ions and collisions with gas molecules in a manner rather similar to CID yet without precursor ion selection. ISD can be used not only for ion fragmentation as an alternative to or to complement MS/MS methods but also for improved desolvation and removal of adducts.26–28 The lack of precursor ion selectivity is balanced by the often efficient ability to decongest mass spectra by eroding the heterogeneous population of solvated ions and adducts. ISD has also been used to cleave both backbone and disulfide bonds in peptides29 as well as cleave glycans from glycopeptides, leaving an N-acetylglucosamine attached to the peptide, thus facilitating localization of the sites of glycosylation in subsequent MS/MS experiments.30 Currently, ISD is mainly utilized in investigations of noncovalent protein complexes.31–35 This technique is routinely employed to disassemble protein complexes prior to subsequent characterization of the subunits or individual proteins by MS/MS.33–35 ISD can also be used to simply desolvate proteins and/or protein complexes, which provides better resolution in the resulting mass spectra. Despite being a nonselective activation method, ISD has proven useful in a variety of applications.
ELECTRON-BASED ACTIVATION
Rivalling collisional activation, electron-based dissociation methods have become a powerful mainstay in proteomic analyses. For a thorough examination of electron-based activation methods, readers are directed to recent reviews that provide an excellent recount of the applications, mechanisms, and evolution of these approaches as well as the instrumental and methodological advances that have fostered widespread adoption of these techniques.10,11 Here, we briefly cover the principles of electron-based activation methods and recent advances.
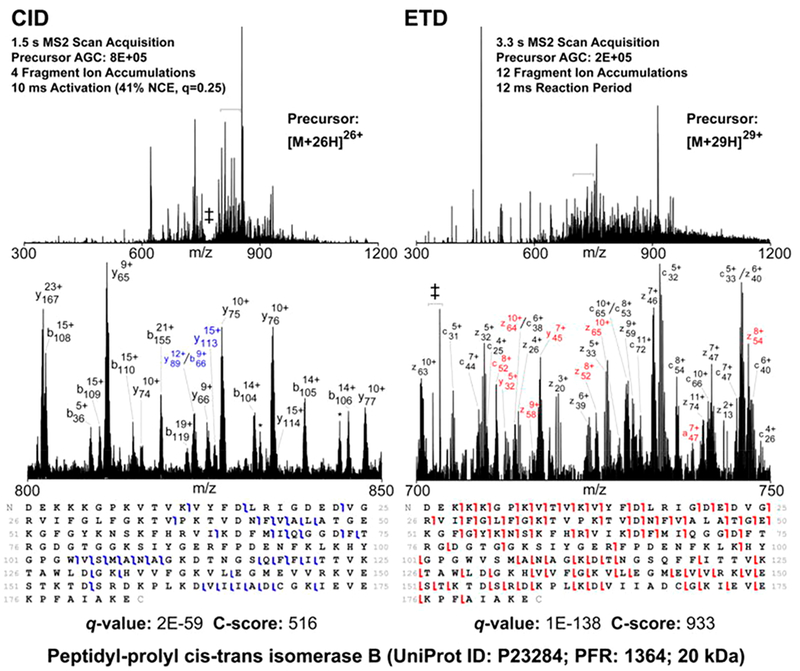
The most mature of these electron-based methods, electron capture dissociation (ECD),36 involves the addition of a low-energy thermal electron to a protein or peptide to generate a charge-reduced radical that may rearrange to induce cleavage of the peptide backbone. Electron transfer dissociation (ETD)37 serves as the ion/ion reaction equivalent of ECD; rather than trapping a free electron, an anionic reagent is introduced to an ion trap mass analyzer holding a population of positively-charged peptides or proteins. When the ions interact, the anion transfers an electron to the peptide or protein to induce both charge reduction and fragmentation of the backbone. Both ECD and ETD lead to preferential cleavage of N–Cα backbone bonds to produce c- and z•-type fragments.36,37 Because the activation/fragmentation process in ETD and ECD occurs with little vibrational energy redistribution, many noncovalent interactions are conserved, and backbone cleavages may reflect protein higher-order structure.38 Conversely, the preservation of noncovalent interactions, particularly for ions with low-charge density, can inhibit separation of the fragment ions, hindering the analysis of protein primary structure and PTM localization. To surpass this hurdle, supplemental excitation has been used to disrupt interactions between fragments, such as via collisional activation39,40 or infrared photoactivation.8–10 A recent breakthrough has been the addition of an IR laser to a quadrupole-Orbitrap-quadrupole linear ion trap hybrid system for the simultaneous irradiation of ETD products with IR photons (known as activated ion-ETD (AI-ETD)) to disrupt higher-order structure and improve sequence coverage.41 Triumphs of AI-ETD spurred by this recent development are discussed in a later section.
Although ECD was originally commercialized on an FTICR mass spectrometer, the widespread implementation of ECD on other platforms has been limited by the need for cotrapping of low-energy electrons and positive ions. Renewed interest in ECD as a viable option for proteomics applications was stoked by the development of a novel electromagnetostatic (EMS) cell42,43 that was utilized recently to equip Orbitrap systems with ECD capabilities.44,45 The cell was designed to require minimal changes to the instrument hardware and to function independently from the instrument’s internal programming. The cell circumvented the need to simultaneously trap positively-charged analyte ions and electrons by placing electrons in the ion flight path. Two ring magnets generate a magnetic field with a “bottle” configuration that axially confines electrons (generated from a heated loop filament in the center of the cell) to be aligned with the trajectory of passing ions. Placement of the EMS cell either prior44 to or after45 the C-trap in a Thermo Q-Exactive Orbitrap mass spectrometer proved successful for ECD of peptides.44,45 Preferential cleavage of disulfide bonds was also observed, pinning potential to this configuration as a tool for disulfide mapping.44 Notable performance of the EMS cell was also demonstrated for top-down analysis of intact proteins.44,45 ECD on the Orbitrap mass spectrometer surpassed the standard higher energy collisional dissociation (HCD) capabilities of this platform, in terms of both sequence coverage and percent matching fragments when applied to myoglobin and ubiquitin.44 Furthermore, this platform afforded the potential to couple ECD with beam-type collisional (EChcD) and photo- (ECuvPD) activation techniques to further enhance protein characterization.45 Although challenges remain toward integrating this ECD mode with data-dependent LC-MS/MS methods, antibody subunits were successfully characterized with high sequence coverage via an LC-MS/MS strategy.45 Overall, this promising development displays the potential to broaden the scope of ECD as a standard tool in proteomics.
Other existing electron-based methods (“hot” ECD (hECD), electronic excitation dissociation (EED), and electron capture-induced dissociation (ECID)) as well as those aimed toward the analysis of anionic peptides (electron-detachment dissociation (EDD), negative electron transfer dissociation (NETD), and negative ion electron capture dissociation (niECD)) have not undergone significant breakthroughs in the last four years in the context of peptide or protein analysis. For detailed descriptions of these methods, readers are directed to a recent review.10
PHOTODISSOCIATION
The use of photons to activate ions has a rich history, in terms of both the diverse photon sources and mass spectrometer platforms utilized for photoactivation and the large array of concomitant applications.13,14,46–53 The implementation of photodissociation requires a suitable source of photons and a means of introducing the photons into the mass spectrometer to intersect or overlap with the ions in a controlled manner. Photon sources successfully used for photodissociation have been cataloged in prior review articles13,14,46–53 and include CO2 lasers, excimer lasers, solid state Nd:YAG lasers, femtosecond titanium sapphire lasers, tunable optical parametric oscillator lasers, gas discharge lamps, synchrotron radiation sources, and even light emitting diodes. Applications of photodissociation have been reported using wavelengths ranging from the infrared (e.g., 10.6 μm) to vacuum ultraviolet. Depending on the mode of photodissociation, the activation process entails absorption of one or more photons resulting in the excitation of vibrational modes (akin to low-energy collisional activation via absorption of multiple IR photons) or access to excited electronic states (via absorption of UV photons). Despite the large and growing number of academic groups that have modified home-built or commercial mass spectrometers for photodissociation, there have been relatively few fully integrated photodissociation-enabled commercial systems. Infrared multiphoton dissociation (IRMPD) is available on a high performance FTICR platform (Bruker Daltonics Inc.), and ultraviolet photodissociation using a 213 nm solid state laser was integrated on an Orbitrap platform (Thermo Fisher Scientific) in 2017.
Readers are directed to other review articles in the past ten years,13,14,46–53 as well as a very recent comprehensive review about ultraviolet photodissociation to be published in 2020,14 that have provided detailed accounts of the instrumentation, mechanisms, and general applications of photodissociation. In recent years, the growing familiarity with the attributes and performance of photodissociation and greater availability of photon sources have motivated the integration of photodissociation with other orthogonal methods, including ion mobility, hydrogen–deuterium exchange, and capillary electrophoresis. Owing to the wide range in the implementation and applications of photoactivation, numerous variations of photodissociation have been developed and explored for the characterization of peptides and proteins, many of which are described throughout this article.13,14,46–53
SURFACE INDUCED DISSOCIATION
Surface induced dissociation (SID) was first reported over 30 years ago and was recognized even then as a method that offered the potential for high-energy deposition owing to the use of a target of enormous mass (a surface) for collisions with ions.54–56 Collision with a surface provides a route for fast, high-energy deposition that opens up new fragmentation pathways not accessible with slower, lower energy methods like CID. SID has been implemented in a wide array of mass spectrometers, including Q-TOF, Orbitrap, and FTICR platforms,57 and a variety of surfaces have been utilized, including graphite, diamond, Langmuir–Blodgett films, fluorocarbon, and functionalized alkanethiolate self-assembled monolayer films. Moreover, SID has proved successful for fragmentation of peptides, proteins, and protein complexes,16,57,58 providing primary sequence and structural information. Recently, it has been shown that SID of protein complexes releases subcomplexes that remain in folded states.16 Additionally, this leads to more symmetric partitioning of charge among the disassembling subunits than observed for CID and UVPD and also allows topologies of protein complexes to be inferred.16 Further, protein–ligand complexes dissociate upon SID to generate fragment ions that contain the bound ligand, yielding insight into the binding site.57,58 With the adoption of SID by a growing number of groups,59,60 its applications and attributes are anticipated to continue to expand, a number of which are described in subsequent sections.61–66
ION/ION PROTON TRANSFER REACTIONS AND PARKING
The analysis of intact proteins by mass spectrometry is challenging due to their high mass, the presence of proteoforms, and the prevalence of multiply charged ions with overlapping charge state envelopes and isotopic profiles, the latter occurring for both precursor ions and fragment ions. Charge state manipulation by ion–ion reactions, a strategy pioneered by the McLuckey group,67,68 can be used to reduce spectral complexity. Ion–ion proton transfer reactions entail the reaction between peptide and protein cations with a proton scavenging anion that extracts protons from the peptides, proteins, or the fragment ions generated after MS/MS, thus causing the reduction of charge states of the ions.67,68 The ions are dispersed through a broader m/z range, thus diminishing the overlap of ions that share the same m/z ratios, ultimately decongesting the spectra. This established approach was adapted to reduce the charge states of histone ions prior to ETD in the recent LC-MS/MS analysis of proteoforms of the H2B histone.69 Charge reduction facilitated the dispersion of isotopic clusters, thus improving the ability to isolate them prior to activation. This strategy allowed one to identify H2B proteoforms that differed by a single amino acid, an analysis that would be difficult to achieve chromatographically.69 A similar approach was used to characterize monoclonal antibodies (mAb), including localization of disulfide bonds.70
During ion/ion proton transfer reactions, it is difficult to avoid sequential charge-reduction reactions that lead to the neutralization of ions. Owing to this reason, the McLuckey group developed a technique based on the application of specific waveforms to the ion trap to allow ions of a specific m/z to be “parked” after undergoing sufficient charge-reduction reactions.71 This manipulation of multiple ions with different m/z values was termed parallel ion parking. More recently, ion parking has been implemented in an Orbitrap platform with PTR and UVPD to analyze intact proteins such as ubiquitin, myoglobin, and carbonic anhydrase72 and determine the charge state dependence of fragmentation by UVPD and HCD. UVPD showed less charge dependence compared to HCD, the latter exhibiting improved performance for intermediate charge states.72 Parallel ion parking was also adapted to control ion–ion proton reactions for the analysis of intact E. coli ribosomal proteins by LC-MS/MS.73 With this approach, the sensitivity of the method was increased by concentrating multiple-charge states into a single-charge state, ultimately allowing the identification of proteins that were below the S/N ratio and not detected in the MS1 survey spectra.73
ION ACTIVATION AND MS/MS APPLICATIONS FOR PEPTIDES, PROTEINS, AND PROTEOMICS
Post-Translational Modifications, Isomeric Amino Acids, and Cyclic Peptides.
The ability to determine minor variations in primary sequences of proteins is a hallmark of tandem mass spectrometry. A goal that transcends basic sequence analysis is the localization of post-translational modifications (PTMs) and differentiation of isomers, an endeavor that has driven advances and innovative applications of separation techniques, hybrid-activation methods, and both top-down and middle-down approaches. This section emphasizes new applications of established tandem mass spectrometry techniques (CAD, electron-based activation, UVPD) to address the challenges of PTM analysis, differentiating isomeric residues, mapping disulfide bridges, and characterizing cyclic peptides.
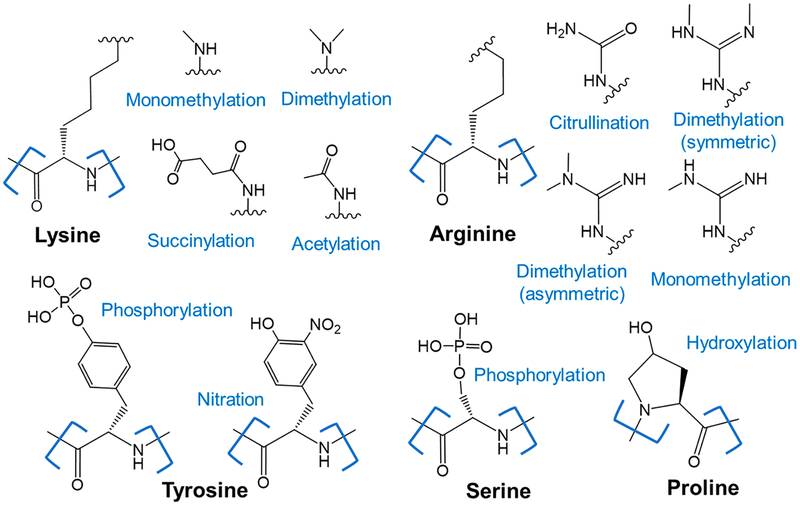
There is an array of different post-translational modifications that result in the covalent attachment of functional groups to side chains of amino acids, as exemplified by the ones shown in Figure 2, and which greatly expand the diversity of the proteome. One of the most common PTMs, protein phosphorylation, regulates almost every basic cellular process including growth, division, and immunity.74 Mass spectrometry-based phosphoproteomics is now an expansive field with a broad suite of innovative tools and methodologies capable of tracking and quantifying protein phosphorylation events at an inspiring scale.2 Readers are directed to recent comprehensive reviews on phosphoproteomics that cover recent advances and discoveries regarding phosphorylation identification/localization strategies, activation methods, and fragmentation mechanisms, with excellent detail regarding benefits and drawbacks of collisional activation, electron activation, and electron activation with supplemental activation.2,3 Many other more specialized reviews on phosphoproteomics and particular diseases have also been published, and thus, this topic will not be covered here.75,76
Figure 2.
Examples of post-translational modifications.
Another important PTM, glycosylation, is a prevalent protein modification that affords particular challenges owing to the variability in the potential sites of modification as well as sizes of the attached glycans, ranging from single sugars to much larger branched polysaccharides. The versatility of separation methods that can be coupled to mass spectrometry and the variety of ion activation methods capable of dissociating the compositionally distinct biopolymers (i.e., oligosaccharides and peptides) have led to significant strides in the applications of mass spectrometry toward glycoproteomics.5,77 Some aspects of glycoprotein analysis are covered in other sections of this review (hybrid activation techniques, top-down, and middle-down proteomics), and are thus not readdressed here. Readers are directed to outstanding recent reviews of glycoproteomics that recount the tandem mass spectrometry-based methodologies adopted by the field.5,77
Exemplifying the maturity of bottom-up proteomics, the ability to characterize a wide array of PTMs showcases the power of currently available mass spectrometry methodologies. In a systematic study spanning common and rare PTMs, the impact of an impressive 21 different types of PTMs on the chromatographic properties and fragmentation behavior of 5000 peptides was evaluated.78 To facilitate this large systematic study, a synthetic peptide library was generated comprising 14 lysine modifications, 4 arginine modifications, 2 tyrosine modifications, and 4-hydroxylation of proline.78 The fragmentation patterns obtained by five MS/MS methods, including HCD, CID, ETD, and hybrid methods EThcD and ETciD, were reported, thus providing one of the largest comparisons of activation methods ever reported.78 Through collection of numerous spectra via LCMS runs and extensive analysis of the large data sets, the effect of each modification on retention time was monitored, while accounting for differences in each peptide, such as peptide length and modification position.78 Scoring for each PTM was also compared across activation methods, with HCD resulting in the highest scores.78 This large study highlighted the potential breadth and utility of multiple ion activation methods for enhancing PTM identification.78 Furthermore, the ability to identify multiple and distinct PTMs aids in unravelling PTM cross-talk, the impact of individual or collections of PTMs on each other and protein function.79 Recently, the identification of both phosphorylated and glycosylated peptides has allowed for the identification of a cross-talk motif for these two common PTMs.79 Through kinetic-based mass spectrometry assays that utilize ETD to localize the PTMs, phosphorylation was determined to hinder O-GlcNAcylation at a (pSp/T)P(V/A/T)(gS/gT) motif. Additionally, it was also found that a common proline-directed kinase phosphorylation site, PX(S/T)P, could not be O-GlcNAcylated, indicating that PTM crosstalk could not occur at this prominent phosphorylation site.79 This revelation serves as an example of the potential biological discoveries now being uncovered as a result of advanced MS/MS methodologies.
Increasingly investigated as a prominent PTM in biotherapeutics, disulfide bridges covalently link cysteine residues to induce proper structural folding and impart protein stability.80–82 In particular, rearrangement or scrambling of the disulfide cross-links in monoclonal antibodies (mAbs) can reduce thermostability and antigen affinity.81 Understanding and mapping disulfide bridges, however, comes with special challenges due to the complexity of pinpointing a PTM that simultaneously links and modifies two distinct residues. Several novel methodologies have been developed to map disulfide linked cysteine residues, such as selective labeling and the cleavage of disulfide bonds via ion/ion reactions.83 The activation of polypeptides complexed with oxidizing reagent anions, such as periodate, resulted in the oxidation of the disulfide to generate a thiosulfinate derivative that rearranged to release and allow identification of the bound peptides.83 Cleavage of some portion of the disulfide linkages can also be achieved by conventional ETD through gas-phase reduction of the disulfide bond. Subsequent HCD of the free peptides generates sequence fragments that can unambiguously identify disulfide bound cysteine pairs. This ETD-HCD strategy was recently applied to completely characterize the previously unknown disulfide structure of murine Meteorin, a newly discovered neurotrophic factor that has shown potential as a drug for neuropathic pain.84 Proteolytic digestion of the protein followed by LC-MS resulted in peptide clusters containing multiple disulfide bonds. Cysteine binding partners were unambiguously identified by employing ETD to reduce some fraction of the disulfide bonds and release free peptides for subsequent identification by HCD, resulting in a complete disulfide map.84 Yet, another method for disulfide bond mapping involves the use of photoactivation. Building from previous work that demonstrated the successful use of UV photons (157 nm85 and 266 nm86) to cleave disulfide bonds in the gas phase, both 193 and 213 nm UVPD have recently exhibited potential as disulfide mapping tools.87–89 193 and 213 nm UVPD induce simultaneous cleavage of disulfide bonds and the peptide backbone.87–89 In one study, 193 nm UVPD was used to release the A- and B-chains of insulin while also producing peptide sequence ions with either reduced or intact intramolecular disulfide bridges.87 Furthermore, partial reduction and alkylation of tryptic peptides using Tris(2-carboxyethyl)phosphine (TCEP, a reducing agent) and N-ethylmaleimide (NEM, an alkylating agent) prior to 193 nm UVPD allowed the four disulfide bonds of lysozyme and the 19 disulfide bonds of serotransferrin to be deciphered in an LC-MS/MS workflow.87 In a different approach, it was shown that 213 nm UVPD of disulfide bound peptides generated diagnostic sets of triplet ions corresponding to fragments that differed by 32 Da, originating from cleavage of the S–S bond and the adjacent C–S bonds.88,89 Exploiting the presence of this triplet ion motif in the resulting MS/MS spectra, a two-tiered data processing approach was implemented to increase confidence and simplify data analysis for cross-linking assignments.89 Although sequence fragments were not as prominent as those observed using 193 nm UVPD, sufficient sequence information was produced by 213 nm UVPD to allow assignment of disulfide-bridged peptides in a peptic digest of mAb Rituximab.89
Distinguishing isomeric amino acids in peptides and proteins is also a demanding endeavor that has prompted the search for new MS/MS strategies incorporating multimodal activation methods. Distinction of the isomeric isoleucine and leucine amino acids has been a key target of interest for antibody de novo sequencing.90 Combinations of several MS/MS workflows, including utilizing HCD MSn and ETD-HCD methods, and integration of these multistage MSn strategies on an LC time scale have been enabled by novel platforms.91,92 Translating a multistage CID into a multistage HCD strategy on an Orbitrap platform simplified and reduced analysis time previously required for differentiation of Ile/Leu by conventional low-energy CID.91,92 The strategy relied on the production of a diagnostic immonium ion of m/z 86 that was not consistently observed upon CID owing to the low-mass cutoff inherent to quadrupole ion trapping instruments.93 Subsequent activation of the m/z 86 ion by HCD leads to production of an ion of m/z 69 that is characteristic for isoleucine.91–93 The presence of multiple Ile/Leu residues in an amino acid may confound identification through this approach. In such cases, HCD may be used to cleave the peptide to generate b/y ions that only contain one such amino acid.91 Subsequent dissociation of these b/y products by HCD also allows the same m/z 86 to m/z 69 transition to be mapped. Alternatively, ETD may be used to generate z ions via backbone cleavage N-terminal to Ile/Leu residues. Subsequent HCD produces side-chain losses of either 43 or 29 Da that identify the residue as either leucine or isoleucine, respectively.91,92,94 Although these multistage strategies have limited utility for some peptides based on their sizes or charge states, these methods are complementary to one another and have been demonstrated on an Orbitrap platform on an LC time scale.91,92 Multienzyme digestion and the described LC-MSn strategies have been used to successfully distinguish Ile and Leu residues in the CDR regions of mAb1, mAb2, and the Waters mAb standard.91,92 Similarly, multistage HCD of dimethylated peptides also resulted in diagnostic a1 product ions for Ile/Leu,95 resulting in the identification of 71% to 94% of the Ile/Leu residues on the heavy and light chains of dimethylated SILuLite and Trastuzumab mAbs, respectively.95 When the differentiation of isomeric peptides was allowed, these new MS/MS methods advanced the feasibility of de novo sequencing through mass spectrometry.
Lacking defined termini, nonlinear peptides like cyclotides and stapled peptides present a unique challenge to tandem mass spectrometry for characterizing primary structure. The production of informative sequence-type fragment ions upon activation of cyclic peptides requires two backbone cleavages, as the first cleavage only opens the ring. Additionally, the interpretation of the resulting tandem mass spectra is complicated by the lack of an anchor point to which other dissociation events can be referenced as the conventional fragment ions (b/y, c/z, a/x) cannot be assigned. Chemical derivatization has been used to circumvent this problem and promote preferential fragmentation at specific residues to allow prediction of one of the cleavage sites and simplify characterization.96 Through selective deguanidination of arginine residues to convert them to ornithines, a selective cleavage site can be incorporated to induce backbone dissociation C-terminal to the modified residue.96 In this manner, both CID and 193 nm UVPD of stapled peptide sequence variants resulted in unique fragments that were used to aid the characterization of cyclic peptides and isobaric stapled peptides.96 Two other investigations utilizing 193 nm UVPD for analysis of cyclic peptides have been reported.97,98 For microcystin peptides, HCD, CID, multistage CID (MS3), and UVPD were used to produce product ions specific to different sequence variants, with UVPD performance comparable to HCD.98 For the cyclotide cyO8, UVPD resulted in spectra containing a diverse array of fragment types that allowed a confident assignment of fragments spanning approximately half of the peptide backbone.97 Although currently limited by the lack of appropriate data analysis tools, UVPD induces cross-ring cleavages and yields rich spectra that could harbor the necessary information for de novo sequencing of these challenging peptides.97
Data Acquisition Methods.
Tandem MS is a powerful tool for the characterization of peptides and proteins; however, the rate of data acquisition is slowed by the common practice of isolating and interrogating individual precursors in a serial manner. The development of various multiplex MS/MS strategies, broadly called data independent acquisition methods, entails the simultaneous activation of broader m/z windows of precursor ions, followed by various postprocessing methods to link fragmentation patterns to specific precursors. These advanced data acquisition strategies allow workflows to be specially tailored for quantitative and/or higher throughput applications in proteomics. Ubiquitous across all proteomic approaches, the conventional data-dependent acquisition (DDA) strategy entails isolation and activation of the most abundant peptides detected in a survey MS1 scan in order to generate high-quality MS/MS spectra. Achieving incredible proteomic depth, this long-standing approach has become the standard workflow for proteome discovery-type applications.99 Exchanging broad proteome coverage for specificity and sensitivity, targeted methods selectively monitor specific peptides, usually to improve upon the quantitative metrics and circumvent the stochastic nature of peptide precursor ion selection of DDA. Regarded as the gold standard for quantitative proteomics, selected reaction monitoring (SRM) entails isolation and activation of selected precursors and monitors specific fragment ions generated from those precursors.100 Together, the precursor and fragment ions comprise transitions that can be tracked with high accuracy, reproducibility, and sensitivity for peptide (and by inference protein) quantification. Parallel reaction monitoring (PRM) offers a targeted approach that maintains some flexibility in postacquisition data analysis by monitoring all fragments generated by the precursor.101 Transitions best suited for quantitative analysis can then be selectively utilized to quantify each peptide. Foregoing precursor isolation, data-independent acquisition (DIA) was introduced as an alternative to DDA with improved quantitative metrics, yet without a limit in the number of transitions that can be monitored in a single experiment.102 Several incarnations of DIA have since arisen, including wide selection ion monitoring (WiSIM-DIA) as an alternative that quantifies peptides on the basis of precursor intensity, rather than from MS2 features, as is done in traditional DIA.103 These landmark data acquisition methods have been the subject of recent reviews.99–102,104
One common DIA strategy, sequential window acquisition of all theoretical mass spectra (SWATH-MS), has gained major interest for deep proteomic analyses. The strategy, along with the implementation and optimization thereof, are expertly detailed in a previous tutorial.104 MS/MS is implemented by simultaneous activation of all ions in a defined m/z window, using multiple windows to cover a wide range of precursors with high efficiency. Typically, a peptide-centric scoring approach is taken where a spectral library containing high-quality MS2 spectra for known peptides is used to parse through the data based on defined precursor-to-fragment ion transitions and expected retention times. Figures-of-merit, including LLOQs, linear dynamic range, and coefficients of variation, of this now widely adopted approach were evaluated in a systematic assessment spanning 11 laboratories.105
Leveraging the reproducibility of this high throughput ion activation strategy, SWATH-MS has been implemented to effectively track quantitative changes in biological systems at the protein and PTM level. When evaluating the extracellular matrix (ECM) and ECM-associated proteins, collectively known as the matrisome, SWATH-MS resulted in more reproducible and comprehensive analysis than conventional DDA.106 SWATH-MS identified 54% more liver matrisomal proteins compared to DDA, without the need for utilizing extensive enrichment strategies.106 SWATH-MS displayed overall lower sensitivity in the analysis of phosphorylated peptides but comparable performance in terms of quantification, while offering the advantage of quantifying unexpected phosphorylations when compared to SRM.107 SWATH has also been adapted to target the protein glycosylation modifications, most notably N-glycosylation based on the alignment of diagnostic fragment ion chromatograms with peptide precursor chromatograms.108,109 This SWATH-MS approach allowed the identification of unique glycoforms for immunoglobulin M in a complex lysate108 as well as unexpected truncated glycopeptides of immunoglobulin G1.109 Furthermore, DIA technologies have been adapted to monitor N-glycoform stoichiometry at the proteome scale by fragmenting both the modified and unmodified peptides, allowing characterization of over 1000 N-glycosites in HEK 293 cells.110
Improvements upon data analysis and acquisition in the traditional SWATH-MS workflow have further enhanced peptide identification and quantification. An experimental approach, EncylcopeDIA, building and searching chromatographic libraries based on the fragment ion chromatographic peak shapes and retention times, demonstrated the detection of 20–25% more peptides in human and yeast cells than traditional DDA libraries.110 Improved demultiplexing algorithms have also enabled the collection of SWATH spectra acquired using overlapping windows to yield an increase in precursor selectivity by almost a factor of 2 with a 64% improvement in sensitivity and 17% increase in number of peptides detected.111
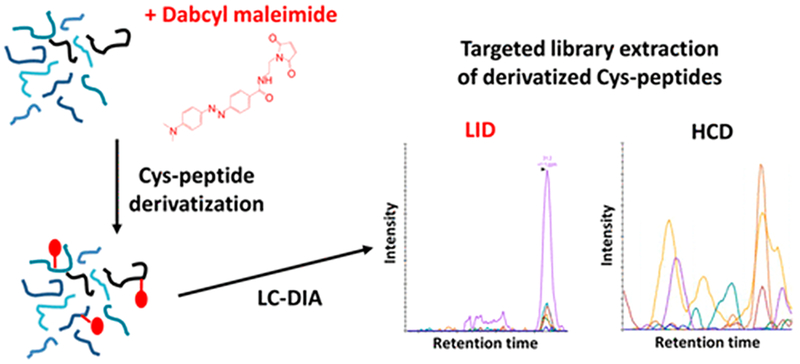
Other SWATH-MS efforts have focused on incorporating alternate activation methods to introduce specificity into peptide fragmentation, effectively reducing the complexity of the chimeric spectra inherent to DIA. Both IRMPD and laser-induced dissociation (LID) have been used to preferentially dissociate cysteine-modified peptides in complex mixtures.112,113 Because sulfoxides have a high IR absorbance at 10.6 μm, IRMPD allowed differentiation of cysteine-containing peptides with S-sulfonylation oxidative modifications from unmodified peptides.112 473 nm of LID of Dabcyl-derivatized peptides was used to preferentially target cysteine-containing peptides, thus providing much greater selectivity in the activation and fragmentation of only those peptides that contained the UV-absorbing Dabcyl tag, as illustrated in Figure 3.113 An expansive LID spectral library consisting of 354 synthetic peptide surrogates for human-related protein kinases was generated to enable the DIA-LID approach. Using the spectral library, DIA-LID led to the identification of 76% of the synthetic peptides when spiked into a complex E. coli lysate.113
Figure 3.
Tagging of cysteine residues with Dabcyl allows selective activation/fragmentation of cysteine-containing peptides using 473 nm laser-induced dissociation in a DIA workflow.113 Reprinted from Garcia, L.; Girod, M.; Rompais, M.; Dugourd, P.; Carapito, C.; Lemoine, J. Data-Independent Acquisition Coupled to Visible Laser-Induced Dissociation at 473 Nm (DIA-LID) for Peptide-Centric Specific Analysis of Cysteine-Containing Peptide Subset. Anal. Chem. 2018, 90, 3928–3935. Copyright 2018 American Chemical Society.
An alternative form of SWATH-MS is broadband DIA in which all ions observed in the MS1 spectra are activated, typically using in-source CID (MSE),114 without any precursor isolation. Although collisional activation is the most common method used for DIA strategies, ETD was recently introduced (MSETD) as an alternative activation mode, aiming to provide a complementary strategy to MSE.115 MSE applied in a top-down manner has also been shown to be feasible on a TOF platform by rapid switching of the voltage used for in-source dissociation in order to record spectra with and without ion activation.116 This method allowed the identification of proteoforms from 14 of the subunits of the 20S proteasome.116
In addition to the vast developments in the implementation and application of DIA strategies, DDA schemes that improve upon the established workflow have also emerged. Advanced peak detection (APD) enhances charge state assignment to target peptide features that would otherwise be missed in a typical DDA LC-MS/MS experiment.117 APD implements an iterative approach to annotate multiple isotope distributions that overlap in m/z space and thus increases the number of unique peptide identifications.117 Another DDA strategy aimed at improving the dynamic range of high throughput proteomics is termed BoxCar acquisition, entailing fractionation of the survey MS1 spectra into separate m/z windows in order to increase S/N of low abundance species that would otherwise be suppressed by more abundant ions.118
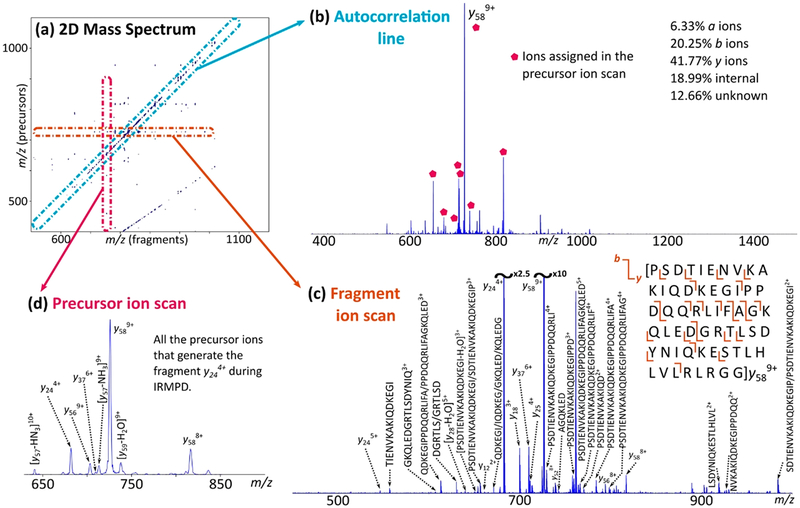
The multiplexing concept for MS/MS data collection has also been implemented on FTICR mass spectrometers.119 All ions in a sample are activated without ion isolation through the use of a more elaborate pulse sequence.119 Ions of different m/z ratios are moved in and out of the ICR cell through modulation of their radii according to ion cyclotron frequency by using identical pulses separated by an encoding time. This encoding time specifies ions of different m/z for selective dissociation, which is performed until the whole m/z range of interest is covered. The two-dimensional (2D) Fourier transform yields a 2D graph correlating the precursor ions with the respective fragment ions. Although initially conceived in 1987,120 the broader application of 2D MS required more advanced algorithms to meet the computational demands of the method. Figure 4 shows the schematic representation of a 2D mass spectrum. Various characteristic lines are observed: the autocorrelation line, fragment ion scans, and precursor ion scans. The y axis represents the precursor ions (m/z), while the x axis represents the fragment ions (m/z).
Figure 4.
CAD-MS/IRMPD-2DMS of ubiquitin in denaturing conditions. (a) Two-dimensional mass spectrum, output of the CAD-MS/IRMPD-2DMS analysis of ubiquitin. The autocorrelation line and horizontal fragmentation patterns are observed. The curved line departing from the bottom of the spectrum constitutes harmonics signal. (b) Extraction of the autocorrelation line, with the percentages of the correctly assigned ions. Labeled ions represent the ions assigned through the precursor ion scan shown in spectrum d; they include ion y589+, whose fragmentation pattern is shown in spectrum c. (c) Fragment ion scan of the ion y589+ (m/z 726.2838) and respective cleavage coverage. Nondiagnostic neutral losses are not labeled for better visualization. (d) Precursor ion scan of the ion y244+ (m/z 682.3820), revealing important information for the assignment of y ions on the autocorrelation line (b).124 Reprinted from Floris, F.; Chiron, L.; Lynch, A. M.; Barrow, M. P.; Delsuc, M.-A.; O’Connor, P. B. Top-Down Deep Sequencing of Ubiquitin Using Two-Dimensional Mass Spectrometry. Anal. Chem. 2018, 90, 7302–7309. Copyright 2018 American Chemical Society.
Two dimensional MS has been used for both bottom-up and top-down analysis of proteins.26,119,121–124 In fact, both approaches were used in the same study for the analysis of calmodulin with IRMPD as the activation method,121 and the results were comparable to those of conventional one-dimensional MS/MS with the advantage of saving considerable data acquisition time. Despite the extreme complexity of the spectra, a good sequence coverage (~40%) was obtained. In a subsequent study, electron-capture dissociation was employed in the 2D MS strategy for the bottom-up analysis of calmodulin.122 The use of ECD resulted in a sequence coverage of ~47%, somewhat higher than the sequence coverage obtained using IRMPD (~40%), and combining both activation methods improved the sequence coverage to ~68%. For further gains in performance, a hybrid method, IR-ECD,123 was implemented to improve de novo peptide sequencing based on extensive production of c/z and b/y ions as demonstrated for cytochrome c. Additionally, IR-ECD facilitated side-chain losses, resulting in wa-type fragments that allowed the differentiation of leucine and isoleucine isomers.123 Recently, an MS3 strategy was used for the top-down analysis of calmodulin and ubiquitin26,124 in which the 2D MS method was employed to interrogate fragment ions produced by an initial stage of CAD. For a small protein like ubiquitin, 97% sequence coverage was obtained.124 The multiplexing capabilities of 2D MS to increase the depth of structural information have many other potential applications that are expected to emerge in the years ahead.
Hybrid Activation Methods.
To circumvent limitations of current modes of activation, different techniques have been combined into a single ion activation step or sequential steps to leverage the benefits of constituent techniques for enhanced analysis of peptides and proteins. Electron-based dissociation in conjunction with collisional or infrared supplemental activation superbly showcases the merits of these methods. For example, despite the success of ETD, a drawback has been the lack of informative sequence fragments upon activation of low-charge density fragments (typically ions in low-charge states). Nondissociative electron transfer (termed ETnoD) predominates due to noncovalent interactions that survive the activation step and hamper the separation and detection of fragment ions. One of the original methods developed to overcome the ETnoD problem entailed utilizing collisional activation to promote disassembly of the products.39 Similarly, implementing broadband HCD of all ETD products (EThcD) not only increased the yield of diagnostic sequence ions but also resulted in production of b/y ions through activation of the unreacted precursors.40 EThcD has garnered extensive use, in large part owing to its commercialization and accessibility. With the widespread availability of EThcD, this hybrid method has been adopted in recent years for identifying and localizing PTMs that are otherwise difficult to characterize, such as phosphorylation,2,125 glycosylation,77,126 ADP-ribosylation,127,128 pyrophosphorylation,129 and disulfide bridges,130 as well as to distinguish isomeric amino acids such as isoleucine/leucine131 and 3-/4-hydroxyproline isomers.132 EThcD has also been shown to be compatible with isobaric chemical tags (like DiLeu and TMT) to enhance data quality for quantitative proteomics.133 Several further applications of EThcD are addressed in recent reviews.2,11,77
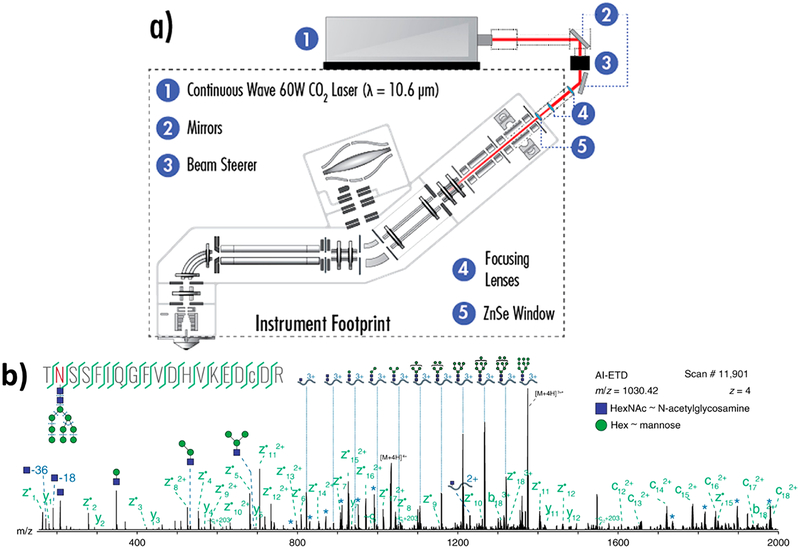
Originally demonstrated on a dual cell linear ion trap,134 activated ion ETD (AI-ETD), a method combining ETD with simultaneous infrared photoactivation, was recently implemented on an Orbitrap platform (Figure 5a).41 Concurrent slow heating of the precursor through absorption of IR photons unfolds the peptide during ETD, which decreases the overall activation time and the extent of hydrogen transfer reactions. As illustrated for shotgun proteomics, AI-ETD almost doubled the number of peptide spectra matches (PSMs) compared to ETD and demonstrated significant gains in performance for low-charge density precursors detected in a mouse brain tryptic digest.41 Another variation of this method, AI-ETD+,41 entailed the photoactivation of AI-ETD products after their transfer to the low-pressure linear ion trap, which enhanced formation of b/y fragment ions. In comparison to AI-ETD and EThcD, AI-ETD+ resulted in a greater number of PSMs and generally provided greater sequence coverage.41 For glycoproteomic analyses, AI-ETD generated c/z fragments that largely retained the intact glycan, aiding in glycopeptide characterization as well as producing Y ions and oxonium ions via glycosidic bond cleavage that allowed characterization of the glycan, as illustrated for one glycopeptide in Figure 5b.135 For phosphoproteomics, AI-ETD identified and localized 24 503 phosphopeptide PSMs, tripling the identifications generated by ETD alone and outperforming ETcaD and EThcD, even though a significant neutral loss of the phosphate group was observed upon photoirradiation.136 AI-ETD yielded higher sequence coverage than ETD, HCD, and EThcD for phosphorylated α-casein, allowing unambiguous localization of the eight phosphorylation modifications.136 When adapted for a top-down workflow, AI-ETD bested ETD, EThcD, and HCD for sequence coverage of both low molecular weight (<20 kDa)137 and high molecular weight (30–70 kDa)138 proteins. AI-ETD was used to boost sequence coverage of proteins possessing disulfide bonds139 and for high throughput top-down proteomics.140 Utilizing AI-ETD, 935 proteoforms were identified from human colorectal cell line HCT116, compared to 1014, 915, and 871 proteoforms identified using HCD, ETD, and EThcD, respectively.140
Figure 5.
(a) Instrument schematic for activated ion-electron transfer dissociation (AI-ETD).41 (b) Annotated single AI-ETD spectrum (i.e., no averaging) of N-glycopeptide TN*SSFIQGFVDHVKEDcDR modified with a high mannose-type glycan [HexNAc(2)Hex(9)]. The red asparagine indicates the site of glycosylation, and the lowercase cysteine indicates carbamidomethylation. Green fragments are products from peptide backbone cleavage; triply-charged y ions are annotated along the top, and b ions include only glycan moieties. Blue asterisks (*) denote doubly- and quadruply-charged y ions (from 1700 to 2000 and 750 to 1000 Th, respectively), each which differ by one hexose residue. Peptide fragments retain the glycan modification unless denoted by a “~”.135 Adapted from Riley, N. M.; Westphall, M. S.; Hebert, A. S.; Coon, J. J. Implementation of Activated Ion Electron Transfer Dissociation on a Quadrupole-Orbitrap-Linear Ion Trap Hybrid Mass Spectrometer. Anal. Chem. 2017, 89, 6358–6366. Copyright 2017 American Chemical Society. Adapted with permission from Riley, N. M.; Hebert, A. S.; Westphall, M. S.; Coon, J. J. Capturing Site-Specific Heterogeneity with Large-Scale N-Glycoproteome Analysis. Nat. Commun. 2019, 10, 1–13. Copyright 2019 Springer Nature.
High-energy 213 nm UV photodissociation has also been coupled with low-energy IR irradiation (termed HiLoPD) to increase the diversity of fragment types achieved by either high-energy or low-energy photons alone.141,142 HiLoPD provided a rich array and even distribution of a/x, b/y, and z ions as well as d, v, and w ions useful for distinguishing isomeric amino acids, such as leucine and isoleucine. For the analysis of ubiquitin, HiLoPD displayed greater sequence coverage of the protein (83%) compared to 213 nm UVPD (76%).141 In the context of mapping post-translational modifications, this method was capable of identifying and localizing modifications such as phospho-tyrosine, phospho-threonine, and sulfo- and glyco-peptides.142
Hybrid ECD methods, particularly ECuvPD and EChcD, were recently implemented on an Orbitrap mass spectrometer equipped with a modular electromagnetostatic ECD cell.45 The performance of ECD was compared to ECuvPD and EChcD in the characterization of proteins like carbonic anhydrase and subunits of monoclonal antibodies, with each method returning good sequence coverage. The best outcome was obtained by combining the results of all three methods, yielding, for example, 89%, 92%, and 76% sequence coverage for the LC, Fc/2, and Fd subunits of the mAbs, respectively.45
Advances in Separations.
Crucial to expanding the power and depth of MS/MS, innovative separation methods have expanded the scale and scope of proteoform analyses. Although the popular use of reversed-phase chromatography coupled with collisional dissociation has become indispensable to standard proteomics workflows,4,6,7 recent innovations have focused on the development of alternative separation techniques, with an emphasis on methods to enhance characterization of PTMs and to facilitate middle-down and top-down analysis. One such technique is size exclusion chromatography (SEC). In SEC, proteins and peptides are separated in MS-friendly mobile phases by size, rather than by hydrophobicity.143,144 For example, SEC has been used to cause coelution of isomers of oxidized peptides, while identification and relative quantification was achieved through ETD.145 The application of SEC to purify large proteins (>200 kDa) prior to limited proteolysis and analysis of the resulting large peptides by ETD enabled the comprehensive characterization (including identification and localization of all PTMs) of the 226 kDa heavy chain of cardiac myosin.146 Formerly limited by low chromatographic resolution, SEC has been shown to produce robust, reproducible, high resolution separations of complex mixtures by the utilization of serial SEC (sSEC).147,148 sSEC is done by combining SEC columns of decreasing pore sizes in series, and it afforded high resolution separations of high MW, intermediate MW, and low MW proteins in complex mixtures over a range of 10–220 kDa. In the first application of this technique, a sarcomeric protein mixture was analyzed in a top-down manner via sSEC-reversed phase chromatography.147 Utilizing three polyhydroxyethyl A columns (with pore sizes of 1000 Å/500 Å/500 Å) in series, the protein mixture was fractionated and each fraction was subsequently separated using a polymeric reversed phase (PLRP) analytical column coupled to a Q-TOF mass spectrometer, resulting in the identification of over 4000 additional proteoforms than a standard reversed-phase LCMS workflow.147 Additionally, sSEC-RPC resulted in a 15-fold increase in the detection of proteoforms greater than 60 kDa and proved especially useful for the analysis of both high MW proteins (>200 kDa) and low abundance proteins containing PTMs.147 Given the excellent separation of high MW proteins in MS compatible solvents, sSEC was used to take full advantage of the ultrahigh resolving power and mass accuracy of FTICR MS for top-down analysis.148 sSEC fractionation of the cytosolic heart proteome improved the sensitivity for the analysis of high MW proteoforms (>30 kDa), facilitating their identification and characterization via ECD.148 Sample dilution, however, has continued to impede more widespread adoption of sSEC as a separation method, in some cases requiring the pooling of multiple fractions to ensure sufficient S/N for the analysis of intact proteins.
Another separation technique that has been adapted for online MS/MS analysis of intact proteins is hydrophobic interaction chromatography (HIC). Complementary to RPC, HIC separates proteins on the basis of hydrophobicity and largely preserves the native structure.149 The elution order is modulated on the basis of interactions between surface exposed hydrophobic residues in the protein tertiary structure and stationary phase ligands of moderate hydrophobicity. Traditional HIC stationary phases, however, require the use of salt gradients that are incompatible with mass spectrometry. To address this challenge, a series of increasingly hydrophobic materials were designed to be compatible with MS-friendly (<1M) levels of ammonium acetate in the mobile phase.150 With these new materials (PolyPENTYL A, PolyHEXYL A, and PolyHEPTYL A) serving as hydrophobic stationary phases, reproducible retention times and well-defined peak shapes were achieved for a standard protein mixture using a diminishing ammonium acetate gradient (1 M to 20 mM) in 50% acetonitrile.150 To showcase the utility of HIC to enhance MS/MS, intact monoclonal antibodies (mAbs) were purified with the aforementioned HIC materials and characterized using ECD on a 12 T FTICR mass spectrometer.151 This strategy yielded impressive 66% and 57% sequence coverages of the light and heavy chains, respectively.151 Although still under development for online proteomics applications, HIC offers a promising complementary separation technique relative to traditional reversed-phase chromatography methods.
Capillary zone electrophoresis (CZE) is another alternative separation technique that has garnered considerable interest for protein analyses, as recently described in a comprehensive review.152 Previous impediments for the application of CZE as a viable tool for top-down proteomics included the low injection volumes permitted by this method and confounding protein adsorption onto the inner capillary wall. The use of a dynamic pH junction technique153 and the development of linear polyacrylamide (LPA)-coated capillaries154 have increased sample loading capacity and reduced sample adsorption, respectively. The combination of these two approaches resulted in the identification of 580 proteoforms, including 180 different proteins, from fractionated yeast via a top-down workflow using CAD as the MS/MS mode.155 The largest bacterial top-down proteomics data set to date was achieved through a multidimensional SEC-RPC-CZE method, again relying on CAD for the identification of 5700 proteoforms in the E. coli proteome.156 Other efforts to enhance top-down characterization of proteins using CZE-MS have focused on the incorporation of alternative ion activation techniques. For example, integrating AI-ETD157 or UVPD158 with SEC-CZE resulted in some of the largest proteomic data sets achieved by each MS/MS method for top-down analysis. SEC-CZE paired with AI-ETD led to the identification of 3028 proteoforms in an E. coli lysate, capturing multiple PTMs including N-terminal acetylation, methylation, S-thiolation, disulfide bonds, and lysine succinylation.157 Furthermore, CZE-AI-ETD performance afforded higher quality spectra displaying greater numbers of sequence informative fragment ions compared to HCD for a number of the identified proteins.157 SEC-CZE-UVPD (213 nm) of proteins extracted from zebrafish brain resulted in the identification of 600 proteoforms, encompassing 369 proteins.158
Other work has focused on using CZE to improve the identification of peptides containing specific PTMs that are otherwise difficult to separate or detect.159–161 As also reported for top-down proteomics, LPA-coated capillaries and the dynamic pH technique have improved CZE for bottom-up applications. For example, a CZE-CID strategy was used for the analysis of peptides generated from proteolysis of antibody drug conjugates159 and both ETD and CID were used to characterize peptides from histones after CZE separation.160 Using an advanced peak determination algorithm (APD),117 a massive data set composed of 4405 identifications was obtained from the CZE-MS/MS analysis of phosphopeptides enriched after LysC proteolysis of a mouse brain extract.161 This large data set was also used to explore the electrophoretic mobilities of phosphopeptides and confirmed the complementarity of CZE-MS/MS and LC-MS/MS for phosphopeptidomics.161
CZE-MS has also recently been adapted for the separation and analysis of nondenatured (native-like) proteins and protein complexes, with HCD for identification.162,163 For the latter, the E. coli proteome was prefractionated using SEC with subsequent analysis of the fractions using CZE-MS/MS. 144 proteins, 672 proteoforms, and 23 protein complexes were identified in this high throughput analysis.163
The examples described above emphasize the benefits of coupling alternative separation techniques with tandem MS for peptide and protein analysis. In a number of the highlighted studies, the most impressive results were achieved by implementing orthogonal fractionation methods to maximize the depth and breadth of analysis,146–148,155–158 and this is likely to become an increasingly popular option for mining more information from complex mixtures in the future.
Structural Proteomics.
Structural proteomic techniques, such as hydrogen–deuterium exchange (HDX),164,165 chemical cross-linking (XL),166,167 and covalent labeling (CL),168,169 induce isotopic or chemical changes that capture the higher-order structure of proteins. Tandem mass spectrometry tracks changes in amino acid composition introduced by each strategy with escalating structural resolution and detail as new developments in the application of ion activation arise. Some of the recent studies that have benefitted from new advances in ion activation are highlighted here.
By encoding solvent accessibility into the isotope distribution, HDX-MS probes the protein structure in solution.164,165 When proteins are diluted in D2O, backbone amide hydrogen atoms are exchanged for deuterium atoms. As only amide hydrogens accessible to the solvent can be displaced, backbone hydrogen bonds, transient interactions, folding, and unfolding events sequester regions of the backbone from the solvent and mediate the rate of H/D exchange, slowing down the process in ordered regions. The rate of H/D exchange can be monitored through high-resolution mass spectrometry by tracking the mass shifts or changes in isotope distributions incurred by the deuterium uptake. Sites of deuterium incorporation are more highly resolved via MS/MS analysis of the labeled peptides (bottom-up or middle-down methods) or proteins (top-down methods). Monitoring the fragmentation patterns of deuterated peptides or proteins is a key method used to obtain site-specific information about isotopic labeling, but H/D scrambling during ion activation nullifies much of the information obtained from the diagnostic fragment ions. H/D scrambling during collisional activation has been reported in many prior studies,170,171 whereas the extent of scrambling has been found to be minimal for electron-activation methods.172 The quest for new activation methods that mitigate hydrogen migration has motivated the exploration of UVPD for HDX studies. 213 nm UVPD exhibited a low degree of scrambling, evidenced by measuring the deuterium content of a/x and c/z fragments from the peptide scrambling probe P1 (sequence HHHHHHIIKIIK2).173 Top-down strategies have also been utilized for HDX-MS, again with an emphasis on finding and utilizing activation methods that minimize scrambling, such as ECD174 and ETD.175,176 213 nm UVPD was reported to proceed without scrambling on the basis of the analysis of deuterium-labeled myoglobin and resulted in sequence fragments complementary to those produced by ETD.177 In this case, each activation method yielded 58% sequence coverage individually but 81% when combined.177
As also noted in the section on PTM analysis, middle-down approaches have been adopted for HDX-MS to study proteins too large to be amenable for top-down workflows.176,178 HDX and limited pepsin digestion prior to disulfide reduction of antibody Herceptin (150 kDa) resulted in the production of large peptides (12–25 kDa) that afforded extensive HDX information on the entire light chain and 95% of the heavy chain via ETD.178 Comparisons between the bottom-up, middle-down, and top-down approaches for the characterization of histone tail dynamics reported that comparable HDX results were achieved by each approach, but inherent to the ETD-based top-down and middle-down approaches, HDX sites could be localized with superior resolution by the MS/MS approaches.176 As the interpretation of and ability to draw meaningful conclusions from HDX data clearly benefits from ion activation methods that minimize scrambling, the increasing availability of platforms suitable for photodissociation and electron dissociation of large peptides and proteins holds great promise for the application of HDX in structural proteomics.
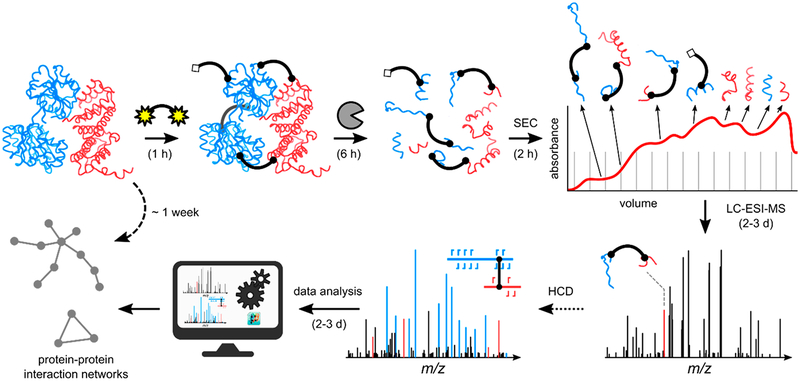
Capable of “freezing” and mapping otherwise transient interactions, protein chemical cross-linking coupled to MS/MS has evolved into a high-throughput method for the elucidation of protein structure and the identification of protein–protein interactions166,167,179 (see the typical workflow in Figure 6). In this technique, a chemical agent of known size and chemistry introduces an inter- or intraprotein covalent bond between amino acid side chains or between an amino acid and another biomolecule.166,167,179 Following the incorporation of the covalent linkages, the proteins are generally proteolyzed and analyzed via bottom-up MS/MS methods to identify the cross-linking sites and reconstruct the spatial arrangements within proteins or interacting proteins. When two peptides are found to be bound using protein chemical cross-linking, it suggests that they are adjacent in the native environment.166,167,179 Previously, a major factor stunting the application of this technique to complex mixtures was the n2 problem.179 That is, the complexity of the search space for peptide combinations increases quadratically as the number (n) of possible peptide candidates increases. The key to overcoming this hurdle is the introduction of gas-phase cleavable cross-linkers to circumvent bioinformatic computational stress by reducing the complexity of data analysis. Some of the original examples of cleavable cross-linkers included the protein interaction reporter (PIR),180 which utilized an MS3 strategy for confident identification of linked peptides, and disuccinimidyl sulfoxide (DSSO) containing two symmetric CID-cleavable sites that resulted in the release of peptides readily identified by product ions with diagnostic mass differences.181 Spurred by these developments, clever data acquisition strategies have surfaced to capitalize on the combination of multiple activation techniques to enhance the cleavable cross-linking concept. For example, one type of cross-linker was designed with both a CID-cleavable and an ETD-cleavable bond, each cleaved in sequential CID and ETD scans.182 For this linker, each activation method yielded distinct signature ions that, when combined, improved cross-link identification. Furthermore, sequential CID and ETD scans have been implemented to take full advantage of the benefits provided by the DSSO cross-linker to achieve proteome-wide cross-link analysis.183–185 Utilizing CID to cleave the cross-linker and ETD to produce c/z-type peptide fragments, this dual CID/ETD approach proved sufficiently robust to identify cross-linked peptides in human cell lysates with high confidence.183 Further incorporation of automated CID-based MS3 strategies led to characterization of 1158 and 3301 unique high confidence cross-links in E. coli and HeLa cell lysates, respectively.184 Detailed comparisons of five activation methods, including CID, HCD, ETD, EThcD, and ETciD, for the analysis of noncleavable cross-linked peptides generated from the bis-(sulfosuccininidyl) suberate reaction of three proteins (human serum albumin, myoglobin, and creatine kinase) have also been presented.186 This latter study reported that EThcD proved most effective for cross-linked peptides with the highest charge densities, whereas HCD gave the best all-around performance for other peptides.186
Figure 6.
Workflow for cross-linking mass spectrometry relies heavily on effective ion activation methods for the production of informative MS/MS spectra. After optimized incubation of proteins with a cross-linking reagent and proteolytic digestion, four types of peptide products may be created. Cross-linked peptides are subjected to ion activation to generate informative MS/MS spectra.190 Reprinted from Götze, M.; Iacobucci, C.; Ihling, C.H.; Sinz, A. A Simple Cross-Linking/Mass Spectrometry Workflow for Studying System-Wide Protein Interactions, Anal. Chem., 2019, 91, 10236–10244. Copyright 2019 American Chemical Society.
The portfolio of activation methods applied for cross-link analysis has been further extended with the use of 193 nm UVPD.187,188 UVPD was used in a top-down strategy to identify intramolecular and intermolecular cross-links, although the method was hampered by the heterogeneity of the cross-linking sites, which impeded resolution of isomeric structures.187 In another application, 193 nm UVPD was used to characterize cross-linked peptides from hemoglobin and the E. coli ribosome, finding some that were complementary to the larger total number of unique ones identified by HCD.188 The release of dityrosine cross-linked peptides via UVPD relieves the n2 problem (i.e., the multiplicative number of potential cross-links for a set of n peptides, which increases the chances of random matches) and results in reporter ions that facilitate automated cross-link peptide identification with readily available MS-cleavable cross-linking software tools.189
The optimization of proteome-wide cross-linking mass spectrometry, including data collection and analysis, is described in an expansive protocol.185 Cross-linking/tandem mass spectrometry workflows have advanced to the point of allowing massive studies at the system-wide level, resulting in compilations of thousands of cross-links and the creation of protein interaction networks for complex systems, as exemplified by a recent study of Drosophilia melanaogaster embryos.190 As summarized in an overview article,191 there has also been considerable progress in applying elegant cross-linking methods for in vivo applications, again underscoring the importance of robust ion activation methods for identification of the cross-links.
Covalent labeling coupled to mass spectrometry provides information about protein surface structure through the modification of solvent accessible amino acid side chains but with fewer bioinformatics hurdles than cross-linking methods.168,169,192 Differential activity of the covalent probes favors the modification of more accessible surface-exposed side chains, while buried residues remain less modified or completely unmodified.168,169,192 Monitoring the extent of modifications, typically via proteolytic digestion of the proteins of interest and quantitative LC-MS/MS analysis of the resulting peptides, provides a snapshot of protein topology.168,169,192 A comparison of the degree of protein modification under different conditions can thus be achieved at the single-residue level to resolve topological changes that occur during folding events and ligand binding as well as modulation of accessibility owing to protein–protein interactions.168,169,192 As summarized in several excellent reviews,168,169,192–197 numerous innovations in covalent labeling have been reported, and these strategies are not even being used to probe protein solvent accessibility in vivo.198 Covalent labeling workflows have rarely departed from the standard bottom-up approach utilizing either collisional activation or electron-activation methods for the identification of peptides, and readers are directed to the review articles cited above to gain more insight into the methods and the conventional use of MS/MS for peptide analysis and the reconstruction of protein structural features.
ION MOBILITY AND MS/MS
Ion mobility (IM) is a collision-based process that separates ions on the basis of their size, charge, and shape. Although not an ion activation method, ion mobility (IM) provides another dimension of separation for characterization of peptides and proteins and has been integrated with many activation methods to provide structural insight about fragment ions199 or to disperse congested populations of ions prior to or after activation.200–202 Commercial mass spectrometers now provide a number of different types of ion mobility methods, including drift-tube ion mobility spectrometry (DTIMS), traveling-wave ion mobility spectrometry (TWIMS), and field-asymmetric ion mobility spectrometry (FAIMS), among others. The description of these methods is beyond the scope of this article, and readers are directed to many recent reviews that have covered ion mobility.17–19,203
IM coupled to MS/MS can aid in the analysis of peptides, proteins, and protein complexes by improving the sensitivity, speed, and limit of detection of the method. IM has also been shown to be able to separate isomers, thus facilitating characterization by MS/MS. Additionally, by adding IM to LC-MS/MS, the additional dimension of separation allows improved peak capacity and the removal of background interferences.200,204–206 FAIMS coupled to LC-MS has shown to improve the separation of precursor ions and thus boost the identification of proteins by 2-fold in a human cell line.207,208 FAIMS has also been used to separate peptide variants and improve the localization of PTMs by ETD.209,210 The same technique was used in a middle-down approach and proved to be efficient in the separation of larger peptides and the localization of PTMs.211
In the past few years, IM has proven to be invaluable in structural proteomics through the measurements of collision cross sections (CCS), an indicator of ion size. These measurements are important to determine the conformation of peptides, proteins, and protein complexes as well as their subunits.203,212,213 Nevertheless, CCS measurements alone are not sufficient to provide information about the stoichiometries, locations of bound ligands, and structures of subunits. Therefore, many studies have combined information from IM measurements with MS/MS to provide greater insight into structures of macromolecular complexes. An example of this multipronged approach was used to decipher the binding sites of cobalt and manganese and confirm significant changes in the structure of α-synuclein, an intrinsically disordered protein, upon metal binding.34 The combination of fragmentation information from ECD and CAD improved the sequence coverage obtained from the analysis of the intact protein, and the complementary CCS values derived from the IM measurements revealed the compaction of the protein after metal binding.34 In a similar manner, ECD and IM were also integrated to elucidate the interaction between a molecular tweezer assembly modulator and the tau protein.214 For this study, ECD was used to determine the binding site, localize phosphorylation sites, and obtain sequence coverage of tau, whereas IM provided insight into the overall changes in tau conformation upon tau binding.214 Another recent study combined ECD and IM to characterize monomers and tetramers of apolipoprotein isoforms.215 ECD of the tetramers caused fragmentation primarily from the N-terminus, whereas evidence of fragmentation of the C-terminus occurred when the tetramers were subjected to collisional activation prior to ECD, suggestive of unfolding.215 In a different approach, electron transfer reactions were used as a tool to reduce the charge states of cytochrome c and myoglobin prior to IM measurement of the protein cross sections in order to determine the impact of charge reduction on the protein structures.216 Significant changes in conformation were observed for high-charge states, an outcome attributed to the disruption of salt bridges, which results in reorganization and unfolding of the protein.216
UVPD has also been coupled to ion mobility to allow the characterization of conformationally selected ions. One of the original studies used 193 nm UVPD to probe the cis/trans isomerization of proline peptide bonds of ubiquitin ions,217 thus demonstrating the insight obtained from combining UVPD with ion mobility. More recently, 266 nm UVPD was used to characterize different conformers of melittin218 and ubiquitin separated by ion mobility.219 For melittin, the fragmentation efficiency and sequence coverage of the more extended conformer was significantly greater, demonstrating the sensitivity of UVPD in the detection of ion conformation.218 213 nm UVPD was also coupled to IM-MS to study the disruption of the tertiary structure based on the conditions used to transfer the ions out of the source region (via in-source collisional activation).219 As shown for ubiquitin in Figure 7 (as well as cytochrome c and myoglobin), compact structures yielded lower photofragmentation efficiencies while unfolded structures yielded higher fragmentation efficiencies.219
Figure 7.
UVPD of two different conformational arrangements of [M + 6H]6+ ubiquitin. (a) UVPD spectra obtained at a cone voltage of 10 V (blue, bottom spectrum) and a cone voltage of 120 V (red, top spectrum). (b) Arrival time distribution as a function of cone voltage ranging from 10 to 120 V. (c) UVPD cleavage sites indicated on the crystal structure (PDB structure 4Z9S) of ubiquitin. Sites with higher relative cleavage abundance in the “soft” spectra are colored blue; those with higher abundance in the “harsh” condition are marked red.219 Reprinted with permission from Theisen, A.; Black, R.; Corinti, D.; Brown, J. M.; Bellina, B.; Barran, P. E. Initial Protein Unfolding Events in Ubiquitin, Cytochrome c and Myoglobin Are Revealed with the Use of 213 Nm UVPD Coupled to IM-MS. J. Am. Soc. Mass Spectrom. 2019, 30, 24–33. Copyright 2019 Springer Nature.
Recently, two-dimensional ion mobility (IMS-IMS) with CAD was used to identify structural changes of the undecapeptide substance P upon collisional activation.220 The CCS measurements and variation in fragmentation pathways allowed the detection of five conformations illustrating differences in the cis/trans isomerization of proline residues.220 The examples described above have demonstrated the significant gains in structural detail obtained by combining ion mobility with MS/MS of peptides and proteins and foretell a promising future capitalizing on the use of the combined techniques in proteomics and structural biology.
Although not a classical ion activation method for MS/MS, collision induced unfolding (CIU) is a specialized method in native MS utilizing collisional heating to unfold proteins and disassemble protein complexes in the gas phase.221 The added energy is sufficient to disrupt noncovalent interactions but not to cause covalent bond cleavage, thus providing an indirect means to evaluate the structure and stability of proteins and protein complexes. CIU is frequently combined with ion mobility mass spectrometry to allow insight into the variations in cross sections (e.g., shape, size) of ions that reflect the conformational changes. Applications of CIU in the arena of structural biology have increased over the years with the more widespread availability of IM-enabled mass spectrometers, but CIU is beyond the scope of this review.
TOP-DOWN AND MIDDLE-DOWN METHODS
Accelerated by improvements in ion activation methods, top-down and middle-down methodologies have evolved to give rise to high-throughput workflows capable of elucidating and profiling proteoforms in complex samples. These workflows have enabled the routine application of top-down and middle-down analysis for the characterization of biotherapeutics as well as the discovery and understanding of important biological pathways.8,9 The activation of intact proteins and polypeptides displays a more comprehensive view of modifications and sequence information, providing a powerful platform for dissecting proteins. With widespread application of these methodologies, only a portion of new studies are emphasized here through the lens of ion activation.
Developments in instrumentation and methods have both increased the performance metrics of top-down methods and enabled facile integration and the combination of ion activation techniques. Several advancements have sought to better resolve or improve the detection of ETD product ions. When the precursor ions were trapped in the center section of a linear ion trap rather than being sequestered and limited to the ion population in the back of the trap, high capacity ETD was introduced and led to an increase in product ion S/N, manifesting as a boost in sequence coverage for intact myoglobin and carbonic anhydrase.222 Another approach to increase S/N for ETD products implemented front-end ETD on a 21 T FTICR mass spectrometer to take full advantage of the high resolution platform.223 Numerous c/z ions identified using this approach led to 87% sequence coverage for carbonic anhydrase, emphasizing the utility of ETD for providing unbiased fragmentation throughout intact proteins.223 Implementing AI-ETD on a quadrupole-Orbitrap-ion trap provided better sequence coverage for intact proteins and alleviated the charge state dependence,137–139 on a time scale suitable for LC.140 ECD implemented on a benchtop Orbitrap mass spectrometer via a modular ECD cell resulted in high sequence coverage (>60%) and fragment identification rates (~70%) for ubiquitin and myoglobin.44 The combination of ECD with HCD and 193 nm UVPD provided even more comprehensive characterization of proteins up to ~30 kDa as well as monoclonal antibody subunits.45 Photo-ECD, a process in which 193 nm photons impinge on a surface and dislodge electrons, was demonstrated to occur simultaneously with UVPD on an FTICR mass spectrometer.224
The development of advanced data acquisition methods for top-down analysis has enabled high-throughput profiling and quantitation of proteoforms in complex mixtures, enabling the differential analysis of disease states. For example, improvements in data acquisition using AUTOPILOT allowed confident quantitation of more than 1100 proteoforms (all less than 30 kDa) in growing and senescent human fibroblasts.225 A modified acquisition strategy, where the MS1 survey scan was replaced with consecutively selected ion monitoring scans, in conjunction with the use of AUTOPILOT to select precursors for tandem mass spectrometry resulted in the identification of almost 2000 proteoforms, including quantitation of some proteins in the 30–60 kDa range.226 When an ultrahigh resolution 21 T FTICR mass spectrometer was employed to characterize human colorectal cancer cell lysate, over 40 data-dependent LC-MS/MS experiments utilizing both CID and ETD led to the identification of more than 3200 proteoforms, including 372 isotopically resolved proteoforms above 30 kDa.227 The examples of CID and ETD mass spectra acquired for peptidyl-prolyl cis–trans isomer B, a 20 kDa protein in the 29+ charge state, are shown in Figure 8, with 51% coverage and higher scoring metrics obtained by ETD compared to CID.227 In addition, 193 nm UVPD was used for high-throughput proteomics, affording increased confidence in proteoform identification and PTM localization in comparison to HCD.228 With the growing adoption of high-throughput methods, tools to accurately calculate false discovery rates (FDR) have been developed to enhance the quality and consistency of top-down proteomic analyses.229 These advancements in high-throughput profiling and proteoform quantitation have been showcased in the comparative analysis of brain proteoforms between mouse strains,230 apolipoprotein A-1 proteoforms between individuals with high- and low high-density-lipoprotein cholesterol efflux,231 sarcomeric proteomes across various species of muscle types,232–234 and the effect of KRAS gene mutations on downstream proteoforms.235 The considerations for high-throughput top-down proteomics are presented in a tutorial on translational top-down proteomics.236
Figure 8.
CID and ETD MS2 spectra of peptidyl-prolyl cis–trans isomerase B. (top) Single-scan CID (left) and ETD (right) MS2 spectra of a precursor identified as peptidyl-prolyl cis–trans isomerase B (UniProt P23284 residues 34–216, PFR 1354). Expanded views (center) of 50 m/z regions (denoted by gray brackets) reveal the wealth of identified fragments. Colored fragment labels indicate multiplets identified by manual inspection only. * = neutral loss of water from a nearby fragment; ‡ = coisolated chemical noise/activation window. Sequence coverage maps and confidence metrics determined by TDPortal are given (bottom).227 Reprinted from Anderson, L. C.; DeHart, C. J.; Kaiser, N. K.; Fellers, R. T.; Smith, D. F.; Greer, J. B.; LeDuc, R. D.; Blakney, G. T.; Thomas, P. M.; Kelleher, N. L.; Hendrickson, C. L. Identification and Characterization of Human Proteoforms by Top-Down LC-21 T FT-ICR Mass Spectrometry. J. Proteome Res. 2017, 16, 1087–1096. Copyright 2017 American Chemical Society.
Middle-down approaches aim to preserve the advantages of top-down approaches by revealing isoforms and positional isomers, while circumventing the challenges in ionization, separation, and dissociation of intact proteins. In an effort to develop a more robust workflow for middle-down proteomics, a standardized protocol from beginning (digestion) to end (activation) of 3–10 kDa peptides was optimized at each step.237 For the activation of these larger peptides, EThcD provided a higher identification rate and higher sequence coverage than HCD and ETD.237 A middle-down strategy was combined with native mass spectrometry to profile glycoforms and localize the modifications.238 Other efforts to enhance middle-down glycoproteomics evaluated collisional activation and electron-based methods to characterize large multi-glycosylated peptides, with hot electron ECD and EThcD providing the most impressive results.239
Complexities of the antibody structure and the stringent need for accurate proteoform characterization for drug efficacy and safety have motivated the use of both top-down and middle-down approaches for comprehensive characterization of monoclonal antibodies (mAbs). For instance, using a middle-down workflow targeting large subunits (~25 kDa), 193 nm UVPD was shown to generate higher sequence coverage than ETD with the localization of glycosylations and the extensive coverage of the critical complementarity determining regions of immunoglobin G1.240 Furthermore, the high sequence coverage achieved by UVPD was leveraged in combination with the high specificity of LysC for unambiguous identification of anti-influenza mAbs, showcasing the use of UVPD for immunoproteomic assays.241 The combination of the results from ETD, EThcD, and 213 nm UVPD provided complementary layers of information, including sequence confirmation and PTM mapping, in both targeted top-down and middle-down analyses of antibodies.242 With this approach, a record ~40% sequence coverage was achieved for top-down analysis of rituximab and up to 90% sequence coverage, for the >25 kDa middle-down subunits.242 Antibody analysis has also been advanced by utilizing ultrahigh resolution FTICR mass spectrometers.243–247 The enhanced sequence coverage obtained on this platform using both ETD and CID allowed de novo sequencing of monoclonal immunoglobulin light chains.247 Recently, middle-down methods have also been extended to the analysis of antibody–drug conjugates (ADCs), showcasing the ability of ETD to characterize conjugation sites, occupancy, and microvariants at the subunit level.248 Advanced data processing via differential analysis of fragment ions was used to accurately distinguish antibody proteoforms by enhancing the assignment of both terminal and internal fragment ions, thereby allowing the localization of deamidation events and single point mutations.249
The achievement of more extensive characterization of heavily modified histone tails has capitalized on the attributes of top-down and middle-down methods to decipher the role of post-translational modifications in the regulation of gene expression. Through the implementation of front-end ETD, ion–ion proton transfer reactions (II-PTR), and parallel ion parking, intact histone proteoforms were identified with up to 81% sequence coverage.69 The charge states of intact histones were reduced via II-PTR and “parked” at a higher m/z to reduce the spectral density and allow more proteoforms to be resolved. To increase the overall S/N of fragment ions, the C-trap was used to store products from multiple iterations of ion–ion reactions.69 For both top-down and middle-down workflows, 193 nm UVPD generated excellent sequence coverage of multiply modified histone proteoforms.250,251 UVPD and ETD were showcased for the analysis of large peptides produced by LysC proteolysis of histones H3 and H4.251 UVPD proved especially useful in the characterization of heavily modified peptides with more than five PTMs.251 With the complexity of unravelling PTM cross-talk, other studies have focused on developing top-down and middle-down methods to accurately quantitate histone variants using both metabolic labeling and label-free approaches.252–255
Given the widespread adoption of top-down analyses, efforts have arisen to promote consistency in the field. Recent efforts include defining proteoform identification,256 notation,257 and lexicon258 as well as enumerating the best practices for high-quality proteomic studies.259
NATIVE PROTEINS AND PROTEIN COMPLEXES
The characterization of protein structures is a critical step in understanding the functions of proteins, elucidating their binding specificity to other molecules, and facilitating the design of new drugs. Tandem mass spectrometry in bottom-up and top-down workflows has been a transformative method for determining primary sequences of proteins and localizing modifications but does not directly reveal secondary, tertiary, and quaternary structural features. The strategy of native MS uses electrospray ionization to transfer proteins from nondenaturing solutions of high ionic strength to the gas phase, thus enhancing the retention of noncovalent interactions and preserving folded states of protein and protein complexes.20–22,260–263 Native MS provides information on the masses of the protein and stoichiometry of subunits in protein complexes. In addition, the activation of protein complexes can further lead to the dissociation and release of subcomplexes or individual subunits, thus providing information about the interactions and organization of the subunits and locations of the binding sites. In fact, it has been shown that the fragmentation pathways of native-like proteins (typically in low-charge states) differ from those of denatured proteins (in high-charge states), and some of the pathways correlate with tertiary structural features of the proteins.264 For example, backbone cleavages occurring from the C-terminal to aspartic acid and between proline and histidine were enhanced for native-like proteins relative to denatured proteins. Moreover, fragmentation was notably enhanced in regions expected to be surface exposed for native-like proteins.264
The CID of protein complexes typically results in protein unfolding and ejection of highly-charged monomers (asymmetric-charge partitioning), regardless of the structure of the complex as supported by molecular dynamics simulations.265 Thus, CID can be used to disassemble complexes and provide general information about strengths of the subunit interfaces, but insight into the structures of the complexes typically cannot be inferred. Nevertheless, CID showed an anomalous behavior when it was used to characterize ring-like and stacked ring peroxiredoxin multimers.266 Symmetric dissociation of cleaved peroxiredoxin rings was observed, resulting in the release of dimers in a manner consistent with the low strengths of the dimer interfaces relative to the monomer–monomer interface of the dimer.266
HCD has also been used to characterize protein complexes, often in an MS3 manner in which multimeric complexes are disassembled on the basis of in-source activation; then, the released proteins are subjected to collisional activation. This multistage method was used to characterize an array of protein complexes derived from mouse heart and human cancer cell lines.267 These mixtures containing proteins and protein complexes were separated using either ion exchange chromatography or native GELFrEE fractionation, and ultimately, 125 protein complexes and 217 proteoforms were identified in total.267 HCD was also used in an MS3 approach to characterize the yeast homotetramer FBP1 complex, thus allowing the localization of phosphorylations.268 Recently, a detailed standard operating procedure for native MS using HCD as an ion activation method was developed to assist others in the acquisition of high quality MS/MS data.269
Membrane proteins are important therapeutic targets but are among the most difficult class of molecules to study by mass spectrometry, owing to their poor solubility in typical ESI-friendly solutions.270 To facilitate analysis by native MS, membrane proteins are usually solubilized with the aid of detergent micelles, liposomes, or nanodiscs.270 CID is particularly valuable as a means to release and characterize the membrane proteins from these stabilization/solubilization cages for further analysis with other activation techniques.271–274 For example, CID was used to disrupt the nanodiscs encapsulating particulate methane monooxygenase (a copper-dependent metalloenzyme) and release the three membrane protein subunits (PmoA, PmoB, PmoC) that were subsequently characterized by HCD to generate b/y ions. These fragment ions were suitable to aid sequence identification and localization of acetylation, methylation shifts, and the Cu(II) binding site.275 Additionally, collisional activation was used to compare the relative lipid binding affinities of soluble and membrane proteins.276 Higher energy was necessary to remove lipid adducts from membrane proteins, indicating the greater stabilities conferred by lipid binding.276 Other techniques such as IRMPD have also been used to successfully eject proteins from micelles.277
Electron-based activation methods are known to cause backbone cleavages of proteins and protein complexes in the regions of greater structural flexibility.12 At the same time, these methods allow preservation of noncovalent interactions, thus producing fragment ions that may retain bound ligands. For example, ECD on an FTICR mass spectrometer was used to characterize a thrombin–aptamer complex and determine the binding site of the 15-nucleotide aptamer.278 ECD yielded predominantly c and z ions, including both apo (aptamer-free) and holo (retaining aptamer) ions, which allowed the identification of the binding site around Lys21.278 Electron ionization dissociation (EID), a higher energy version of ECD that causes both ionization via electron detachment and electronic excitation, was used to characterize human carbonic anhydrase I and superoxide dismutase dimers. This study afforded some insight into the impact of single point mutations on the structures, suggesting potential variations in the locations of protons on the protein.279 Native electron capture dissociation (NECD) was also used to study ferritin, a complex containing 24 protein molecules that allows storage of up to 4500 iron atoms.280 NECD occurred for metal-containing proteins as they traversed the heated capillary used for electrospray ionization, resulting in the cleavage of backbone bonds adjacent to residues interacting with the heme group and the formation of c/z ions. Changes in the yields of these specific fragment ions were correlated with structural changes in the higher order structures of heme-containing proteins. For ferritin, NECD provided information about the locations of two iron-binding channels.280
UVPD (193 nm) has also proven to be an effective activation method to study the secondary and tertiary structures of proteins, provide sequence information, identify binding sites, and probe conformational changes induced by ligand binding or sequence variations. Past studies have indicated that the extent of backbone cleavage induced by UVPD was enhanced in regions with greater flexibility and/or fewer stabilizing noncovalent interactions,281–283 in many ways similar to the behavior observed by using electron activation methods. The preservation of noncovalent interactions results in the production of fragment ions that retain bound ligands, termed holo fragments. For example, the impact of single point mutations at Gly12 in the K-Ras protein caused variations in the pattern of backbone cleavages observed upon UVPD, suggestive of some degree of conformational reorganization of the protein induced by the replacement of even just a single amino acid for another.284 The impact of single point mutations on protein fragmentation was also echoed in a study that used UVPD to characterize two clinically relevant variants of dihydrofolate reductase found in trimethoprim-resistant E. coli.285 UVPD was also used to identify conformational changes of phosphotransferase adenylate kinase during each stage of its multistep catalytic cycle.286 Differences in the UVPD patterns, particularly evident in the regions spanning three α-helices and the adenosine binding site, were attributed to conformational rearrangement related to cofactor and ligand binding.286
For characterization of multimeric protein complexes, UVPD results both in production of sequence-type ions and disassembly into subunits with symmetrical-charge partitioning that is consistent with the quaternary organization of the complexes.287,288 One study found that HCD resulted in less symmetric partitioning of charges among the separated monomers of dimeric conconavalin A owing to unfolding of complexes during activation, whereas UVPD led to a more symmetric distribution of charges indicative of less unfolding.288 It was also shown that the outcome of UVPD depended on the size of the protein complex and the stability of the intersubunit interface.288 Recently, the impact of charge state on the fragmentation pathways of an array of multimeric protein complexes was assessed using 193 nm UVPD.289 UVPD using higher energy laser powers resulted in more symmetric fragmentation of the complexes and exhibited little dependence on the precursor charge state, as well as yielding far greater sequence coverage of the constituent proteins in the complexes.289 This outcome was reflected for several multimeric complexes in a range of charge states, as illustrated in Figure 9 for dimeric superoxide dismutase, tetrameric streptavidin, tetrameric transthyretin, tetrameric hemoglobin, and pentameric C-reactive protein.289 As an alternative to UV photodissociation, IR photoactivation had been explored for the characterization of membrane protein assemblies.290 The employment of a pulsed IR laser allowed the removal of membrane proteins from their lipid micelles or the dissociation of proteins after they were released from the micelles by collisional activation.290
Figure 9.
Percent sequence coverage of all of the charge states generated from supercharging, standard, and charge-reducing solutions for (a) superoxide dismutase (SOD), (b) streptavidin (SA), (c) transthyretin (TTR), (d) hemoglobin (Hb), and (e) C-reactive protein (CRP). A single pulse of 3 mJ was used for all UVPD measurements. HCD energy was optimized for each protein and ranged from 1400 to 2500 eV of lab frame collision energy. Error bars are equal to the standard deviation of replicate data. Charge states with insufficient abundances for MS/MS analysis are not included.289 Reprinted with permission from Sipe, S. N.; Brodbelt, J. S. Impact of Charge State on 193 nm Ultraviolet Photodissociation of Protein Complexes. Phys. Chem. Chem. Phys. 2019, 21, 9265–9276, with permission of The Royal Society of Chemistry.
Recently, pseudo MS3 based on the combination of nonselective in-source dissociation (ISD) followed by precursor isolation and UVPD was used to characterize the enzyme branched-chain amino acid transferase 2 (BCAT2).291 ISD afforded a means to break the dimeric protein into its constituent proteins, allowing the determination of the mass of each protein and subunit stoichiometry. Localization of amino acid mutations and mapping of binding sites of the cofactors were subsequently achieved using UVPD.291
Surface induced dissociation (SID) has proven to be one of the most valuable activation methods to study protein complexes, as it produces fragment ions that are reflective of the quaternary structure. It has been previously shown that this technique results in symmetric-charge partitioning for protein assemblies owing to its fast, high-energy deposition.16,57 Several of the more recent applications of SID in the arena of structural biology are described here. SID was used to evaluate structural changes of protein complexes as large as 237 kDa after confinement in a trapping region for extended periods (1–60 s).292 No significant changes were observed in the fragmentation patterns, suggesting that proteins undergo minimal structural changes even in the absence of stabilizing solvent interactions. Enabling SID in a FTICR mass spectrometer allowed higher resolution measurements for improved confidence in assigning fragment ions as well as the resolution of metal adducted ions, as illustrated for protein complexes ranging from 53 to 85 kDa.62 SID was further explored for the analysis of multicopper oxidase protein complex (211 kDa), providing information on the metal binding stoichiometries and confirming previously uncharacterized modifications of the protein.63 This activation method has also proven versatile for localization of binding sites and the determination of the architectures of a number of other large protein complexes including the C-reactive protein (CRP) pentamer and cholera toxin B (CTB) pentamer,27 transthyretin tetramers,60 and dodecameric assemblies293 as well as integral membrane protein complexes (AmtB and AqpZ).65 Additionally, SID has been implemented on an Orbitrap platform to further extend the success of this activation method for the characterization of protein complexes between 53 and 336 kDa.64 Recently, SID was coupled with computational modeling to predict the structures of protein complexes.66,67
Using multiple activation methods in an integrated manner can provide even broader and deeper characterization of protein complexes. One of the most extensive studies to date reported the use of multiple MS/MS methods, including CAD, ECD, AI-ECD, and IRMPD, to elucidate sequence and structural information about 11 protein complexes.31 The scope of this study illustrated the strategic benefits of combining insight from several complementary ion activation techniques.31
The analysis of even larger macromolecular complexes, such as ribosomes, proteasomes, and viral particles, is especially challenging owing to the size and heterogeneity of such assemblies. Nevertheless, improvements in the performance metrics of mass spectrometers, such as increased resolution, sensitivity, and improved transmission of larger molecules, as well as advances in the utilization of ion activation methods have facilitated the characterization of macromolecular complexes as large as 50 MDa.262 For example, CID and HCD were used to disassemble intact ribosomes294,295 and brome mosaic and cowpea chlorotic mottle viruses,296 while SID has been used to dissect the 20S proteasome from M. thermophila.297 UVPD was employed to characterize an array of macromolecular complexes, including B-phycoerythrin (265 kDa), CRISPR-Cas Csy ribonucleoprotein complex (347 kDa), and a virus-like particle termed AaLS (~1 MDa).298 Using lower laser powers for UVPD allowed the dissociation of the complexes and release of its subunits, whereas using higher laser powers for UVPD resulted in backbone cleavages of the subunits, yielding sufficient sequence coverage to identify proteoforms as shown in Figure 10 for the wild-type AaLS virus-like nanocontainer.298
Figure 10.
(a) Native top-down mass spectrometry of wt-AaLS virus-like nanocontainers (Mw > 1 MDa) using UVPD ([95+ to 87+], 0.7 mJ per pulse, 3.7 10–10 mbar N2 readout, and resolution of 4350 at m/z 200). (b) Some low mass peptide and (c) intact wt-AaLS subunits resulting from the dissociation of the (d) precursor ions of the VLPs are detected together with (e) larger fragments. (f) Structural model of wt-AaLS (PDB ID: 5MPP) with a pore formed by a pentamer of subunits highlighted in yellow.298 Reprinted with permission from Greisch, J.-F.; Tamara, S.; Scheltema, R. A.; Maxwell, H. W. R.; Fagerlund, R. D.; Fineran, P. C.; Tetter, S.; Hilvert, D.; Heck, A. J. R. Expanding the Mass Range for UVPD-Based Native Top-down Mass Spectrometry. Chem. Sci. 2019, 10, 7163–7171, with permission of The Royal Society of Chemistry.
CONCLUSIONS
In the context of tandem mass spectrometry, ion activation is a core concept that continues to amplify the impact of mass spectrometry as an analytical method for the characterization of peptides, proteins, and protein complexes. The accurate measurement of molecular masses provides important information about elemental composition, but it is the fragmentation patterns that give extraordinary insight into sequences, modifications, and greater structural details. The obtainment of the molecular signatures provided by ion fragmentation is what has continued to motivate the development of new activation methods as well as the design of innovative data acquisition strategies for countless applications in proteomics, structural biology, and protein science in general. Since the prior edition of this review in 2016,1 few new ion activation methods have emerged, and instead, the focus has shifted toward the greater integration of ion activation with auxiliary methods like ion mobility as well as new separation techniques to evaluate complex mixtures in a higher throughput manner. In the realm of applications, a fast growing area that has relied heavily on the full suite of ion activation methods is structural biology. The ability to decipher protein structures, ranging from insoluble membrane proteins to noncovalent protein–ligand complexes to ultrahigh molecular weight macromolecular protein assemblies, poses a major analytical challenge and represents one of the newest frontiers for mass spectrometry.15,262,299 Virtually every current ion activation method has been used to characterize protein assemblies; another rising trend is the utilization of multiple activation modes and the exploration of hybrid activation methods, both aimed at increasing the information content obtained from MS/MS spectra. Along with these endeavors, there have been concerted efforts to extract more information from the fragmentation patterns generated by ion activation, thus improving the ability to elucidate the most intricate structural features of peptides and proteins, such as multiple modifications and isomeric residues. All of these inroads have placed increasing emphasis on the design and implementation of smarter data acquisition algorithms, superior data processing and analysis tools, and cutting edge bioinformatics strategies.
ACKNOWLEDGMENTS
Financial support from the National Science Foundation (CHE1402753), the National Institutes of Health (R01-GM103655 and R01-GM121714), and the Welch Foundation (F-1155) is gratefully acknowledged.
Biographies
Luis A. Macias earned his B.S. in chemistry from Houston Baptist University in 2016 and is currently a Ph.D. candidate in Prof. Jennifer S. Brodbelt’s research lab. His research focuses on the application of ultraviolet photodissociation mass spectrometry for structural biology.
Inês C. Santos is a postdoctoral fellow in the Department of Chemistry at the University of Texas at Austin. She obtained her Ph.D. degree in Biotechnology with a specialization in Chemistry at the Faculty of Biotechnology of the Portuguese Catholic University (ESB-UCP) in 2015. She has an M.Sc. and a bachelor degree from ESB-UCP in Microbiology. For the past two years, she has worked on the development of new gas chromatography–vacuum ultraviolet absorption detection and matrix-assisted laser desorption/ionization–mass spectrometry methods to study the microbiome in environmental samples. Her research is currently focused on the mass spectrometric analysis of biological molecules such as proteins, protein complexes, and nucleic acids.
Jennifer S. Brodbelt earned her B.S. in chemistry from the University of Virginia and her Ph.D. from Purdue University. After a postdoctoral stint at the University of California at Santa Barbara, she joined the faculty at the University of Texas at Austin where she is currently a professor and the Rowland Pettit Centennial Chair in Chemistry. She serves as an associate editor for the Journal of the American Society for Mass Spectrometry. Her research focus is on the development of ultraviolet photodissociation and ion trap mass spectrometry for characterization of peptides, proteins, protein complexes, and lipids.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Brodbelt JS Anal. Chem 2016, 88, 30–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Potel CM; Lemeer S; Heck AJR Anal. Chem 2019, 91, 126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Riley NM; Coon JJ Anal. Chem 2016, 88, 74–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhang Y; Fonslow BR; Shan B; Baek M-C; Yates JR Chem. Rev 2013, 113, 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ruhaak LR; Xu G; Li Q; Goonatilleke E; Lebrilla CB Chem. Rev 2018, 118, 7886–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gillet LC; Leitner A; Aebersold R Annu. Rev. Anal. Chem 2016, 9, 449–472. [DOI] [PubMed] [Google Scholar]
- (7).Mayne J; Ning Z; Zhang X; Starr AE; Chen R; Deeke S; Chiang C-K; Xu B; Wen M; Cheng K; Seebun D; Star A; Moore JI; Figeys D Anal. Chem 2016, 88, 95–121. [DOI] [PubMed] [Google Scholar]
- (8).Toby TK; Fornelli L; Kelleher NL Annu. Rev. Anal. Chem 2016, 9, 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen B; Brown KA; Lin Z; Ge Y Anal. Chem 2018, 90, 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lermyte F; Valkenborg D; Loo JA; Sobott F Mass Spectrom. Rev 2018, 37, 750–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Riley NM; Coon JJ Anal. Chem 2018, 90, 40–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Qi Y; Volmer DA Mass Spectrom. Rev 2017, 36, 4–15. [DOI] [PubMed] [Google Scholar]
- (13).Brodbelt JS Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brodbelt JS; Morrison LJ; Santos I Chem. Rev 2020, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kaur U; Johnson DT; Chea EE; Deredge DJ; Espino JA; Jones LM Anal. Chem 2019, 91, 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Stiving AQ; VanAernum ZL; Busch F; Harvey SR; Sarni SH; Wysocki VH Anal. Chem 2019, 91, 190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gabelica V; Marklund E Curr. Opin. Chem. Biol 2018, 42, 51–59. [DOI] [PubMed] [Google Scholar]
- (18).Ben-Nissan G; Sharon M Curr. Opin. Chem. Biol 2018, 42, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zheng X; Wojcik R; Zhang X; Ibrahim YM; Burnum-Johnson KE; Orton DJ; Monroe ME; Moore RJ; Smith RD; Baker ES Annu. Rev. Anal. Chem 2017, 10, 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hopper JTS; Robinson CV Mass Spectrometry of Intact Protein Complexes In Proteomics for Biological Discovery; John Wiley & Sons, Ltd, 2019; pp 145–173. [Google Scholar]
- (21).Chen F; Gülbakan B; Weidmann S; Fagerer SR; Ibańñez AJ; Zenobi R Mass Spectrom. Rev 2016, 35, 48–70. [DOI] [PubMed] [Google Scholar]
- (22).Leney AC; Heck AJR J. Am. Soc. Mass Spectrom 2017, 28, 5–13. [DOI] [PubMed] [Google Scholar]
- (23).Roepstorff P; Fohlman J Biomed. Mass Spectrom 1984, 11, 601. [DOI] [PubMed] [Google Scholar]
- (24).Paizs B; Suhai S Mass Spectrom. Rev 2005, 24, 508–548. [DOI] [PubMed] [Google Scholar]
- (25).Mitchell Wells J; McLuckey SA Collision-Induced Dissociation (CID) of Peptides and Proteins In Methods in Enzymology; Biological Mass Spectrometry; Academic Press, 2005; Vol. 402, pp 148–185. [DOI] [PubMed] [Google Scholar]
- (26).Floris F; Chiron L; Lynch AM; Barrow MP; Delsuc M-A; O’Connor PB J. Am. Soc. Mass Spectrom 2018, 29, 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Busch F; VanAernum ZL; Ju Y; Yan J; Gilbert JD; Quintyn RS; Bern M; Wysocki VH Anal. Chem 2018, 90, 12796–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Leney AC J. Am. Soc. Mass Spectrom 2019, 30, 1389–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Li X; Yang X; Hoang V; Liu Y-H J. Am. Soc. Mass Spectrom 2019, 30, 519–528. [DOI] [PubMed] [Google Scholar]
- (30).Zhao J; Song E; Zhu R; Mechref Y Electrophoresis 2016, 37, 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Li H; Nguyen HH; Ogorzalek Loo RR; Campuzano IDG; Loo JA Nat. Chem 2018, 10, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kaldmäe M; Sahin C; Saluri M; Marklund EG; Landreh M Protein Sci 2019, 28, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Vincent D; Mertens D; Rochfort S Molecules 2018, 23, 2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wongkongkathep P; Han JY; Choi TS; Yin S; Kim HI; Loo JA J. Am. Soc. Mass Spectrom 2018, 29, 1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kilpatrick LE; Kilpatrick EL J. Proteome Res 2017, 16, 3255–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zubarev RA; Kelleher NL; McLafferty FW J. Am. Chem. Soc 1998, 120, 3265–3266. [Google Scholar]
- (37).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF Proc. Natl. Acad. Sci. U. S. A 2004, 101, 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Breuker K; Oh H; Horn DM; Cerda BA; McLafferty FW J. Am. Chem. Soc 2002, 124, 6407–6420. [DOI] [PubMed] [Google Scholar]
- (39).Swaney DL; McAlister GC; Wirtala M; Schwartz JC; Syka JEP; Coon JJ Anal. Chem 2007, 79, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Frese CK; Altelaar AFM; van den Toorn H; Nolting D; Griep-Raming J; Heck AJR; Mohammed S Anal. Chem 2012, 84, 9668–9673. [DOI] [PubMed] [Google Scholar]
- (41).Riley NM; Westphall MS; Hebert AS; Coon JJ Anal. Chem 2017, 89, 6358–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Voinov VG; Deinzer ML; Barofsky DF Rapid Commun. Mass Spectrom 2008, 22, 3087–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Voinov VG; Deinzer ML; Barofsky DF Anal. Chem 2009, 81, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Fort KL; Cramer CN; Voinov VG; Vasil’ev YV; Lopez NI; Beckman JS; Heck AJR J. Proteome Res 2018, 17, 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Shaw JB; Malhan N; Vasil’ev YV; Lopez NI; Makarov A; Beckman JS; Voinov VG Anal. Chem 2018, 90, 10819–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Brodbelt JS; Wilson JJ Mass Spectrom. Rev 2009, 28, 390–424. [DOI] [PubMed] [Google Scholar]
- (47).Reilly JP Mass Spectrom. Rev 2009, 28, 425–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ly T; Julian RR Angew. Chem., Int. Ed 2009, 48, 7130–7137. [DOI] [PubMed] [Google Scholar]
- (49).Brodbelt JS J. Am. Soc. Mass Spectrom 2011, 22, 197–206. [DOI] [PubMed] [Google Scholar]
- (50).Baer T; Dunbar RC J. Am. Soc. Mass Spectrom 2010, 21, 681–693. [DOI] [PubMed] [Google Scholar]
- (51).Antoine R; Dugourd P Phys. Chem. Chem. Phys 2011, 13, 16494–16509. [DOI] [PubMed] [Google Scholar]
- (52).Cismesia AP; Bailey LS; Bell MR; Tesler LF; Polfer NC J. Am. Soc. Mass Spectrom 2016, 27, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zeng HJ; Yang N; Johnson MA Faraday Discuss 2019, 217, 8–33. [DOI] [PubMed] [Google Scholar]
- (54).Cooks RG; Ast T; Beynon JH Int. J. Mass Spectrom. Ion Phys 1975, 16, 348–352. [Google Scholar]
- (55).Cooks RG; Terwilliger DT; Ast T; Beynon JH; Keough TJ Am. Chem. Soc 1975, 97, 1583–1585. [Google Scholar]
- (56).Mabud Md. A.; Dekrey MJ; Graham Cooks R Int. J. Mass Spectrom. Ion Processes 1985, 67, 285–294. [Google Scholar]
- (57).Wysocki VH; Joyce KE; Jones CM; Beardsley RL J. Am. Soc. Mass Spectrom 2008, 19, 190–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zhou M; Wysocki VH Acc. Chem. Res 2014, 47, 1010–1018. [DOI] [PubMed] [Google Scholar]
- (59).Donor MT; Mroz AM; Prell JS Chem. Sci 2019, 10, 4097–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Shirzadeh M; Boone CD; Laganowsky A; Russell DH Anal. Chem 2019, 91, 2345–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Yan J; Zhou M; Gilbert JD; Wolff JJ; Somogyi Á;Pedder RE; Quintyn RS; Morrison LJ; Easterling ML; Paša-Tolić L; Wysocki VH Anal. Chem 2017, 89, 895–901. [DOI] [PubMed] [Google Scholar]
- (62).Zhou M; Yan J; Romano CA; Tebo BM; Wysocki VH; Paša-Tolić L J. Am. Soc. Mass Spectrom 2018, 29, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).VanAernum ZL; Gilbert JD; Belov ME; Makarov AA; Horning SR; Wysocki VH Anal. Chem 2019, 91, 3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Harvey SR; Liu Y; Liu W; Wysocki VH; Laganowsky A Chem. Commun 2017, 53, 3106–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Harvey SR; Seffernick JT; Quintyn RS; Song Y; Ju Y; Yan J; Sahasrabuddhe AN; Norris A; Zhou M; Behrman EJ; Lindert S; Wysocki VH Proc. Natl. Acad. Sci. U. S. A 2019, 116, 8143–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Seffernick JT; Harvey SR; Wysocki VH; Lindert S ACS Cent. Sci 2019, 5, 1330–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).McLuckey SA; Huang T-Y Anal. Chem 2009, 81, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).McLuckey SA; Mentinova MJ Am. Soc. Mass Spectrom 2011, 22, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Anderson LC; Karch KR; Ugrin SA; Coradin M; English AM; Sidoli S; Shabanowitz J; Garcia BA; Hunt DF Mol. Cell. Proteomics 2016, 15, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Zhang L; English AM; Bai DL; Ugrin SA; Shabanowitz J; Ross MM; Hunt DF; Wang W-H Mol. Cell. Proteomics 2016, 15, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).McLuckey SA; Reid GE; Wells JM Anal. Chem 2002, 74, 336–346. [DOI] [PubMed] [Google Scholar]
- (72).Holden DD; McGee WM; Brodbelt JS Anal. Chem 2016, 88, 1008–1016. [DOI] [PubMed] [Google Scholar]
- (73).Ugrin SA; English AM; Syka JEP; Bai DL; Anderson LC; Shabanowitz J; Hunt DF J. Am. Soc. Mass Spectrom 2019, 30, 2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Ubersax JA; Ferrell EF Jr. Nat. Rev. Mol. Cell Biol 2007, 8, 530–541. [DOI] [PubMed] [Google Scholar]
- (75).Arrington JV; Hsu C-C; Elder SG; Tao WA Analyst 2017, 142, 4373–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ramroop JR; Stein MN; Drake JM Front. Oncol 2018, 8,28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Reiding KR; Bondt A; Franc V; Heck AJR TrAC, Trends Anal. Chem 2018, 108, 260–268. [Google Scholar]
- (78).Zolg DP; Wilhelm M; Schmidt T; Médard G; Zerweck J;Knaute T; Wenschuh H; Reimer U; Schnatbaum K; Kuster B Mol. Cell. Proteomics 2018, 17, 1850–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Leney AC; El Atmioui D; Wu W; Ovaa H; Heck AJR Proc. Natl. Acad. Sci. U. S. A 2017, 114, E7255–E7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Hogg PJ Trends Biochem. Sci 2003, 28, 210–214. [DOI] [PubMed] [Google Scholar]
- (81).Liu H; May K mAbs 2012, 4, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Fass D Annu. Rev. Biophys 2012, 41, 63–79. [DOI] [PubMed] [Google Scholar]
- (83).Pilo AL; McLuckey SA Anal. Chem 2016, 88, 8972–8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Wen D; Xiao Y; Vecchi MM; Gong BJ; Dolnikova J; Pepinsky RB Anal. Chem 2017, 89, 4021–4030. [DOI] [PubMed] [Google Scholar]
- (85).Fung YME; Kjeldsen F; Silivra OA; Chan TWD; Zubarev RA Angew. Chem 2005, 117, 6557–6561. [DOI] [PubMed] [Google Scholar]
- (86).Agarwal A; Diedrich JK; Julian RR Anal. Chem 2011, 83, 6455–6458. [DOI] [PubMed] [Google Scholar]
- (87).Quick MM; Crittenden CM; Rosenberg JA; Brodbelt JS Anal. Chem 2018, 90, 8523–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Talbert LE; Julian RR J. Am. Soc. Mass Spectrom 2018, 29, 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Bonner J; Talbert LE; Akkawi N; Julian RR Analyst 2018, 143, 5176–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Castellana NE; McCutcheon K; Pham VC; Harden K; Nguyen A; Young J; Adams C; Schroeder K; Arnott D; Bafna V; Grogan JL; Lill JR Proteomics 2011, 11, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Xiao Y; Vecchi MM; Wen D Anal. Chem 2016, 88, 10757–10766. [DOI] [PubMed] [Google Scholar]
- (92).Bagal D; Kast E; Cao P Anal. Chem 2017, 89, 720–727. [DOI] [PubMed] [Google Scholar]
- (93).Armirotti A; Millo E; Damonte GJ Am. Soc. Mass Spectrom 2007, 18, 57–63. [DOI] [PubMed] [Google Scholar]
- (94).Lebedev AT; Damoc E; Makarov AA; Samgina T Yu. Anal. Chem 2014, 86, 7017–7022. [DOI] [PubMed] [Google Scholar]
- (95).Maibom-Thomsen S; Heissel S; Mørtz E; Højrup P; Bunkenborg J Anal. Chem 2018, 90, 9055–9059. [DOI] [PubMed] [Google Scholar]
- (96).Crittenden CM; Parker WR; Jenner ZB; Bruns KA; Akin LD; McGee WM; Ciccimaro E; Brodbelt JS J. Am. Soc. Mass Spectrom 2016, 27, 856–863. [DOI] [PubMed] [Google Scholar]
- (97).Parsley NC; Kirkpatrick CL; Crittenden CM; Rad JG; Hoskin DW; Brodbelt JS; Hicks LM Phytochemistry 2018, 152, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Attard TJ; Carter MD; Fang M; Johnson RC; Reid GE J. Am. Soc. Mass Spectrom 2018, 29, 1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Aebersold R; Mann M Nature 2016, 537, 347–355. [DOI] [PubMed] [Google Scholar]
- (100).Vidova V; Spacil Z Anal. Chim. Acta 2017, 964, 7–23. [DOI] [PubMed] [Google Scholar]
- (101).Rauniyar N Int. J. Mol. Sci 2015, 16, 28566–28581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Venable JD; Dong M-Q; Wohlschlegel J; Dillin A; Yates JR Nat. Methods 2004, 1, 39–45. [DOI] [PubMed] [Google Scholar]
- (103).Koopmans F; Ho JTC; Smit AB; Li KW Proteomics 2018, 18, 1700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Ludwig C; Gillet L; Rosenberger G; Amon S; Collins BC; Aebersold R Mol. Syst. Biol 2018, 14, e8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Collins BC; Hunter CL; Liu Y; Schilling B; Rosenberger G; Bader SL; Chan DW; Gibson BW; Gingras A-C; Held JM; Hirayama-Kurogi M; Hou G; Krisp C; Larsen B; Lin L; Liu S; Molloy MP; Moritz RL; Ohtsuki S; Schlapbach R; Selevsek N; Thomas SN; Tzeng S-C; Zhang H; Aebersold R Nat. Commun 2017, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Krasny L; Bland P; Kogata N; Wai P; Howard BA; Natrajan RC; Huang PH J. Proteomics 2018, 189, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Schmidlin T; Garrigues L; Lane CS; Mulder TC; van Doorn S; Post H; de Graaf EL; Lemeer S; Heck AJR; Altelaar AFM Proteomics 2016, 16, 2193–2205. [DOI] [PubMed] [Google Scholar]
- (108).Pan K-T; Chen C-C; Urlaub H; Khoo K-H Anal. Chem 2017, 89, 4532–4539. [DOI] [PubMed] [Google Scholar]
- (109).Lin C-H; Krisp C; Packer NH; Molloy MP J. Proteomics 2018, 172, 68–75. [DOI] [PubMed] [Google Scholar]
- (110).Searle BC; Pino LK; Egertson JD; Ting YS; Lawrence RT; MacLean BX; Villén J, MacCoss M J. Nat. Commun 2018, 9, 5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Amodei D; Egertson J; MacLean BX; Johnson R; Merrihew GE; Keller A; Marsh D; Vitek O; Mallick P; MacCoss MJ J. Am. Soc. Mass Spectrom 2019, 30, 669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Borotto NB; McClory PJ; Martin BR; Håkansson K. Anal. Chem 2017, 89, 8304–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Garcia L; Girod M; Rompais M; Dugourd P; Carapito C; Lemoine J Anal. Chem 2018, 90, 3928–3935. [DOI] [PubMed] [Google Scholar]
- (114).Silva JC; Denny R; Dorschel C; Gorenstein MV; Li G-Z; Richardson K; Wall D; Geromanos SJ Mol. Cell. Proteomics 2006, 5, 589–607. [DOI] [PubMed] [Google Scholar]
- (115).Cramer CN; Brown JM; Tomczyk N; Nielsen PK; Haselmann KF J. Am. Soc. Mass Spectrom 2017, 28, 384–388. [DOI] [PubMed] [Google Scholar]
- (116).Lakshmanan R; Loo JA Int. J. Mass Spectrom 2019, 435, 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Hebert AS; Thöing C; Riley NM; Kwiecien NW; Shiskova E; Huguet R; Cardasis HL; Kuehn A; Eliuk S; Zabrouskov V; Westphall MS; McAlister GC; Coon JJ Anal. Chem 2018, 90, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Meier F; Geyer PE; Winter SV; Cox J; Mann M Nat. Methods 2018, 15, 440. [DOI] [PubMed] [Google Scholar]
- (119).van Agthoven MA; Lam YPY; O’Connor PB; Rolando C; Delsuc M-A Eur. Biophys. J 2019, 48, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Pfandler P; Bodenhausen G; Rapin J; Houriet R; Gäumann T Chem. Phys. Lett 1987, 138, 195–200. [Google Scholar]
- (121).Floris F; van Agthoven M; Chiron L; Soulby AJ; Wootton CA; Lam YPY; Barrow MP; Delsuc M-A; O’Connor PB J. Am. Soc. Mass Spectrom 2016, 27, 1531–1538. [DOI] [PubMed] [Google Scholar]
- (122).Floris F; van Agthoven MA; Chiron L; Wootton CA; Lam PYY; Barrow MP; Delsuc M-A; O’Connor PB J. Am. Soc. Mass Spectrom 2018, 29, 207–210. [DOI] [PubMed] [Google Scholar]
- (123).van Agthoven MA; Lynch AM; Morgan TE; Wootton CA; Lam YPY; Chiron L; Barrow MP; Delsuc M-A; O’Connor PB Anal. Chem 2018, 90, 3496–3504. [DOI] [PubMed] [Google Scholar]
- (124).Floris F; Chiron L; Lynch AM; Barrow MP; Delsuc M-A; O’Connor PB Anal. Chem 2018, 90, 7302–7309. [DOI] [PubMed] [Google Scholar]
- (125).Penkert M; Hauser A; Harmel R; Fiedler D; Hackenberger CPR; Krause EJ Am. Soc. Mass Spectrom 2019, 30, 1578. [DOI] [PubMed] [Google Scholar]
- (126).Yu Q; Wang B; Chen Z; Urabe G; Glover MS; Shi X; Guo L-W; Kent KC; Li LJ Am. Soc. Mass Spectrom 2017, 28, 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Bilan V; Leutert M; Nanni P; Panse C; Hottiger MO Anal. Chem 2017, 89, 1523–1530. [DOI] [PubMed] [Google Scholar]
- (128).Hendriks IA; Larsen SC; Nielsen ML Mol. Cell. Proteomics 2019, 18, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Penkert M; Yates LM; Schümann M; Perlman D; Fiedler D; Krause E Anal. Chem 2017, 89, 3672–3680. [DOI] [PubMed] [Google Scholar]
- (130).Zhao X; Shen Y; Tong W; Wang G; Chen DDY Analyst 2018, 143, 817–823. [DOI] [PubMed] [Google Scholar]
- (131).Zhokhov SS; Kovalyov SV; Samgina TY; Lebedev AT J. Am. Soc. Mass Spectrom 2017, 28, 1600–1611. [DOI] [PubMed] [Google Scholar]
- (132).Ma F; Sun R; Tremmel DM; Sackett SD; Odorico J; Li L Anal. Chem 2018, 90, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Yu Q; Shi X; Feng Y; Kent KC; Li L Anal. Chim. Acta 2017, 968, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Ledvina AR; McAlister GC; Gardner MW; Smith SI; Madsen JA; Schwartz JC; Stafford GC; Syka JEP; Brodbelt JS; Coon JJ Angew. Chem., Int. Ed 2009, 48, 8526–8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Riley NM; Hebert AS; Westphall MS; Coon JJ Nat. Commun 2019, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Riley NM; Hebert AS; Dürnberger G; Stanek F; Mechtler K; Westphall MS; Coon JJ Anal. Chem 2017, 89, 6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Riley NM; Westphall MS; Coon JJ J. Proteome Res 2017, 16, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Riley NM; Westphall MS; Coon JJ J. Am. Soc. Mass Spectrom 2018, 29, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Rush MJP; Riley NM; Westphall MS; Coon JJ Anal. Chem 2018, 90, 8946–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Riley NM; Sikora JW; Seckler HS; Greer JB; Fellers RT; LeDuc RD; Westphall MS; Thomas PM; Kelleher NL; Coon JJ Anal. Chem 2018, 90, 8553–8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Halim MA; Girod M; MacAleese L; Lemoine J; Antoine R; Dugourd PJ Am. Soc. Mass Spectrom 2016, 27, 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Halim MA; MacAleese L; Lemoine J; Antoine R; Dugourd P; Girod MJ Am. Soc. Mass Spectrom 2018, 29, 270–283. [DOI] [PubMed] [Google Scholar]
- (143).Simpson DC; Ahn S; Paša-Tolić L; Bogdanov B; Mottaz HM; Vilkov AN; Anderson GA; Lipton MS; Smith RD Electrophoresis 2006, 27, 2722–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Doucette AA; Tran JC; Wall MJ; Fitzsimmons S Expert Review of Proteomics 2014, 8, 787–800. [DOI] [PubMed] [Google Scholar]
- (145).Xie B; Sharp JS J. Am. Soc. Mass Spectrom 2016, 27, 1322–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Jin Y; Wei L; Cai W; Lin Z; Wu Z; Peng Y; Kohmoto T; Moss RL; Ge Y Anal. Chem 2017, 89, 4922–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Cai W; Tucholski T; Chen B; Alpert AJ; McIlwain S; Kohmoto T; Jin S; Ge Y Anal. Chem 2017, 89, 5467–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Tucholski T; Knott SJ; Chen B; Pistono P; Lin Z; Ge Y Anal. Chem 2019, 91, 3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Queiroz JA; Tomaz CT; Cabral JMS J. Biotechnol 2001, 87, 143–159. [DOI] [PubMed] [Google Scholar]
- (150).Chen B; Peng Y; Valeja SG; Xiu L; Alpert AJ; Ge Y Anal. Chem 2016, 88, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Chen B; Lin Z; Alpert AJ; Fu C; Zhang Q; Pritts WA; Ge Y Anal. Chem 2018, 90, 7135–7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Shen X; Yang Z; McCool EN; Lubeckyj RA; Chen D; Sun L TrAC, Trends Anal. Chem 2019, 120, 115644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Britz-McKibbin P; Chen DDY Anal. Chem 2000, 72, 1242–1252. [DOI] [PubMed] [Google Scholar]
- (154).Zhao Y; Sun L; Knierman MD; Dovichi NJ Talanta 2016, 148, 529–533. [DOI] [PubMed] [Google Scholar]
- (155).Zhao Y; Sun L; Zhu G; Dovichi NJ J. Proteome Res 2016, 15, 3679–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).McCool EN; Lubeckyj RA; Shen X; Chen D; Kou Q; Liu X; Sun L Anal. Chem 2018, 90, 5529–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).McCool EN; Lodge JM; Basharat AR; Liu X; Coon JJ; Sun LJ Am. Soc. Mass Spectrom 2019, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).McCool EN; Chen D; Li W; Liu Y; Sun L Anal. Methods 2019, 11, 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Said N; Gahoual R; Kuhn L; Beck A; Francois Y-N; Leize-Wagner E Anal. Chim. Acta 2016, 918, 50–59. [DOI] [PubMed] [Google Scholar]
- (160).Faserl K; Sarg B; Maurer V; Lindner HH Journal of Chromatography A 2017, 1498, 215–223. [DOI] [PubMed] [Google Scholar]
- (161).Zhang Z; Hebert AS; Westphall MS; Coon JJ; Dovichi NJ J. Proteome Res 2019, 18, 3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Belov AM; Viner R; Santos MR; Horn DM; Bern M; Karger BL; Ivanov AR J. Am. Soc. Mass Spectrom 2017, 28, 2614–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Shen X; Kou Q; Guo R; Yang Z; Chen D; Liu X; Hong H; Sun L Anal. Chem 2018, 90, 10095–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Konermann L; Pan J; Liu Y-H Chem. Soc. Rev 2011, 40, 1224–1234. [DOI] [PubMed] [Google Scholar]
- (165).Oganesyan I; Lento C; Wilson DJ Methods 2018, 144, 27–42. [DOI] [PubMed] [Google Scholar]
- (166).O’Reilly FJ; Rappsilber J Nat. Struct. Mol. Biol 2018, 25, 1000–1008. [DOI] [PubMed] [Google Scholar]
- (167).Sinz A Angew. Chem., Int. Ed 2018, 57, 6390–6396. [DOI] [PubMed] [Google Scholar]
- (168).Wang L; Chance MR Anal. Chem 2011, 83, 7234–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Limpikirati P; Liu T; Vachet RW Methods 2018, 144, 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Rand KD; Jørgensen TJD Anal. Chem 2007, 79, 8686–8693. [DOI] [PubMed] [Google Scholar]
- (171).Jørgensen TJD; Gårdsvoll H; Ploug M; Roepstorff PJ Am. Chem. Soc 2005, 127, 2785–2793. [DOI] [PubMed] [Google Scholar]
- (172).Rand KD; Zehl M; Jensen ON; Jørgensen TJD Anal. Chem 2009, 81, 5577–5584. [DOI] [PubMed] [Google Scholar]
- (173).Mistarz UH; Bellina B; Jensen PF; Brown JM; Barran PE; Rand KD Anal. Chem 2018, 90, 1077–1080. [DOI] [PubMed] [Google Scholar]
- (174).Pan J; Han J; Borchers CH; Konermann LJ Am. Chem. Soc 2009, 131, 12801–12808. [DOI] [PubMed] [Google Scholar]
- (175).Abzalimov RR; Kaplan DA; Easterling ML; Kaltashov IA J. Am. Soc. Mass Spectrom 2009, 20, 1514–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Karch KR; Coradin M; Zandarashvili L; Kan Z-Y; Gerace M; Englander SW; Black BE; Garcia BA Structure 2018, 26, 1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Brodie NI; Huguet R; Zhang T; Viner R; Zabrouskov V; Pan J; Petrotchenko EV; Borchers CH Anal. Chem 2018, 90, 3079–3082. [DOI] [PubMed] [Google Scholar]
- (178).Pan J; Zhang S; Chou A; Borchers CH Chem. Sci 2016, 7, 1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Rappsilber JJ Struct. Biol 2011, 173, 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Tang X; Munske GR; Siems WF; Bruce JE Anal. Chem 2005, 77, 311–318. [DOI] [PubMed] [Google Scholar]
- (181).Kao A; Chiu C; Vellucci D; Yang Y; Patel VR; Guan S; Randall A; Baldi P; Rychnovsky SD; Huang L Mol. Cell. Proteomics 2011, 10, M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Chakrabarty JK; Naik AG; Fessler MB; Munske GR; Chowdhury SM Anal. Chem 2016, 88, 10215–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Liu F; Rijkers DTS; Post H; Heck AJR Nat. Methods 2015, 12, 1179–1184. [DOI] [PubMed] [Google Scholar]
- (184).Liu F; Lössl P; Scheltema R; Viner R; Heck AJR Nat. Commun 2017, 8, 15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Klykov O; Steigenberger B; Pektas S; Fasci D; Heck AJR; Scheltema RA Nat. Protoc 2018, 13, 2964–2990. [DOI] [PubMed] [Google Scholar]
- (186).Kolbowski L; Mendes ML; Rappsilber J Anal. Chem 2017, 89, 5311–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Cammarata MB; Brodbelt JS ChemistrySelect 2016, 1, 590–593. [Google Scholar]
- (188).Cammarata MB; Macias LA; Rosenberg J; Bolufer A; Brodbelt JS Anal. Chem 2018, 90, 6385–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Mukherjee S; Kapp EA; Lothian A; Roberts AM; Vasil’ev YV; Boughton BA; Barnham KJ; Kok WM; Hutton CA; Masters CL; Bush AI; Beckman JS; Dey SG; Roberts BR Anal. Chem 2017, 89, 6136–6145. [DOI] [PubMed] [Google Scholar]
- (190).Götze M; Iacobucci C; Ihling CH; Sinz A Anal. Chem 2019, 91, 10236–10244. [DOI] [PubMed] [Google Scholar]
- (191).Sinz A cels 2018, 6, 10–12. [DOI] [PubMed] [Google Scholar]
- (192).Mendoza VL; Vachet RW Mass Spectrom. Rev 2009, 28, 785–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Niu B; Gross ML Mass Spectrometry-Based Chemical Proteomics 2019, 363–416. [Google Scholar]
- (194).Johnson DT; Di Stefano LH; Jones LM Fast Photochemical Oxidation of Proteins(FPOP): A Powerful Mass Spectrometry Based Structural Proteomics Tool. J. Biol. Chem 2019, DOI: 10.1074/jbc.REV119.006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Wang L; Chance MR Mol. Cell. Proteomics 2017, 16, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Kiselar J; Chance MR Annu. Rev. Biophys 2018, 47, 315–333. [DOI] [PubMed] [Google Scholar]
- (197).Xie B; Sood A; Woods RJ; Sharp JS Sci. Rep 2017, 7, 4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Espino JA; Jones LM Anal. Chem 2019, 91, 6577–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Ewing MA; Glover MS; Clemmer DE Journal of Chromatography A 2016, 1439, 3–25. [DOI] [PubMed] [Google Scholar]
- (200).Zinnel NF; Pai P-J; Russell DH Anal. Chem 2012, 84, 3390–3397. [DOI] [PubMed] [Google Scholar]
- (201).Distler U; Kuharev J; Navarro P; Levin Y; Schild H; Tenzer S Nat. Methods 2014, 11, 167–170. [DOI] [PubMed] [Google Scholar]
- (202).Uetrecht C; Rose RJ; van Duijn E; Lorenzen K; Heck AJR Chem. Soc. Rev 2010, 39, 1633–1655. [DOI] [PubMed] [Google Scholar]
- (203).Lanucara F; Holman SW; Gray CJ; Eyers CE Nat. Chem 2014, 6, 281–294. [DOI] [PubMed] [Google Scholar]
- (204).Kanu AB; Dwivedi P; Tam M; Matz L; Hill HH J. Mass Spectrom 2008, 43, 1–22. [DOI] [PubMed] [Google Scholar]
- (205).D’Atri V; Causon T; Hernandez-Alba O; Mutabazi A; Veuthey J-L; Cianferani S; Guillarme DJ Sep Sci 2018, 41, 20–67. [DOI] [PubMed] [Google Scholar]
- (206).Thomas A; Görgens C; Guddat S; Thieme D; Dellanna F; Schänzer W; Thevis MJ Sep Sci 2016, 39, 333–341. [DOI] [PubMed] [Google Scholar]
- (207).Hebert AS; Prasad S; Belford MW; Bailey DJ; McAlister GC; Abbatiello SE; Huguet R; Wouters ER; Dunyach J-J; Brademan DR; Westphall MS; Coon JJ Anal. Chem 2018, 90, 9529–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Pfammatter S; Bonneil E; McManus FP; Prasad S; Bailey DJ; Belford M; Dunyach J-J; Thibault P Mol. Cell. Proteomics 2018, 17, 2051–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Baird MA; Shvartsburg AA J. Am. Soc. Mass Spectrom 2016, 27, 2064–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Shliaha PV; Baird MA; Nielsen MM; Gorshkov V; Bowman AP; Kaszycki JL; Jensen ON; Shvartsburg AA Anal. Chem 2017, 89, 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Garabedian A; Baird MA; Porter J; Jeanne Dit Fouque K; Shliaha PV; Jensen ON; Williams TD; Fernandez-Lima F; Shvartsburg AA Anal. Chem 2018, 90, 2918–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Chen S-H; Russell DH J. Am. Soc. Mass Spectrom 2015, 26, 1433–1443. [DOI] [PubMed] [Google Scholar]
- (213).Stojko J; Fieulaine S; Petiot-Bécard S; Van Dorsselaer A; Meinnel T; Giglione C; Cianférani S Analyst 2015, 140, 7234–7245. [DOI] [PubMed] [Google Scholar]
- (214).Nshanian M; Lantz C; Wongkongkathep P; Schrader T; Klärner F-G; Blümke A; Despres C; Ehrmann M; Smet-Nocca C; Bitan G; Loo JA J. Am. Soc. Mass Spectrom 2019, 30, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Wang H; Eschweiler J; Cui W; Zhang H; Frieden C; Ruotolo BT; Gross ML J. Am. Soc. Mass Spectrom 2019, 30, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Jhingree JR; Beveridge R; Dickinson ER; Williams JP; Brown JM; Bellina B; Barran PE Int. J. Mass Spectrom 2017, 413, 43–51. [Google Scholar]
- (217).Warnke S; Baldauf C; Bowers MT; Pagel K; von Helden GJ Am. Chem. Soc 2014, 136, 10308–10314. [DOI] [PubMed] [Google Scholar]
- (218).Theisen A; Yan B; Brown JM; Morris M; Bellina B; Barran PE Anal. Chem 2016, 88, 9964–9971. [DOI] [PubMed] [Google Scholar]
- (219).Theisen A; Black R; Corinti D; Brown JM; Bellina B; Barran PE J. Am. Soc. Mass Spectrom 2019, 30, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Conant CR; Fuller DR; Zhang Z; Woodall DW; Russell DH; Clemmer DE J. Am. Soc. Mass Spectrom 2019, 30, 932–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Dixit SM; Polasky DA; Ruotolo BT Curr. Opin. Chem. Biol 2018, 42, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Riley NM; Mullen C; Weisbrod CR; Sharma S; Senko MW; Zabrouskov V; Westphall MS; Syka JEP; Coon JJ J. Am. Soc. Mass Spectrom 2016, 27, 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Weisbrod CR; Kaiser NK; Syka JEP; Early L; Mullen C; Dunyach J-J; English AM; Anderson LC; Blakney GT; Shabanowitz J; Hendrickson CL; Marshall AG; Hunt DF J. Am. Soc. Mass Spectrom 2017, 28, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Shaw JB; Robinson EW; Paša-Tolić L Anal. Chem 2016, 88, 3019–3023. [DOI] [PubMed] [Google Scholar]
- (225).Durbin KR; Fornelli L; Fellers RT; Doubleday PF; Narita M; Kelleher NL J. Proteome Res 2016, 15, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Fornelli L; Durbin KR; Fellers RT; Early BP; Greer JB; LeDuc RD; Compton PD; Kelleher NL J. Proteome Res 2017, 16, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Anderson LC; DeHart CJ; Kaiser NK; Fellers RT; Smith DF; Greer JB; LeDuc RD; Blakney GT; Thomas PM; Kelleher NL; Hendrickson CL J. Proteome Res 2017, 16, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Cleland TP; DeHart CJ; Fellers RT; VanNispen AJ; Greer JB; LeDuc RD; Parker WR; Thomas PM; Kelleher NL; Brodbelt JS J. Proteome Res 2017, 16, 2072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).LeDuc RD; Fellers RT; Early BP; Greer JB; Shams DP; Thomas PM; Kelleher NL Mol. Cell. Proteomics 2019, 18, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Davis RG; Park H-M; Kim K; Greer JB; Fellers RT; LeDuc RD; Romanova EV; Rubakhin SS; Zombeck JA; Wu C; Yau PM; Gao P; van Nispen AJ; Patrie SM; Thomas PM; Sweedler JV; Rhodes JS; Kelleher NL Anal. Chem 2018, 90, 3802–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Seckler H. d. S.; Fornelli L; Mutharasan RK; Thaxton CS; Fellers R; Daviglus M; Sniderman A; Rader D; Kelleher NL; Lloyd-Jones DM; Compton PD; Wilkins JT J. Proteome Res 2018, 17, 2156–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Lin Z; Wei L; Cai W; Zhu Y; Tucholski T; Mitchell SD; Guo W; Ford SP; Diffee GM; Ge Y Mol. Cell. Proteomics 2019, 18, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Jin Y; Diffee GM; Colman RJ; Anderson RM; Ge YJ Am. Soc. Mass Spectrom 2019, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Cai W; Hite ZL; Lyu B; Wu Z; Lin Z; Gregorich ZR; Messer AE; McIlwain S; Marston SB; Kohmoto T; Ge YJ Mol. Cell. Cardiol 2018, 122, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Ntai I; Fornelli L; DeHart CJ; Hutton JE; Doubleday PF; LeDuc RD; van Nispen AJ; Fellers RT; Whiteley G; Boja ES; Rodriguez H; Kelleher NL Proc. Natl. Acad. Sci. U. S. A 2018, 115, 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Toby TK; Fornelli L; Srzentić K; DeHart CJ; Levitsky J; Friedewald J; Kelleher NL Nat. Protoc 2019, 14, 119–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Cristobal A; Marino F; Post H; van den Toorn HWP; Mohammed S; Heck AJR Anal. Chem 2017, 89, 3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Yang Y; Liu F; Franc V; Halim LA; Schellekens H; Heck AJR Nat. Commun 2016, 7, 13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (239).Khatri K; Pu Y; Klein JA; Wei J; Costello CE; Lin C; Zaia JJ Am. Soc. Mass Spectrom 2018, 29, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Cotham VC; Brodbelt JS Anal. Chem 2016, 88, 4004–4013. [DOI] [PubMed] [Google Scholar]
- (241).Cotham VC; Horton AP; Lee J; Georgiou G; Brodbelt JS Anal. Chem 2017, 89, 6498–6504. [DOI] [PubMed] [Google Scholar]
- (242).Fornelli L; Srzentić K; Huguet R; Mullen C; Sharma S; Zabrouskov V; Fellers RT; Durbin KR; Compton PD; Kelleher NL Anal. Chem 2018, 90, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Tran BQ; Barton C; Feng J; Sandjong A; Yoon SH; Awasthi S; Liang T; Khan MM; Kilgour DPA; Goodlett DR; Goo YA J. Proteomics 2016, 134, 93–101. [DOI] [PubMed] [Google Scholar]
- (244).He L; Anderson LC; Barnidge DR; Murray DL; Hendrickson CL; Marshall AG J. Am. Soc. Mass Spectrom 2017, 28, 827–838. [DOI] [PubMed] [Google Scholar]
- (245).Jin Y; Lin Z; Xu Q; Fu C; Zhang Z; Zhang Q; Pritts WA; Ge Y mAbs 2019, 11, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).van der Burgt YEM; Kilgour DPA; Tsybin YO; Srzentić K; Fornelli L; Beck A; Wuhrer M; Nicolardi S Anal. Chem 2019, 91, 2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).He L; Anderson LC; Barnidge DR; Murray DL; Dasari S; Dispenzieri A; Hendrickson CL; Marshall AG Anal. Chem 2019, 91, 3263–3269. [DOI] [PubMed] [Google Scholar]
- (248).Chen B; Lin Z; Zhu Y; Jin Y; Larson E; Xu Q; Fu C; Zhang Z; Zhang Q; Pritts WA; Ge Y Anal. Chem 2019, 91, 11661–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Griaud F; Denefeld B; Kao-Scharf C-Y; Dayer J; Lang M; Chen J-Y; Berg M Anal. Chem 2019, 91, 8845–8852. [DOI] [PubMed] [Google Scholar]
- (250).Greer SM; Brodbelt JS J. Proteome Res 2018, 17, 1138–1145. [DOI] [PubMed] [Google Scholar]
- (251).Greer SM; Sidoli S; Coradin M; Schack Jespersen M; Schwämmle V; Jensen ON; Garcia BA; Brodbelt JS Anal. Chem 2018, 90, 10425–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Zhou M; Paša-Tolić L; Stenoien DL J. Proteome Res 2017, 16, 599–608. [DOI] [PubMed] [Google Scholar]
- (253).Sidoli S; Lu C; Coradin M; Wang X; Karch KR; Ruminowicz C; Garcia BA Epigenet. Chromatin 2017, 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Dang X; Singh A; Spetman BD; Nolan KD; Isaacs JS; Dennis JH; Dalton S; Marshall AG; Young NL J. Proteome Res 2016, 15, 3196–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Holt MV; Wang T; Young NL J. Am. Soc. Mass Spectrom 2019, 1–12. [DOI] [PubMed] [Google Scholar]
- (256).Smith LM; Thomas PM; Shortreed MR; Schaffer LV; Fellers RT; LeDuc RD; Tucholski T; Ge Y; Agar JN; Anderson LC; Chamot-Rooke J; Gault J; Loo JA; Paša-Tolić L; Robinson CV; Schlüter H; Tsybin YO; Vilaseca M; Vizcaíno JA; Danis PO; Kelleher NL Nat. Methods 2019, 16, 939–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).LeDuc RD; Schwämmle V; Shortreed MR; Cesnik AJ; Solntsev SK; Shaw JB; Martin MJ; Vizcaino JA; Alpi E; Danis P; Kelleher NL; Smith LM; Ge Y; Agar JN; Chamot-Rooke J; Loo JA; Paša-Tolić L; Tsybin YO J. Proteome Res 2018, 17, 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (258).Lermyte F; Tsybin YO; O’Connor PB; Loo JA J. Am. Soc. Mass Spectrom 2019, 30, 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (259).Donnelly DP; Rawlins CM; DeHart CJ; Fornelli L; Schachner LF; Lin Z; Lippens JL; Aluri KC; Sarin R; Chen B; Lantz C; Jung W; Johnson KR; Koller A; Wolff JJ; Campuzano IDG; Auclair JR; Ivanov AR; Whitelegge JP; Paša-Tolić L; Chamot-Rooke J; Danis PO; Smith LM; Tsybin YO; Loo JA; Ge Y; Kelleher NL; Agar JN Nat. Methods 2019, 16, 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Hilton GR; Benesch JLPJR Soc., Interface 2012, 9, 801–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (261).Mehmood S; Allison TM; Robinson CV Annu. Rev. Phys. Chem 2015, 66, 453–474. [DOI] [PubMed] [Google Scholar]
- (262).Lössl P; van de Waterbeemd M; Heck AJ EMBO J 2016, 35, 2634–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Chait BT; Cadene M; Olinares PD; Rout MP; Shi YJ Am. Soc. Mass Spectrom 2016, 27, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (264).Haverland NA; Skinner OS; Fellers RT; Tariq AA; Early BP; LeDuc RD; Fornelli L; Compton PD; Kelleher NL J. Am. Soc. Mass Spectrom 2017, 28, 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (265).Popa V; Trecroce DA; McAllister RG; Konermann LJ Phys. Chem. B 2016, 120, 5114–5124. [DOI] [PubMed] [Google Scholar]
- (266).Yewdall NA; Allison TM; Pearce FG; Robinson CV; Gerrard JA Chem. Sci 2018, 9, 6099–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Skinner OS; Haverland NA; Fornelli L; Melani RD; Do Vale LHF; Seckler HS; Doubleday PF; Schachner LF; Srzentić K; Kelleher NL; Compton PD Nat. Chem. Biol 2018, 14, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Ben-Nissan G; Belov ME; Morgenstern D; Levin Y; Dym O; Arkind G; Lipson C; Makarov AA; Sharon M Anal. Chem 2017, 89, 4708–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (269).Schachner LF; Ives AN; McGee JP; Melani RD; Kafader JO; Compton PD; Patrie SM; Kelleher NL J. Am. Soc. Mass Spectrom 2019, 30, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (270).Calabrese AN; Radford SE Methods 2018, 147, 187–205. [DOI] [PubMed] [Google Scholar]
- (271).Landreh M; Marklund EG; Uzdavinys P; Degiacomi MT; Coincon M; Gault J; Gupta K; Liko I; Benesch JLP; Drew D; Robinson CV Nat. Commun 2017, 8, 13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Yen H-Y; Hopper JTS; Liko I; Allison TM; Zhu Y; Wang D; Stegmann M; Mohammed S; Wu B; Robinson CV Science Advances 2017, 3, e1701016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Susa AC; Lippens JL; Xia Z; Loo JA; Campuzano IDG; Williams ER J. Am. Soc. Mass Spectrom 2018, 29, 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).Lippens JL; Nshanian M; Spahr C; Egea PF; Loo JA; Campuzano IDG J. Am. Soc. Mass Spectrom 2018, 29, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Ro SY; Schachner LF; Koo CW; Purohit R; Remis JP; Kenney GE; Liauw BW; Thomas PM; Patrie SM; Kelleher NL; Rosenzweig AC Nat. Commun 2019, 10, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Landreh M; Costeira-Paulo J; Gault J; Marklund EG; Robinson CV Anal. Chem 2017, 89, 7425–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (277).Campuzano IDG; Li H; Bagal D; Lippens JL; Svitel J; Kurzeja RJM; Xu H; Schnier PD; Loo JA Anal. Chem 2016, 88, 12427–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (278).Zhang J; Loo RRO; Loo JA J. Am. Soc. Mass Spectrom 2017, 28, 1815–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Li H; Sheng Y; McGee W; Cammarata M; Holden D; Loo JA Anal. Chem 2017, 89, 2731–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (280).Skinner OS; McAnally MO; Van Duyne RP; Schatz GC; Breuker K; Compton PD; Kelleher NL Anal. Chem 2017, 89, 10711–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).O’Brien JP; Li W; Zhang Y; Brodbelt JS J. Am. Chem. Soc 2014, 136, 12920–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Cammarata MB; Thyer R; Rosenberg J; Ellington A; Brodbelt JS J. Am. Chem. Soc 2015, 137, 9128–9135. [DOI] [PubMed] [Google Scholar]
- (283).Cammarata MB; Brodbelt JS Chem. Sci 2015, 6, 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (284).Cammarata MB; Schardon CL; Mehaffey MR; Rosenberg J; Singleton J; Fast W; Brodbelt JS J. Am. Chem. Soc 2016, 138, 13187–13196. [DOI] [PubMed] [Google Scholar]
- (285).Cammarata M; Thyer R; Lombardo M; Anderson A; Wright D; Ellington A; Brodbelt JS Chem. Sci 2017, 8, 4062–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).Mehaffey MR; Cammarata MB; Brodbelt JS Anal. Chem 2018, 90, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (287).Morrison LJ; Brodbelt JS J. Am. Chem. Soc 2016, 138, 10849–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Tamara S; Dyachenko A; Fort KL; Makarov AA; Scheltema RA; Heck AJR J. Am. Chem. Soc 2016, 138, 10860–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Sipe SN; Brodbelt JS Phys. Chem. Chem. Phys 2019, 21, 9265–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (290).Mikhailov VA; Liko I; Mize TH; Bush MF; Benesch JLP; Robinson CV Anal. Chem 2016, 88, 7060–7067. [DOI] [PubMed] [Google Scholar]
- (291).Mehaffey MR; Sanders JD; Holden DD; Nilsson CL; Brodbelt JS Anal. Chem 2018, 90, 9904–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (292).Harvey SR; Yan J; Brown JM; Hoyes E; Wysocki VH Anal. Chem 2016, 88, 1218–1221. [DOI] [PubMed] [Google Scholar]
- (293).Sahasrabuddhe A; Hsia Y; Busch F; Sheffler W; King NP; Baker D; Wysocki VH Proc. Natl. Acad. Sci. U. S. A 2018, 115, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (294).van de Waterbeemd M; Fort KL; Boll D; Reinhardt-Szyba M; Routh A; Makarov A; Heck AJR Nat. Methods 2017, 14, 283–286. [DOI] [PubMed] [Google Scholar]
- (295).van de Waterbeemd M; Tamara S; Fort KL; Damoc E; Franc V; Bieri P; Itten M; Makarov A; Ban N; Heck AJR Nat. Commun 2018, 9, 2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (296).van de Waterbeemd M; Snijder J; Tsvetkova IB; Dragnea BG; Cornelissen JJ; Heck AJR J. Am. Soc. Mass Spectrom 2016, 27, 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (297).Ma X; Zhou M; Wysocki VH J. Am. Soc. Mass Spectrom 2014, 25, 368–379. [DOI] [PubMed] [Google Scholar]
- (298).Greisch J-F; Tamara S; Scheltema RA; Maxwell HWR; Fagerlund RD; Fineran PC; Tetter S; Hilvert D; Heck AJR Chem. Sci 2019, 10, 7163–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (299).Liko I; Allison TM; Hopper JT; Robinson CV Curr. Opin. Struct. Biol 2016, 40, 136–144. [DOI] [PubMed] [Google Scholar]