Summary paragraph
Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) cause severe respiratory diseases in infants and elder adults1. Neither a vaccine nor an effective antiviral therapy exists to control RSV or HMPV infections. During viral genome replication and transcription, the tetrameric phosphoprotein P serves as a crucial adaptor between the nucleoprotein-RNA (N-RNA) template and the L protein, which has RNA-dependent RNA polymerase (RdRp), GDP polyribonucleotidyltransferase (PRNTase) and cap-specific methyltransferases (MTases) activities2,3. How P interacts with L and mediates association with the free form of N and with the ribonucleoprotein (RNP) is not clear for HMPV or other prominent human pathogens including measles, Ebola and rabies viruses. Here, we report a cryo-EM reconstruction showing the ring-shaped structure of the polymerase and capping domains of HMPV L, bound with a tetramer of P. The connector and MTase domains are mobile with respect to the core. The putative priming loop important for initiation of RNA synthesis is fully retracted, leaving space in the active-site cavity for RNA elongation. P interacts extensively with the N-terminal region of L, burying more than 4,016 Å2 of molecular surface area in the interface. Two of the four helices forming the coiled-coil tetramerization domain of P, and long C-terminal extensions projecting from these two helices, wrap around the L protein like tentacles. The structural versatility of the four P protomers, which are largely disordered in their free state, demonstrates an example of a “folding-upon-partner-binding” mechanism for carrying-out P adaptor functions. The structure shows that P has the potential to modulate multiple functions of L and should accelerate the design of specific antiviral drugs.
The 13 kb non-segmented negative-strand (NS) HMPV RNA genome encodes nine proteins1. Following virus entry, N-RNA is released into the host-cell cytoplasm and serves as template for the viral polymerase to produce sub-genomic capped and polyadenylated viral mRNAs, and for genome replication through an anti-genome intermediate. A cap 1 structure is formed at the 5’ end of viral mRNAs by the capping and MTase domains of L2,3. Structures are available for segmented NS virus polymerases such as influenza4, which acquire a capped primer by “cap snatching”. For polymerases of the Mononegavirales order, which includes several major human pathogens, the structure of the rhabdovirus vesicular stomatitis virus (VSV) L protein was determined first3 and a crystal structure of the MTase domain of HMPV-L was reported5.
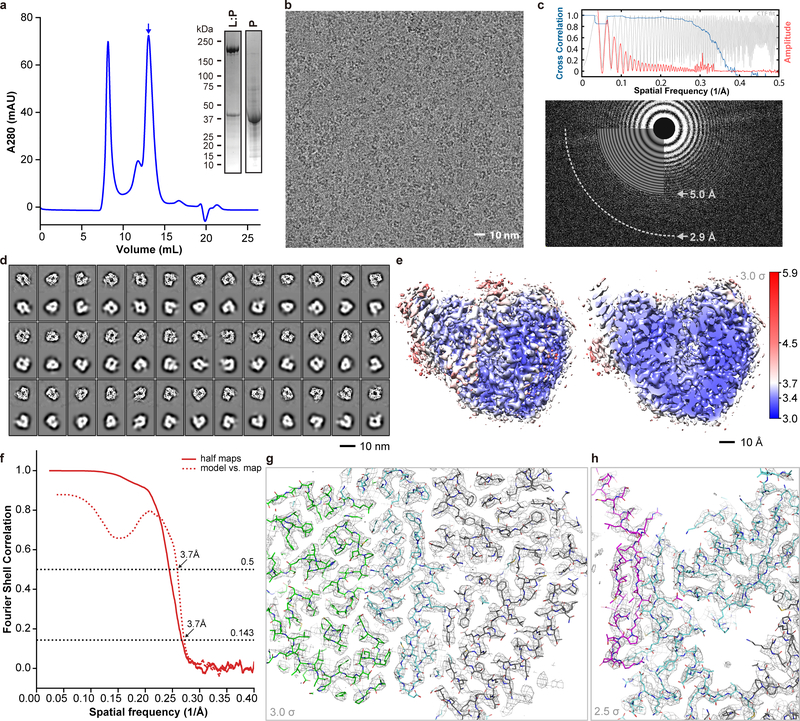
Co-expressing the L and P proteins (Fig. 1a, b and Extended Data Fig. 1a) from HMPV in Sf9 insect cells yielded a stoichiometric preparation suitable for functional and structural studies (Fig. 1c–e). A primer elongation assay demonstrated that LWT:P complexes had polymerase activity (Fig. 1c). LWT:P complexes were also tested for de novo RNA synthesis activity using a template consisting of a sequence from the 3’ end of the HMPV genome (“le14”). The major band detected in reactions containing LWT:P, all four NTPs and radiolabeled UTP as a tracer was 12 nt in length, consistent with position 3C being the dominant initiation site (Fig. 1d). This product was not detectable in a reaction mix containing only radiolabeled UTP and no other NTPs, or in a reaction containing a chain terminator, confirming that it was the product of de novo initiation at the promoter. LD745A:P, in which Asp745 from the conserved “GDNQ” motif C was mutated to Ala, was devoid of polymerase activity, ruling out the possibility that a contaminating polymerase was responsible for the observed activity. Thus, recombinant L:P is active for RNA polymerization (Fig. 1c–d and Extended Data Fig. 2).
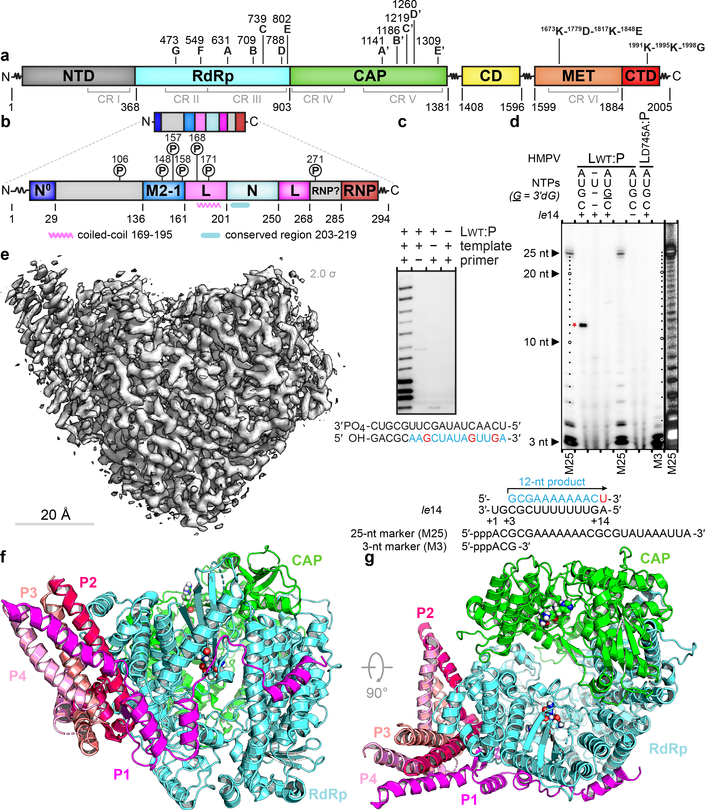
Figure 1. Overall structure of the HMPV L:P complex.
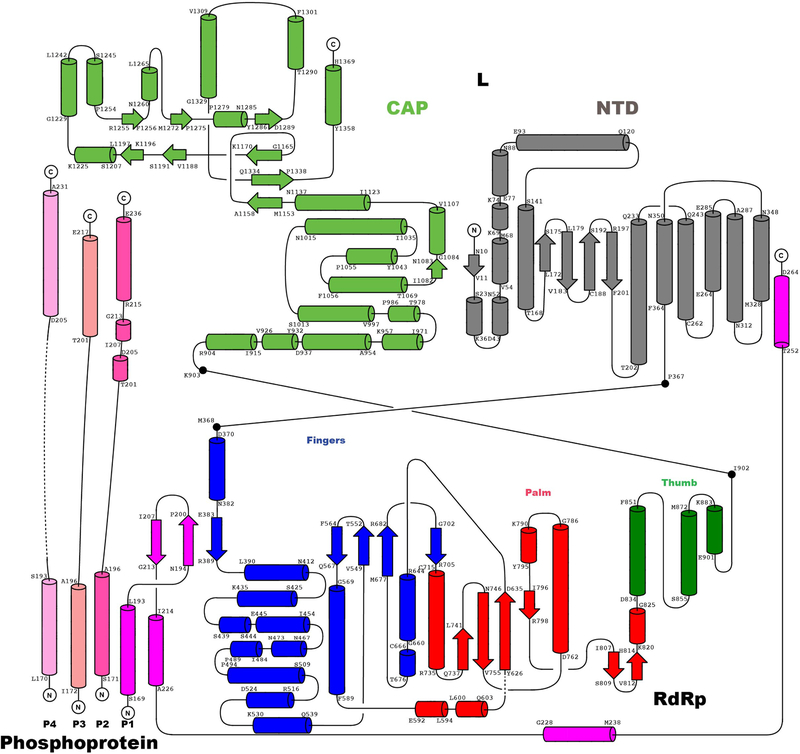
Domain organization of L (a) and P (b) outlining conserved regions and motifs. Regions of HMPV-P predicted to interact with N0, M2–1, L and RNP, and phosphorylation sites are indicated11. (c) Primer elongation assay. Sequences of the 18 nt RNA template and for the 5 nt primer are shown with nascent RNA in blue and radiolabel incorporation sites in red. This experiment was performed a total of four times with two different buffer conditions. (d) RdRp activity assay using the “le14” RNA template. Sequences for the 5’-triphosphorylated 25 nt and 3 nt markers and the 12 nt product are indicated. The 3’dGTP chain terminator is labeled “G”. Radiolabeled UMP is in red. Data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1. (e) Overview of the HMPV-L:P cryo-EM 3D reconstruction (f) Overview of the L:P atomic model. RdRp: cyan, Capping domain: green, phosphoprotein tetramer subunits P1 magenta, P2 hot pink, P3 salmon and P4 pink. Atoms from the “GDNQ” motif in the RdRp and from the “HR” motif in the capping domains are shown as colored spheres (g) Rotated view of the L:P atomic model.
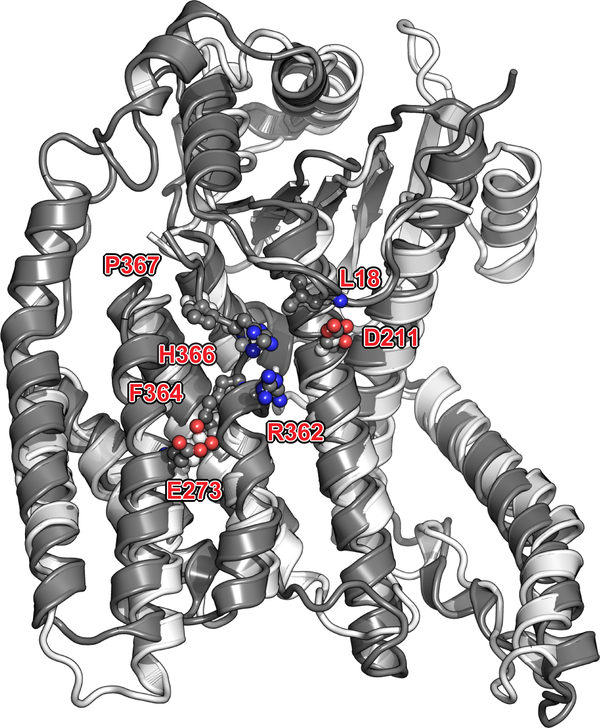
We determined the structure of the L:P complex to a resolution of 3.7 Å using cryo-EM (Fig. 1e, Supplementary Video 1, Extended Data Fig. 1). The final 3D reconstruction (Extended Data Fig. 3) allowed us to build an atomic model for residues 8 to 1380 of L and for the bound P tetramer (Fig. 1f–g, Extended Data Fig. 3–5). Residues 1381 to the C-terminus are not visible, suggesting flexibility of the peripheral appendage comprising CR-VI+5. A comparison of HMPV-L with VSV-L3 returns an r.m.s.d. of 2.8 Å, illustrating the preservation of the core L structure during Mononegavirales evolution (Supplementary Fig. 2). This similarity extends to the N-terminal subdomain (NTD) of VSV-L, which is essential for viral transcription (Extended Data Fig. 6)6. The RdRp folds into the canonical fingers-palm-thumb subdomains observed in many RNA polymerase structures (Fig. 2a)3. Nucleoside inhibitors have been developed targeting RSV and HMPV RdRp activity7. The 5’ triphosphate metabolites of ALS-8176/ALS8112 inhibit HMPV and RSV in non-human primates with similar potency and resistant RSV mutants were sequenced7. We mapped the locations of RSV-L resistance mutations7 to helix α29 of the present HMPV-L structure (Extended Data Fig. 7).
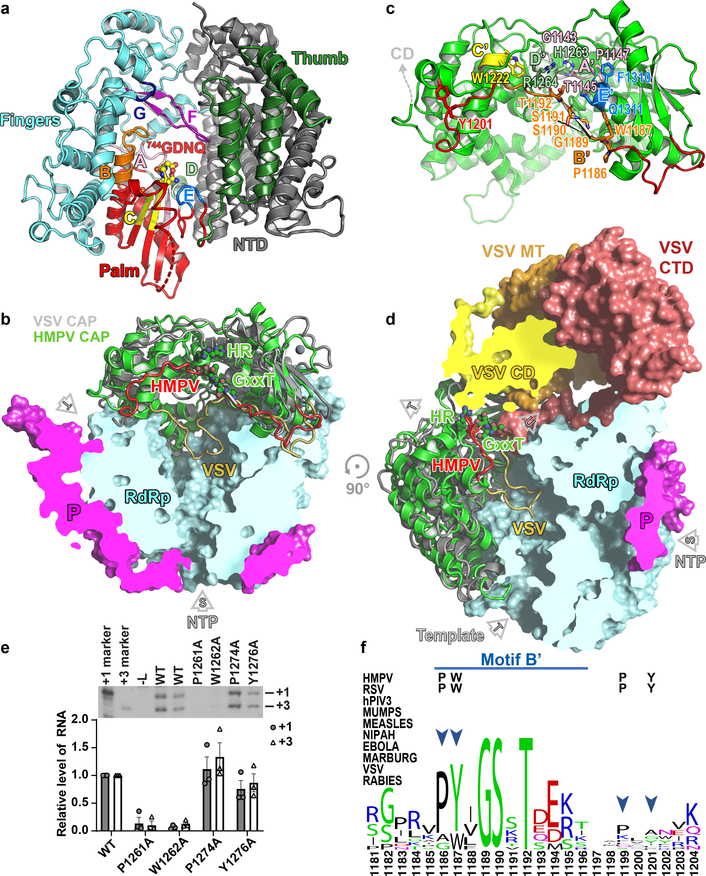
Figure 2. Structure of the HMPV L protein.
(a) The RdRp viewed in its “front” orientation. Fingers subdomain: blue; palm: red and thumb: green, NTD: grey. RdRp motifs A-G are shown. (b) The capping domain of L (green ribbons) of HMPV-L. A superposition of HMPV-L with VSV-L3 (grey ribbons) highlights the difference between conformations of the putative priming loops: putative priming loop of HMPV-L (including Thr1192 of the GxxT motif): red and of VSV-L (1157–1173): gold. P: magenta. (c) RNA capping motifs A’-E’.2 (d) Overall view of HMPV-L with VSV-L superimposed, highlighting the mobile appendage (displayed in the VSV-L orientation, but disordered in the present structure) and rNTP (S) entry, template (T) and nascent (U) RNA tunnels. (e) Impact of substitutions in aromatic and proline residues in the putative priming loop of RSV-L on RNA production from the +1 and +3 sites. The mean and standard error of three independent experiments are indicated. (f) Sequence conservation in the putative priming loop of L. The four residues that were mutated (panel e) are indicated by arrows. A proline residue (P1186 in HMPV-L) important for an early stage of RNA synthesis is conserved in Pneumoviridae, Paramyxoviridae and Filoviridae but not in Rhabdoviridae.
As in VSV-L, the capping domain of HMPV-L (Fig. 2b), containing the “HR” catalytic motif2 has a large interface with the RdRp, accounting for the rigidity of the “doughnut”. Arg1264 is exposed to the solvent, due to the mobility of the C-terminal appendage. A feature shared by RdRps that do not need a polynucleotide primer to initiate polymerization, is a “priming loop” (Fig. 2c, d) that supports the initiating rNTP8. In the influenza polymerase4, a proline at the tip of a β-hairpin of PB1 plays a key role for replication initiation. In rhabdoviruses (VSV and rabies virus), the corresponding loop is in the capping domain and contains residues important both for terminal de novo initiation and for mRNA capping10, with a Trp residue in the capping domain essential for de novo initiation10, indicating it is the priming residue. In sequence alignments, a Tyr residue lies at a similar position in HMPV-L (Tyr1201) and RSV-L (Tyr1276), which could provide base-stacking interactions required for priming (Supplementary Fig. 2). We tested this possibility using the RSV polymerase, which initiates at positions 1 and 3 on its promoter. A Tyr1276Ala substitution did not hamper RSV initiation from either site, but Ala substitution of either Pro1261 or Trp1262, corresponding to Pro1186 and Trp1187 in HMPV-L in motif B’ of the capping domain (Fig. 2c), inhibited an early stage of RNA synthesis within the promoter region (Fig. 2e, f), suggesting that either this Pro or Trp residue might be responsible for priming de novo RNA synthesis. However, we cannot exclude the possibility that the N-terminal region of the loop plays a structural role not directly related to HMPV de novo initiation.
In VSV-L, the priming loop largely occludes the central RNA binding cavity (Fig. 2d), precluding the formation of a long dsRNA duplex and indicating a pre-initiation conformation3. The putative priming loop in our structure of HMPV-L retracts completely into a cavity in the capping domain, leaving ample space for several base-pairs of a primer-template dsRNA intermediate (Fig. 2b, d). Modeling an elongation complex in the HMPV L tunnel shows that an RNA duplex can indeed be accommodated in the central cavity (Fig. 3a–d).
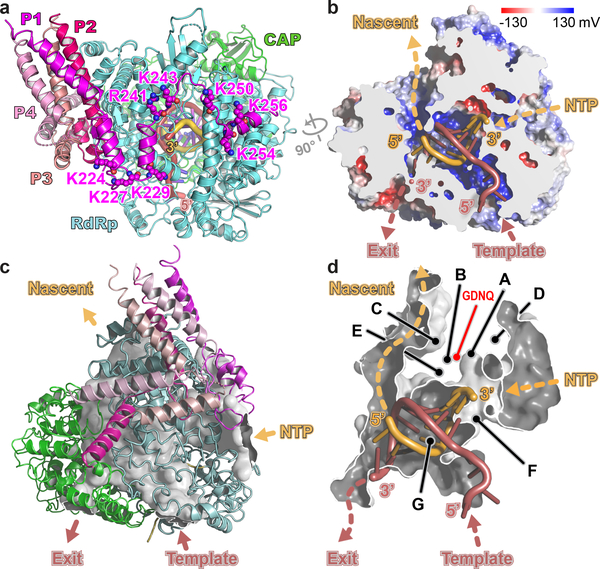
Figure 3. Model for RNA elongation by L:P from Pneumoviridae.
(a) Model for HMPV L:P elongation complex. Template strand: firebrick, nascent strand: orange. Residues from L (Lys307, Arg313, Arg788) and from the C-terminus of subunit P1 (Lys at positions 224, 227, 229, 243, 250, 254 and 256 and Arg241) form a positively-charged arch attracting rNTPs to the rNTP entry tunnel. (b) Cut-out view of L showing electrostatic surfaces (blue: positive and red: negative) and paths followed by the template and nascent RNA strands. (c) Tunnels and interior cavities of L depicted as a white surface enclosed by the L:P complex (ribbons). Cut open are various tunnel openings leading to the exterior. (d) Magnified view of the tunnel that traverses the L protein. Functional motifs A-G and catalytic residues 744GDNQ are indicated.
In the preinitiation state of VSV-L, the connector domain (CD) and MTase domain have a defined orientation with respect to the core, being locked in place by the addition of peptide 35–106 from the VSV-P protein3. In HMPV-L, one linker region located approximately at residues 1381–1407 just upstream of the CD and another at residues 1596–1598 before the CR-VI+ domain (Fig. 1a), likely account for their flexibility. Mobility of the C-terminal domain, which has nucleoside 5’-triphosphatase, nucleoside-2’-O- and guanine-N7 MTase activities5, might be necessary to approach the 5’ end of nascent mRNA for cap addition. Thus, it is possible that N-terminal residues of HMPV-P (which are disordered here) are a latch that keeps the appendage in a fixed orientation for initiation and releases it during RNA elongation.
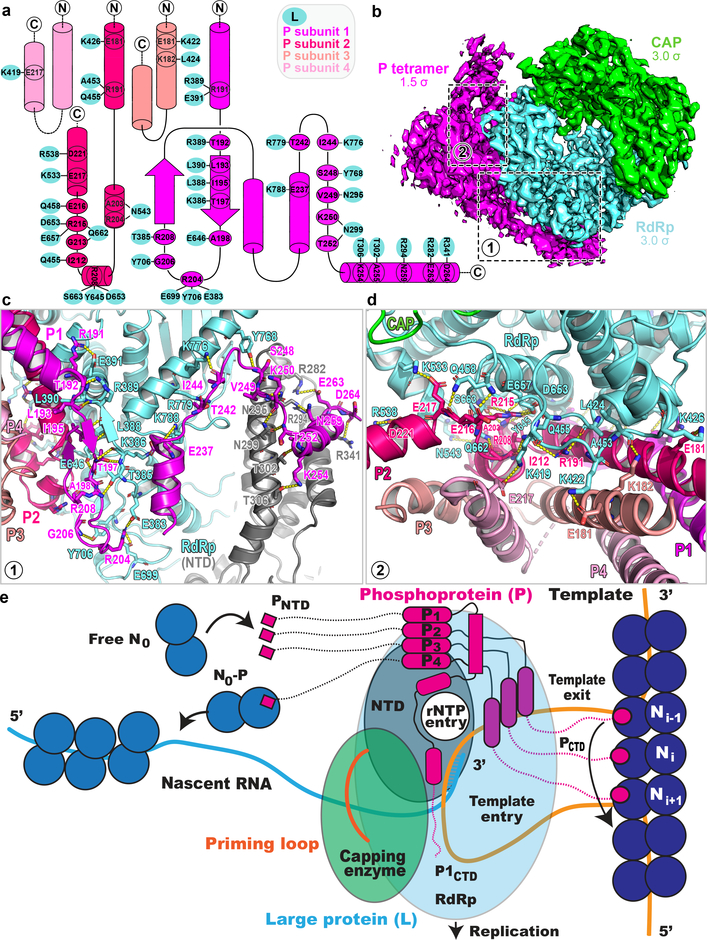
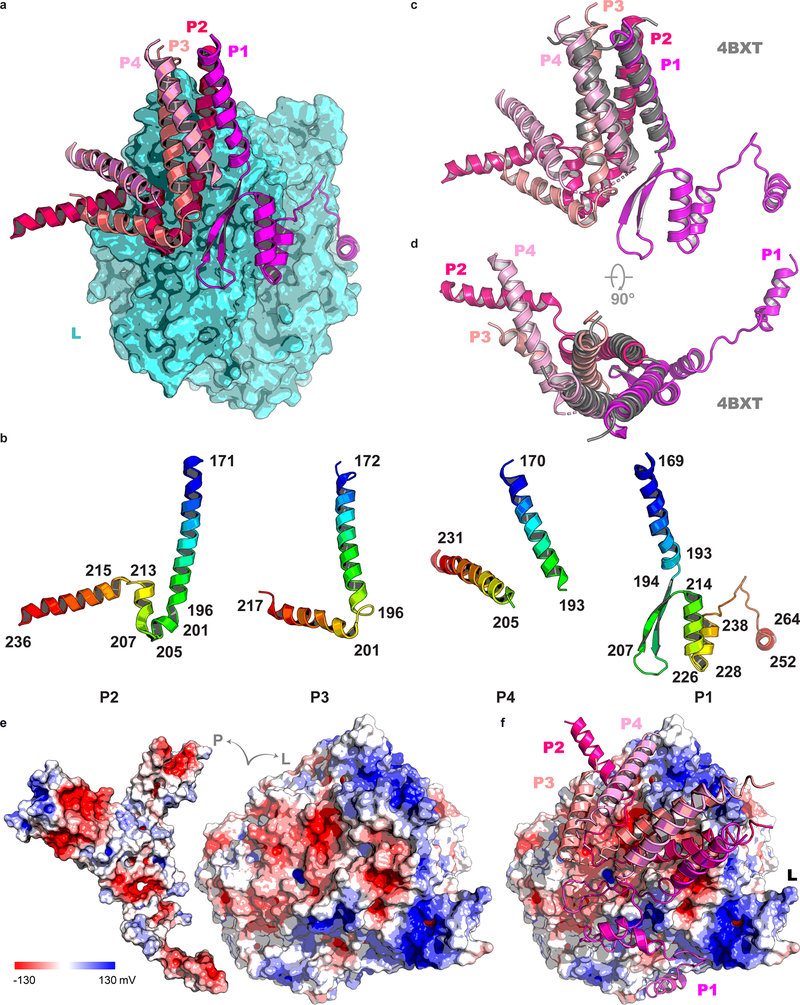
Clear density corresponding to a coiled coil of four helices that constitute the P oligomerization region11 allowed tracing of residues 169–266 and 171–236 for the two proximal P subunits P1 and P2 in close contact with L, and of residues 168–219 and 169–231 for the two distal P subunits P3 and P4 (Fig. 1f, g and Fig. 4a). Much of P appears to fold upon binding to L, as shown by the presence of several extended arms typical of large molecular assemblies such as ribosomes or viral particles (Fig. 1, Fig. 4b and Supplementary Video 2). A large molecular surface area (Extended Data Fig. 4) becomes buried in the interaction, mostly involving residues from P1 and P2, while P3 and P4 provide few interactions (Fig. 4a). Previous structural studies mapped the tetramerization and L-binding regions of P to the four parallel α-helices (spanning residues 171–193 of P) that could be visualized using X-ray crystallography11. In contrast, the N-terminal M2–1 binding regions of P12, and the C-terminal regions of P that bind to the RNP were found to be “intrinsically disordered” in the absence of their respective binding partners11. While the tetramerization region of P appears as a rigid unit largely unaltered upon binding L, a disorder-to-order transition in the C-terminal regions of the two subunits proximal to L takes place upon binding: residues 194 to 213 of P1 fold into a β-hairpin, such that a three-stranded antiparallel β-sheet is formed with residues 383–391 of the fingers subdomain of L (Fig. 4c). This β-hairpin is followed by a long tentacle-like structure composed of three consecutive α-helices that encircle the rear aperture, through which rNTPs gain access to the active site chamber (Fig. 3a). Residues at the C-terminus of the adjacent coiled-coil-forming helix (from P2) project in the opposite direction towards the capping domain (Fig. 4d) with residues 199–205 and 207–213 making two short α-helices and 215–236 forming an extended helical arm (Fig. 4a, d). Only residues Glu181 and Lys182 from P3 make contact with Lys422 and Leu424 from the finger subdomain of L and a single contact is seen between L and P4 (Fig. 4d). Finally, the helix-forming residues 215–236 of P2, together with two long and mobile helices- presumably belonging to residues 201–217 and 205–231 of P3 and P4 respectively form a three-helix bundle largely exposed to solvent. In agreement with the present observations, earlier studies suggested that residues downstream of the coiled-coil region tend to form helices and constitute a “molecular recognition element”11.
Figure 4. The P homo-tetramer and L:P interactions.
(a) Interactions between residues from P1 (magenta), P2 (hotpink), P3 (salmon), P4 (pink) of P and the RdRp (cyan). (b) The electron density map (RdRp: cyan, capping domain: green and P: magenta). (c) A β-hairpin of P1 forms an antiparallel β-sheet with residues 383–391 of the fingers subdomains of L (cyan). Polar contacts between P1 and L are indicated by dashes. (d) interactions between the coiled-coil of P and other parts of P2, P3 and P4 with L. (e) Model for RNA replication by L:P. Genomic viral RNA, extruded from three adjacent N units from the RNP, inserts as a loop of ~27 nts into the RNA entry tunnel and back through the template exit tunnel. Genomic RNA displacement is achieved by three PCTD of phosphoprotein subunits P2, P3 and P4 (while P1 plays a structural role). Concomitantly, four PNTD helices, flexibly linked to the tetramerization domain, bring four new free N0 subunits towards the nascent RNA exit tunnel for encapsidation.
Phosphorylation by cellular kinases regulates the transcription modulation activity of VSV-P13. Analysis of recombinant HMPV-P using tandem mass-spectrometry (Extended Data Fig. 8) showed that P residues targeted by phosphorylation are largely solvent-exposed and changes in phosphorylation should only affect contacts of P with the RNP and M2–1 protein, not complex formation with L (Fig. 4a).
In the Mononegavirales, the genome forms a helical nucleocapsid, with RNA bound in a crevice between the two N proteins subdomains14. P acts as a chaperone by preventing non-specific aggregation of the N protein in the presence of non-viral RNA, preserving a pool of monomeric RNA-free N protein (N0) for RNP assembly15. The P-N0 interaction involves an α-helix from HMPV-P (residues 12–28) that inserts into a hydrophobic groove in the CTD of the N0 protein15. Here the entire regions N-terminal to the coiled coil of the four P subunits, including the N-terminal α-helix (PNTD), are disordered. The tetrameric coiled-coil appears to be anchored to the L protein at a location such that these helices, at the tips of flexible polar linkers spanning residues 29–135, can bind and deliver N0 to the RNA exit tunnel for encapsidating nascent RNA (Fig. 4e). The amino-acid sequence 29–135 of P (Extended Data Fig. 9) bears physico-chemical properties expected of a linker that evolved to ensure the spatial search of nascent RNA exiting L, by N0 bound to PNTD. In the present structure, the C-terminal regions of P are in non-equivalent positions (Fig. 1f, g). These segments have been proposed to dislodge RNA from the RNP complex and feed it into the template tunnel (Fig. 4e)15. Their structural asymmetry probably reflects different roles for the C-terminal domains of each subunit: while the C-terminal end of subunit P2 approaches the RNA template entry tunnel (Fig. 3c) and is well placed to bind N-RNA, the corresponding end of P1 is too distant to play a similar role (Fig. 3a). Moreover, the N-terminus of L (residues 8–24) wraps around the template entry tunnel (Fig. 3c). Thus, in addition to the C-terminal helix of P2, the NTD of L could help displace N to allow naked RNA to feed into the template tunnel (Fig. 3c). In summary, the tetrameric phosphoprotein binds to the monomeric L and fulfills its various adaptor functions by adopting an asymmetric structure (Fig. 4e) through (i) “conformational switching”, whereby each of the four P subunits folds into a different structure, depending on its position in the molecular assembly (ii) spatially confining structurally important N- and C-terminal helical elements (PNTD and PCTD) to the free and RNA-bound N protein respectively, through long flexible regions tethered to the rigidly anchored coiled-coil (Fig. 4e).
While this manuscript was under revision, the structure of RSV L:P was reported16. RSV and HMPV L and P proteins have only 48% and 36% overall amino acid identity respectively, but the structures are similar (r.m.s.d. of 1.34 Å over 1501 residues). Moreover, the putative RSV-L priming loop retracts to a similar position as seen here for HMPV-L16. In RSV, hydrophobic residues critical for L: P interactions (L216, L223 and L227 of RSV-P) have been shown to be critical for polymerase function17. The corresponding region, residues 251–265, of subunit P1 of the HMPV phosphoprotein contains several residues that contact L (Fig. 4c). The overall conservation of interactions between L and P within the family Pneumoviridae, suggests the possibility of broad-spectrum inhibitors disrupting the L: P interface16. We note that dimeric P proteins from rabies virus that interact with the C-terminal region of L18 clearly differ from tetrameric P proteins as for HMPV, that bind to the N-terminal part of L. Whether the mode of interactions between L and P observed here for HMPV and for RSV16, is also found in more distant Mononegavirales viral families such as the Filoviridae must await further structural studies.
Methods
Cloning and expression
HMPV L and P genes (from hMPV isolate CAN97–83, Genbank: AY297749.1)19 codon-optimized for expression in Spodoptera frugiperda 9 (Sf9) cells, were chemically synthesized and cloned into modified pFastBacDual (Thermo Fischer Scientific) by Bio Basic Inc. (Singapore). HMPV L (protein sequence Genbank: AAQ67700.1) was inserted downstream of a Strep-tag-II followed by a TEV cleavage sequence. Strep-TEV-HMPV L was inserted downstream of polyhedrin promoter on pFastBacDual. HMPV P (protein sequence Genbank: AAQ67693.1) was inserted downstream of an 8× His-tag sequence, followed by a TEV cleavage sequence. 8×His-TEV-HMPV P was inserted downstream of p10 promoter on pFastBacDual. We mutated pFastBacDual_HMPV_L:P by plasmid PCR to generate pFastBacDual_HMPV_LD745A:P using forward primer 5’-GACGTGTCCAAGCCTGTGAAGCTGTC and reverse primer 5’-GATGGACTGGTTCGCGCCGTTGAGC. The mutation was confirmed by sequencing. Generation and isolation of bacmid of HMPV_L:P and HMPV_LD745A:P followed bac to bac expression system instructions manual (Invitrogen). Viral stocks generated from purified bacmids were amplified and used for protein expression.
Protein purification
Sf9 cells pellet was resuspended in 1/10 of cultured volume in cold HMPV L:P lysis buffer, 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 15 mM imidazole, 1 mM MgCl2, 0.5 mM TCEP and lysed by sonication. Cell debris was pelleted by centrifugation at 50,000 g for 45 min at 4 °C. Supernatant was incubated with 3 mL pre-equilibrated nickel-NTA beads (Thermo Fisher) per liter of cell pellet for 2 hours at 4 °C. The beads were collected by centrifugation at 500 g for 3 min at 4 °C. The pelleted beads were subjected to washes at 15 mM, 25 mM and 50 mM imidazole with 40, 30 and 20 bed volumes, respectively. Washed beads were transferred into a gravity flow column for elution. HMPV L:P complex was eluted in HMPV L:P His-trap elution buffer containing 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 500 mM imidazole, 1 mM MgCl2, 0.5 mM TCEP, through gravity flow at 4 °C. Eluted fractions were analyzed using SDS PAGE and stained with InstantBlue (Expedeon) to visualize the HMPV L and P proteins (Extended data Fig. 1a). Fractions containing the HMPV L:P complex were pooled with the addition of 1/10 (mg/ml) of TEV and subjected to dialysis against HMPV L:P lysis buffer with no imidazole at 4 °C overnight. Cleaved HMPV L:P protein was incubated with 1 mL nickel-NTA beads at 4 °C for 1 h. Flow-through and wash fractions containing L:P were pooled and subjected to heparin chromatography: a 5 mL HiTrap Heparin (GE healthcare) column was equilibrated with 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP and protein sample was loaded, washed with the same buffer, and eluted using a buffer containing 20 mM Na Hepes, pH 7.4, 1 mM MgCl2, 1 M NaCl, 0.5 mM TCEP. Fractions containing pure L:P were pooled, concentrated and loaded onto a Superose 6 increase 10/300 column (GE healthcare) pre-equilibrated with 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP. Fractions collected were subjected to SDS PAGE and instant blue staining. Fractions containing L:P complex were pooled and concentrated using Vivaspin15R with a 100 kDa cutoff concentrator (Sartorius) to a concentration of 0.85 mg/mL for cryo-EM. The final yield for the L:P complex used for cryo EM was 0.045 mg/L. During the heparin chromatographic step, a large excess of P was observed to flow through the heparin column. This fraction was further purified by size exclusion using a superdex 200 10/300 column (GE healthcare) pre-equilibrated with 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP and the pure P was finally concentrated in a Vivaspin15R concentrator (10 kDa cut-off). The yield of free P was 0.23 mg/L. In order to obtain protein for the polymerization assay, both the wild-type L:P and LD745A:P mutant were purified following the same procedure, but without the heparin chromatographic step. The L protein in the L:P preparations was quantified by separation on SDS-PAGE and detection using colloidal blue staining. The molar concentration of L:P was determined with respect to L.
RdRp activity assays
PAGE-purified RNA oligo-ribonucleotides representing residues 1–14 (Fig. 1d) or 1–25 (Extended data Fig. 2b) of the le sequence purchased from Sigma were used as a template for in vitro RNA synthesis. Purified recombinant HMPV L:P at a concentration of 13 nM was incubated with 1 μM RNA oligonucleotide in a total reaction volume of 20 μL in vitro transcription buffer (50 mM Tris-HCl, pH 7.5, 40 mM KCl, 5.0 mM MgCl2, 0.2 U/μL murine RNase inhibitor, 0.01% (v/v) Triton X-100 and 1 mM DTT). The reactions were pre-equilibrated on ice for 30 min. Transcription reactions were initiated by the addition of rNTPs (1 mM each of rATP, rGTP and rCTP, 20 μM rUTP) and 20 nM [α-32P] rUTP (3,000 Ci/mmol, Perkin Elmer) or as indicated in the legends and incubated at 30° C for 3 hours. Reactions were terminated by addition of an equal volume of deionized formamide. After addition of proteinase K, the reaction mixtures were further incubated at 37° C for 20 min. The reaction products were heat-denatured for 10 min at 95° C and ¼ of the reaction volume was resolved by denaturing urea PAGE (20% acrylamide, 0.5× TBE). Gels were subsequently dried onto a positively charged Hybond N+ nylon membrane (GE Healthcare) and imaged using a phosphorimager. Data are representative of three independent experiments.
The length of the RNA products of the RdRp activity assay was determined by comparison with 3-nt and 25-nt long 5’-triphospho-oligo-RNAs synthesised in vitro using T7 RNA polymerase and [α-32P] rATP incorporation. Succinctly, annealed complementary DNA oligonucleotides representing a class II T7 phage promoter followed by the anticipated 25-nt Tr RNA product sequence were incubated with T7 RNA polymerase (New England Biolab) and [α-32P] rATP according to the manufacturer’s instructions. To generate the 3-nt long 5’-triphospho-oligo-RNA marker (5’-pppACG), reaction was performed under the same experimental conditions except that rGTP was substituted with 3ʹ-dGTP chain terminator.
To characterize the 3′ extension activity of HMPV polymerase on the RNA template, the le25 template was labelled with E. coli poly(A) polymerase (New England Biolab) in the presence of [α-32P] rATP and excess of the chain-terminating analogue cordycepin 3′-triphosphate (3′-dATP). A quantity of 1 μM RNA was incubated (37° C, 60 min) with 0.25 U/μL E. coli poly(A) polymerase in reaction buffer (50 mM Tris-HCl (pH 7.9), 250 mM NaCl, 10 mM MgCl2) supplemented with 20 μM 3ʹ-dATP and 100 nM [α-32P] rATP (3,000 Ci/mmol, Perkin Elmer).
Primer elongation reactions contained 2 μM template (containing a 3´phosphate group), 20 μM primer (containing a 5´- hydroxyl group) and 20 μM each rATP, rCTP, rGTP and rUTP, with 60 nM [α-32P] rGTP tracer in a buffer containing 50 mM Tris-HCl, pH 7.4, 8 mM MgCl2, 5 mM DTT and 10% glycerol. Primer elongation was initiated by the addition of HMPV L:P to a concentration of 20 nM in a total reaction volume of 25 μL. Reactions were allowed to proceed for 2 h at 30˚C. After heat inactivation for 3 min at 90˚C, reactions were heat denatured for 5 min at 95˚C in an equal volume of 10 mM EDTA in deionized formamide with 0.02% each bromophenol blue and xylene cyanol dyes. 7 μL of the 50 μL stopped reaction mix were subjected to denaturing urea PAGE (20% acrylamide, 7 M urea, 1× TBE). The gel was dried onto 3MM chromatography paper and the radiolabelled products were detected by phosphorimager analysis.
Cryo Electron Microscopy
We glow-discharged Quantifoil R 1.2/1.3 holey carbon 200-mesh copper grids (Quantifoil Micro Tools, Großlöbichau, Germany) at 20 mA for 45 seconds. To vitrify the sample, we applied 3.5 μL of the HMPV L:P complex (both diluted to 0.8 mg/mL) to each grid, and plunge-froze in liquid ethane using a Vitrobot Mark IV (FEI) after a 4.0-second blotting (offset -2) with filter papers pre-saturated for 15 minutes under 100 % humidity. We screened the sample homogeneity and cryo preparation in an Oxford-style CT3500 cryo-specimen holder (Gatan Inc, Pleasanton, CA) on a liquid nitrogen cooling Tecnai F20 electron microscope (FEI, Lausanne, Switzerland), operated at 200 kV and equipped with a K2 summit direct detector (Gatan Inc., Pleasanton, CA). We screened for thin and clean ice with maximum distribution of non-overlapping protein particles. Sample homogeneity was judged by visual observation and by 2D classification (Extended data Fig. 1). We optimized and prepared two sets of cryo-grids, one with the purified L:P sample and the other with purified L:P supplemented with the free P sample at a 1:0.4 molar ratio.
We automated data acquisition using SerialEM20 on a K2 summit detector (Gatan Inc., Pleasanton, CA) equipped to a Tecnai Polara electron microscope (FEI, Lausanne, Switzerland) operated at 300 kV and at a nominal magnification of 31,000×. The calibrated physical pixel size was 1.24 Å corresponding to a calibrated magnification of 40,323× on the camera sensor plane. We confirmed beam parallelization by collecting a few movies on carbon after comma-free and objective astigmatism alignments and prior to the actual data collection; the Thon rings of carbon images consistently extended beyond 2.9–2.7 Å and were of excellent stigmation (Extended data Fig. 1). For all datasets, we recorded 40 frames/movie in super-resolution mode (0.62 Å/pixel) using a dose rate of 9.2 electrons/physical pixel/second (1.24 Å/pixel), and a total exposure of 48 electrons/Å2. We recorded a total of 7,438 and 1,100 dose-fractionated images for the L:P sample and L:P supplemented with P sample, respectively.
Image processing
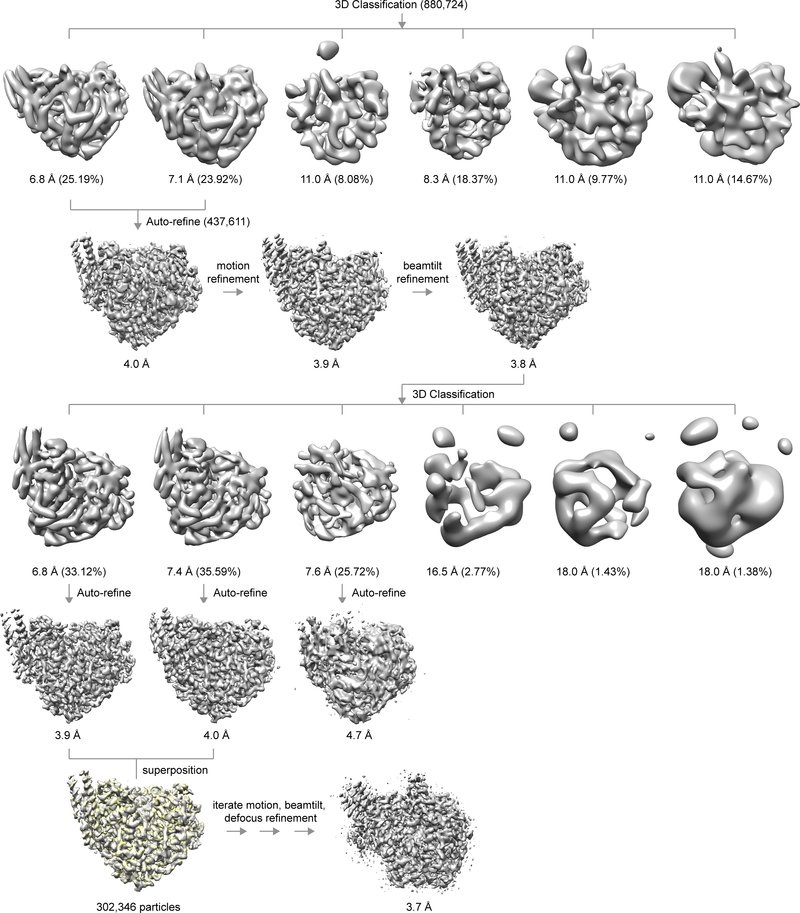
We aligned the super resolution movie frames using MotionCor221 with dose filtering and 5 by 5 patched sub-frame alignment; the frames were binned over 2 by 2 super-resolution “pixels” in Fourier space, yielding the physical pixel size of 1.24 Å of the saved micrographs. We binned the motion-corrected micrographs by a factor of 4 (4.96 Å/pixel) and low-pass filtered to 15 Å for visual examination. We discarded those of thick ice, abnormal defocus, or large carbon areas. For particle picking and 2D classification, we down-sampled the micrographs to 2.48 Å/pixel in Fourier space using resample_mp.exe22 to facilitate computation. We manually picked particles for 10 representative micrographs using e2boxer.py23 and used these boxes to train the auto-picking model in crYOLO24. We then used the trained model to automatically pick particles from all micrographs with the crYOLO Phosaurus network. We visually reviewed the results and manually improved the automatically picked boxes for the 10 micrographs, while incorporating missed representative micrographs for the next round training. We repeated the crYOLO training/autopicking procedures until all particles were accurately picked and well centered from all micrographs. The final L:P data contained a total of 5,381,943 particles, while that of L:P supplemented with P contained 606,410 particles.
We determined the defocus values and defocus angles of the unbinned (1.24 Å/pixel) micrographs using ctffind425. The Thon ring fitting resolution (reported by ctffind4) of most selected micrographs consistently fell into the 2.9~3.3 Å range (Extended data Fig. 1), indicating good data quality. We performed 2D classification in Relion26 until all high-quality 2D class images became apparent (within 50 iterations). We only selected the best 2D classes judged by visual examination and by the resolution and other statistics reported by Relion, retaining 880,724 particles for 3D analysis.
To speed-up computation, we started the initial 3D analysis with the data downsampled to 2.48 Å/pixel. We generated the ab initio model (3 classes) using Relion, and selected the best class as a reference model for further data analysis. We performed one round of mode 3 alignment for the 2D class images against the 3D initial model in frealign (version 9.11)22, with CTF correction turned off. We then calculated projection matching using frealign (version 9.11)22 to cross-validate the 3D model and to confirm the 2D class selection as a self-consistency test. We performed 3D classification straightly with 6 classes in Relion until convergence (within 50 iterations). Two classes were clearly of high quality and superimposed well, with a cross correlation coefficient higher than 99%. We combined these two classes, yielding a total of 437,611 particles. The handedness of the maps was confirmed to be correct as judged by well-defined α-helices. From this point on, we scaled the alignment parameters to 1.24 Å/pixel and used the full resolution data for further analysis. Relion auto-refinement of these particles produced a map of 4.0-Å resolution. We further improved the resolution to 3.9-Å after refining the per-particle motion in Relion and to 3.8 Å after beam tilt refinement by treating all particles as one group. Refining the defocus of individual particles did not yield obvious improvement. We confirmed convergence of motion correction by another round of per-particle motion refinement, which yielded no apparent improvement. Nonetheless, the quality of this map at 3.8-Å resolution was excellent, as judged by main-chain continuity and side-chain densities (Extended data Fig. 1g, h). We further classified the 437,611 particles into six classes and discarded four junk classes, yielding a total of 302,346 particles. We auto-refined the two good classes individually or combined and again they overlapped nearly perfectly. We then gave the data more weights by adjusting the T number in Relion and later performed masked alignment, while correcting FSC for effects of the solvent mask. We visually examined the results and found out that a T number of 4 produced the best map without apparent signs of over-fitting. This combined with another round of per-particle motion refinement, beam tilt refinement, and per particle defocus refinement eventually yielded a map at 3.7 Å. In an effort to improve the density associated with P, we generated masks for P and L using Chimera segger27 and performed multi-body refinement using Relion, with the orientation of P relative to L fixed. We also subtracted the L signal from the 2D images, excised P from the 2D images, and performed 3D classification without alignment. Neither approach improved the P density, perhaps due to insufficient signal considering the small size of P. The final map was at 3.7 Å resolution and was sharpened with a B factor value of −188.25 Å2. The procedure is summarized in Extended Data Fig. 3.
We followed the same data processing paradigm with minor adjustments for the dataset of the L:P preparation supplemented with P. The classification results were consistent with the L:P dataset, leaving 273,574 and 65,398 particles for the good classes after 2D classification and a first round of 3D classification, respectively. The final map after a second round of 3D classification was at 4.1-Å resolution from 29,376 particles and was nearly identical to the map from the L:P dataset. Hence, we mainly used the map from the L:P dataset for map interpretation, except for two less well-defined C-terminal helices of two P subunits (P3 and P4).
Model building and refinement
We built the atomic model using the graphics programs O28 and Coot29. First, we fitted the RdRp and capping enzymatic domains of the VSV-L model (PDB access code: 5A22) into the map as well as the coiled-coil domain of P (PDB access code: 4BXT) and then used these as a guide for finding main-chain connectivity. Local resolution analysis indicated that the majority of the core portion of the complex falls within a resolution range of 3.0–3.6 Å, with excellent main-chain connectivity and side chain densities throughout the molecules (Extended Data Fig 1g, h). The majority of the electron density showed excellent continuity giving confidence for tracing the α–carbon chain. Moreover, in most regions, clear electron density for side chains allowed us to unambiguously assign the sequence register. We only observed weak and disordered extra density accounting for a portion of the CD and CRVI+ domains in maps sharpened with lower B factors and/or low-pass filtered with lower spatial frequency, suggesting that these domains are highly flexible in the complexes we examined. The appendage of HMPV-L might become fixed with respect to the ring-shaped core domain upon binding to a peptide encompassing residues 35–106 of the VSV-P used in the VSV-L study. Indeed, a recent study of the rabies virus L protein in complex with the counterpart of this P peptide demonstrates such contacts between P and L (Josh Horwitz and Stephen C. Harrison, personal communication). Although there were some stretches of weaker densities for the P protein, long side-chains of arginine and lysine residues ensured the correct polarity and register assignment of the sequence. We further confirmed that the sequence was in register by comparing the model with the coiled-coil interface of the known structure of the phosphoprotein tetramerization region (PDB code: 4BXT). Overall, we traced the entire NTD subdomain and RdRp domain spanning residues 8–892, except for mobile residues 1–7 (predicted to be disordered using the PrDOS prediction server http://prdos.hgc.jp/cgi-bin/top.cgi), 607–625 (603–625 are predicted to be disordered), the complete capping enzyme domain (residues 893–1380) and the four P subunits (residues 169–266, 171–236, 168–219, and 169–193/204–231 of subunits P1, P2, P3 and P4, respectively). Residues 194–203 of subunit P4 are disordered. While the sequences could be assigned unambiguously for P1 and P2 residues as well as for the coiled coil region of P3 and P4, the sequence assignment for the C-terminal ends of P3 and P4 that are predicted to form helices is less certain. These segments might become fully ordered only in the presence of the ribonucleoprotein (see main text). Model building was interspersed with refinement using the real space refinement option in the package PHENIX30 (rigid_body, minimization_global, local grid searching and ADP) for the coordinates and B factors. We applied Ramachandran, rotamer and secondary structure restraints throughout the refinement, in addition to the standard geometry and B factor restraints. Over 40 rounds of manual rebuilding and phenix real space refinement were carried out until we had high confidence of the sequence register, density fitting, model geometry and local chemical environments. We analyzed the final model with MolProbity31 and statistics are given in Extended Data Table 1. We deposited the refined coordinates to wwPDB (PDB access code: 6U5O) and the electron density map to EMDB (EMDB access code: EMD-20651).
Modeling of the polymerase: RNA elongation complex
We fitted a rotavirus in situ elongation complex (PDB ID 6OJ3) to the HMPV L:P complex by superimposing the CA atoms of the 5 functional polymerase motifs in the program O28 and refined the alignment using the O command lsq_improve. The relatively long RNA allowed us to easily identify the NTP entry, template entry, template exit, and transcript exit tunnels in the HMPV L structure. Modeling using elongation complexes of other polymerases such as influenza virus PB1 (PDB code: 6QCT), the HCV NS5 (PDB code: 4WTG) and reovirus λ3 elongation complex (PDB code 1N35) confirmed the results.
Tunnel identification
We identified the tunnels in the structure using voidoo, mama and mapman of the Uppsala Software Factory34. We tested values for the probe radius in the range of 1.4–2.0 Å and 1.8 Å yielded the most accurate tunnels. We generated pseudo-atoms to define the range of the calculation and displayed the tunnel along with the models in PyMOL (https:pymol.org). We performed object-specific clipping of the tunnel mask using PovRay (http://www.povray.org) to display its interior along with the modeled RNA.
Tandem Mass spectrometry
We performed tandem mass spectrometry data collection and analysis (Extended Data Fig. 8) for both the purified L:P complex and free P following a standard protocol35. Briefly, we digested 2 μg of each sample over night at 37°C with trypsin (1:100 enzyme:sample molar ratio). Digests were carried out in 200 mM EPPS pH 8.5 in presence of 2% acetonitrile (v/v) with trypsin (Promega #V5111). After peptide purification by reversed phase C18 chromatography 1 μg of peptides was separated with a 4-hour acetonitrile gradient by HPLC prior to injection. Data were collected using an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) system (Thermo Fisher Scientific). The 75 μm inner diameter capillary column used was packed with C18 resin (Accucore 2.6 μm). The scan sequence started with collection of an MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400–1400 Th). MS2 analysis followed collision-induced dissociation (CID, CE=35) with a maximum ion injection time of 250 ms and an isolation window of 1.2 Da. Neutral loss of phosphate was taken into account by using a multi-stage activation (MSA) method with a neutral loss of 97.9763 Da. Peptides were searched with a SEQUEST-based algorithm against an insect database containing L and P protein sequences and common contaminants in addition. Searches were performed with a target decoy database strategy and a false discovery rate (FDR) of 1% set for peptide-spectrum matches followed by linear discriminant analysis (LDA) filtering and a final collapsed protein-level FDR of 1%. We dynamically searched for oxidized methionine (+15.9949146221 Da) and phospho-serine, -threonine and -tyrosine (+79.9663304104 Da). Phosphorylation site localization used a modified Anova score (ModScore). We repeated the experiments twice and the results were consistent.
RSV priming loop experiments
Mutations in codon-optimized RSV L (strain A2) were generated in a fragment of L ORF subcloned into a pGEM T easy vector (Promega). The W1262A substitution was generated by site-directed mutagenesis by Genscript, the P1261A, P1274A, Y1276A substitutions were generated using Q5 site directed mutagenesis (NEB). RSV P1261A was generated using forward primer 5′-ACCTACTAAGGCCTGGGTCGG-3′ and reverse primer 5′-CCCCTCTCGCCCCTGGTC-3′. RSV P1274A was generated using forward primer 5′-CCAGGAAAAAAAGACCATGGCCGTCTACAACAGG -3′ and reverse primer 5′- GTGGAGGAGCCGACCCAGGGCTTAGTAGG -3′. RSV Y1276A was generated using forward primer 5′-CATGCCCGTCGCCAACAGGCAG-3′ and reverse primer 5′-GTCTTTTTTTCCTGGGTG-3′. The mutated ORF fragments were sequenced and then substituted in the codon optimized L ORF contained in a T7 expression vector (pTM1) by restriction digest and ligation. All plasmids were sequenced to confirm the presence of mutations, and to ensure that no other changes were introduced during PCR-amplification steps. BSR-T7 cells, a BHK-derived cell line that constitutively expresses T7 polymerase46 (provided by Dr Karl Conzelmann), in six well dishes were transfected with pTM1 plasmids expressing the following proteins: RSV (A2 strain) N (400 ng/well), P (200 ng/well), M2–1 (100 ng/well), and L (or L mutant; 100 ng/well), and a plasmid expressing a minigenome RNA containing nt 1–36 of the trailer (tr) promoter at its 3´ end (200 ng/well). The minigenome was limited to synthesizing only the first strand of replication, by the lack of a promoter specific sequence at the 3´ end of the replicative intermediate. The cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 40–48 h post transfection, cells were harvested and RNA was purified using Trizol, according to the manufacturer’s instructions, except that the RNA was subjected to a further round of purification by extraction with phenol-chloroform, followed by ethanol precipitation. RNA products initiated within the tr promoter were analyzed by primer extension. RNA representing one-tenth of a well of cells were subjected to reverse transcription at 37˚C using the Sensiscript RT kit (Qiagen) and a radiolabeled primer (5´-TACGAGATATTAGTTTTTGAGAC-3´). One half the primer extension reaction was subjected to electrophoresis in 8% polyacrylamide gels containing 7 M urea in 1× TBE. Radiolabeled oligonucleotides corresponding in sequence to cDNA representing initiation from positions +1 and +3 of the tr promoter were used as markers (+1, 5´-TACGAGATATTAGTTTTTGAGACTTTTTTTCTCGT-3´; +3, 5´- TACGAGATATTAGTTTTTGAGACTTTTTTTCTC-3´´). The same RNA samples, RNA representing one-tenth of a well of cells, were also subjected to denaturing gel electrophoresis on a 1.5% formaldehyde-agarose gel and transferred to nitrocellulose by northern blotting. The input minigenome template was detected using a radiolabeled strand-specific riboprobe. Primer extension products were visualized by autoradiography and the primer extension products and input minigenome were quantified using phosphorimager analysis. The levels of input minigenome were used to normalize for transfection efficiency. The data were normalized to the mean of the two WT samples included in each experiment. With the caveat that the sample sizes were an n of 3, statistical analysis using ANOVA showed that the levels of +1 and +3 RNA products for the P1261A and W1262A L variants were significantly different than those from wt L (p values of 0.002 and 0.003, respectively for +1; 0.004 for both variants for +3), whereas the levels of RNA produced from P1274A and Y1276A L variants were not statistically different (p values of 0.0385 and 0.549, respectively for +1; 0.227 and 0.518, respectively for +3).
Figure preparation
We prepared the figures and videos using PyMOL (Schrödinger LLC), Chimera27, python matplotlib, gnuplot, CCP4 TopDraw36, and PovRay (http://www.povray.org). Multiple sequence alignment was performed with Clustal Omega37 (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the alignment displayed with ESPript38.
Data availability
Structure coordinates are available from the RCSB Protein Data Bank (PDB) under accession code 6U5O and the electron density map from the Electron Microscopy Data Bank (EMDB) under accession code EMD-20651. All other data generated or analyzed in this work are available from the corresponding authors upon reasonable request.
Extended Data
Extended Data Fig. 1 |. Purification of HMPV L:P and structure determination using cryo-EM.
a, Representative size exclusion chromatogram of the L:P complex (these experiments were repeated more than 5 times). Fractions indicated by an arrow were collected and concentrated to 0.85 mg/mL and used for cryo EM analysis. Inset: SDS PAGE followed by Coomassie blue staining of the purified samples. Also shown: free P protein separated from L:P complex by heparin chromatography (For gel source data, see Supplementary Fig. 1). b, Raw micrograph of HMPV-L:P particles recorded in vitreous ice. Scale bar 10 nm. c, Power spectrum of the image shown in panel (b). We limited the high resolution for fitting to a spatial frequency of 1/5.0 Å and 1/2.9 Å marks the highest spacing to which CTF rings were successfully fit. d, 2D classes and “self-consistency check” for the cryo-EM 3D reconstruction. In each box over the three rows, the upper panel shows one 2D class average, whilst the lower panel shows the corresponding projection from the initial 3D model. e, Local resolution of the cryo-EM density map. Variations in local resolution are color- coded from blue (3.0 Å) to red (5.9 Å), computed with Resmap.39 f, Fourier Shell Correlation (FSC) of the cryo-EM map as a function of the spatial frequency. The gold standard resolution is 3.7 Å based on the FSC=0.143 criterion, consistent with the model to map correlation (0.5 criterion). g, Example of the electron density map that allowed model building. The region shown is at an interface between the RdRp and capping domain. The map is shown as a gray mesh, contoured at a level of 3 σ. The atomic model is shown as sticks with residues from RdRp colored in cyan (NTD in grey) and in green for the capping domain. h, The region shown is the three-stranded β-sheet at the interface between the RdRp (cyan sticks) and the phosphoprotein (magenta sticks). The map is shown as a gray mesh, contoured at a level of 2.5 σ. We observed a nearly identical structure of the L:P complex in a reconstruction obtained by premixing the L:P complex with fully phosphorylated P, indicating that potential exchange of P affected neither the formation nor the structure of the L:P complex.
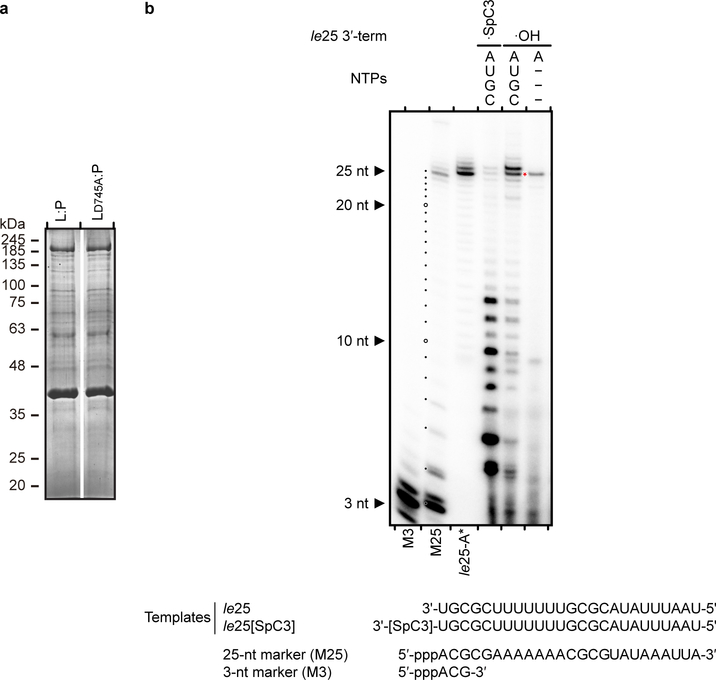
Extended Data Fig. 2 |. RdRp activity assay.
a, SDS PAGE of HMPV wild type L:P, LD745A:P purified for RdRp activity assays. Proteins were purified by metal affinity, TEV cleavage of Histidine-tag followed by reverse His-tag affinity purification and size exclusion chromatography. b, Analysis of the 3′ extension activity of HMPV polymerase using the le25 RNA template. Reactions were performed with rNTPs (0.5 mM each of rUTP, rGTP and rCTP), 20 μM rATP and 20 nM [α-32P] rATP. When a 3’-modified le25 (le25[SpC3], three-carbon spacer group linked to the 3’ extremity) was used as a template, synthesis of products greater than 25 nt was greatly reduced compared to le25. When only [α-32P]rATP and no other rNTP was supplied, only a product with size greater than 25 nt was observed. This result shows that the L:P complex was capable of modifying the 3’ terminus of the template, in addition to engaging in de novo initiation at the promoter. The radiolabeled RNA products were visualized by phosphorimaging. Data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 |. Flow-chart depicting structure determination using cryo-EM.
Please see methods sections for details.
Extended Data Fig. 4 |. Phosphoprotein tetramer in complex with L.
a, The L protein (cyan) is represented as a molecular surface and the tetrameric P protein subunits are represented as ribbons, following the color codes in Figs. 1–3 (P1 in magenta, P2 in hot pink, P3 in salmon and P4 in pink). b, Structures adopted by the four individual P subunits bound to L, colored as a blue to red “rainbow” from the N- to the C- terminal ends. Secondary structures boundaries are noted for each subunit. c and d, Superposition of the tetramerization helices in the context of the L:P complex and the free P protein. Structures are represented as colored ribbons with the free phosphoprotein coiled-coil (PDB access code 4BXT) colored in gray and the four P subunits reported in this work colored according to Fig. 1 (P1 in magenta; P2, hot pink; P3, salmon and P4, pink). The r.m.s.d. of the superimposition is 1.13 Å over 88 α-carbon atoms. e, View of the complex where L and P have been pulled apart to display electrostatic surfaces. f, Overall view of the L:P complex with P shown as ribbons and L as electrostatic surface. The P tetramer consists of subunits P1(magenta), P2 (hotpink), P3 (salmon), and P4 (pink).
Extended Data Figure 5 |. Topology of the L:P complex.
Topological depiction of the secondary structure elements of L and P. Helices are depicted as tubes and strands as arrows. The color code is the same as in Fig. 1. The RdRp domain and its subdomains and the capping domain are colored as in Figs. 1 and 2: NTD in grey, finger in blue, palm in red, thumb in dark green and CAP in green. The four subunits of the phosphoprotein P1, P2, P3 and P4 are colored as in Fig. 1. Secondary structure boundaries are indicated.
Extended Data Fig. 6 |. View of the N-terminal domain (NTD).
NTD is displayed as grey ribbons (following Fig. 2a colors), with evolutionary conserved residues clustered near the rNTP entry tunnel playing a role in transcription, represented as sticks and labeled. Colored in lighter grey is the equivalent region of the VSV_L superimposed to HMPV_L.
Extended Data Fig. 7 |. Model for an elongation complex stalled by the addition of ALS-8176 5’-triphosphate.
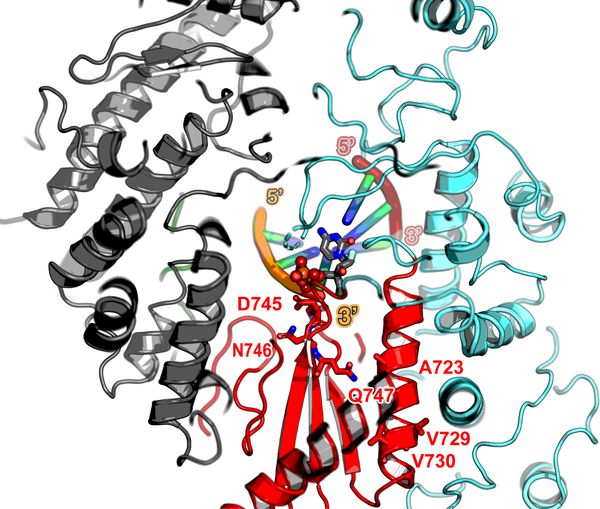
ALS-8176 5’-triphosphate is a nucleoside triphosphate analog against RSV and HMPV currently in phase 2 clinical trials. The 744GDNQ catalytic motif and positions (A789V, L795I and I796V) of which mutations conferred resistance (identified by passaging RSV) are mapped onto the HMPV-L structure (respectively corresponding to A723, V729 and V730) and displayed as sticks. These conservative mutations probably affect inhibitor binding by inducing a slight repositioning of the helix, due to altered hydrophobic contacts with neighboring helices. Please refer also to the sequence alignment displayed in Supplementary Fig. 2. Protein is colored according to Fig. 2a, and the template and nascent RNA strands according to Fig. 3a.
Extended Data Fig. 8 |. MS2 spectrum of the Ser148 P phosphopeptide.
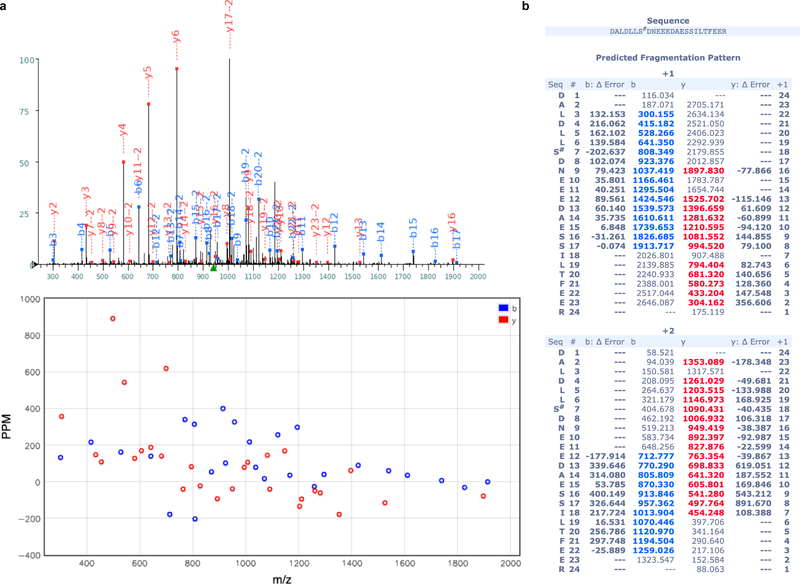
(a) One MS2 spectrum used for identification of the phosphorylated P peptide 142DALDLLS#DNEEEDAESSILTFEER is displayed. Tandem mass spectrum (top) and deviation (bottom) allowed detection of phosphorylation (symbol #) at site Ser148. Peptides fragmented from the N-terminus (b-fragments) and C-terminus (y-fragments) are colored in blue and red, respectively. (b) y and b ion series m/z identified in the spectrum (a) and their deviation from theoretical m/z are displayed in the Table. The present pattern of phosphorylation agrees with observations showing that phosphorylation of the peptide comprising residues 100–120 (ref 44) of RSV-P - in particular phosphorylation of Thr108 (ref 45) corresponding to Ser148 of HMPV-P (Extended Data Fig. 9)) - controls its interaction with the M2–1 protein.
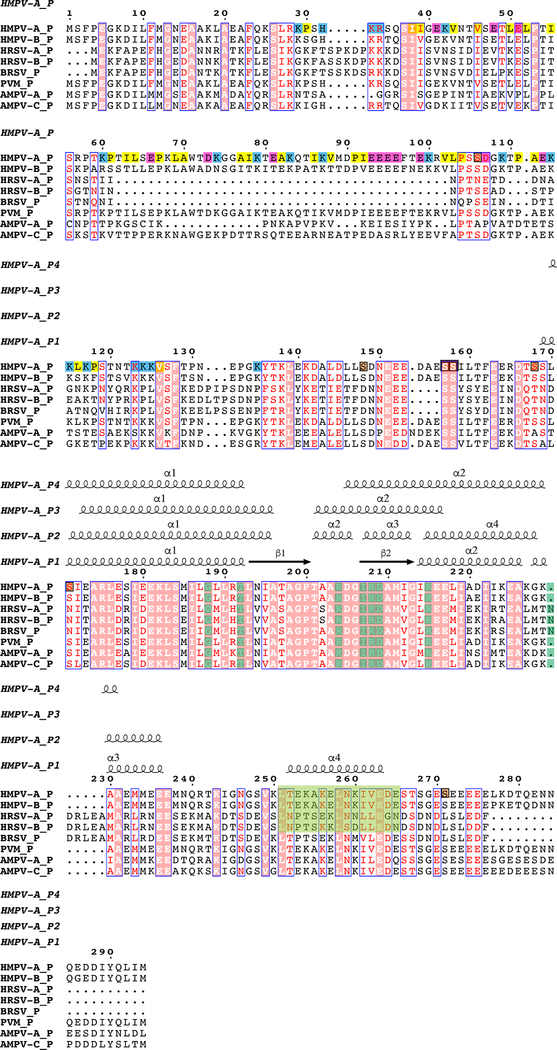
Extended Data Fig. 9 |. Structure-based sequence alignment of the phosphoprotein from HMPV (labeled HMPV-A, strain CAN97-83) and other known pneumoviral P proteins:
HMPV-B, human metapneumovirus subgroup B; HRSV-A and B, human respiratory syncytial virus subgroup A and B respectively; BRSV, bovine respiratory syncytial virus; PVM, pneumonia virus of mice; AMPV-A and AMPV-C, avian metapneumovirus subgroup A and C respectively. Sequences accession codes for the alignment HMPV-A: AAQ67693.1 (used in this work), HMPV-B: AAQ67684.1, HRSV-A: AAX23990.1, HRSV-B: AAR14262.1, BRSV: AAL49395.1, PVM: AAW79177.1, AMPV-A: AAT68644.1 and AMPV-C: AAT86110.1.
The secondary structure of HMPV_P subunit P1 (this work) is displayed above the alignment. Phosphorylation sites are highlighted in brown. Positively-charged residues of HMPV_P are shaded in blue, negatively charged residues in magenta and hydrophobic residues 29 to 135 in yellow. The conserved region containing hydrophobic residues critical for L:P interactions are highlighted in green. Structural alignment of P from HMPV and RSV16 showed similar overall tetramer organization. However, differences are observed in subunit P1 with an r.m.s.d. of 2.24 Å over 82 residues. Although P is in general more mobile with weaker densities and higher B factors compared to L, the region following the beta-hairpin (residues 175–215 in HMPV) does adopt a slightly different conformation compared to RSV P1. Subunit P3 has an r.m.s.d. of 1.94 Å over 45 residues due to a slightly tilted C-terminal helix compared to RSV. Subunit P2 is most similar with an r.m.s.d. of 0.92 Å over 56 residues. Subunit P4 has an r.m.s.d. of 1.33 Å over 47 residues. The eight residues of HRSV-P, that are crucial for interacting with HRSV-L and whose substitutions impair viral replication, are shaded in dark green (data from reference 16). With the exception of Asn189 (HRSV-P) where a deletion is present in HMPV-P, these residues are conserved in HMPV-P and other known pneumoviral P proteins.
Extended Data Table 1 |.
Cryo-EM data collection, structure refinement and model statistics.
| HMPV L:P complex (EMDB-20651) (PDB 6U50) | |
|---|---|
| Data collection and processing | |
| Magnification (nominal/calibrated) | 31,000/40,322.58 |
| Voltage (kV) | 300 |
| Electron exposure (e-/Å2) | 48 |
| Defocus range (μm) | 1.5–3.0 |
| Pixel size (Å) | 1.24 |
| Symmetry imposed | Cl |
| Initial particle images (no.) | 5,381,943 |
| Final particle images (no.) | 302,346 |
| Map resolution (Å) | 3.7 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | ∞-3.7 |
| Refinementa | |
| Initial model used (PDB code) | - |
| Model resolution (Å) | 3.7 |
| FSC threshold | 0.5 |
| Model resolution range (Å) | ∞-3.7 |
| Map sharpening B factor (Å2) | −188.25 |
| Model composition | |
| Non-hydrogen atoms | 12,977 |
| Protein residues | 1,623 |
| Ligands | - |
| B factors (Å2) | |
| Protein | 15.62 (5.27–52.43) |
| Ligand | - |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.717 |
| Validationb | |
| MolProbity scorec | 1.31 (98th percentile) |
| Clashscore | 2.13 |
| Poor rotamers (%) | 0.00 |
| Ramachandran plot | |
| Favored (%) | 100.00 |
| Allowed (%) | 0.00 |
| Disallowed (%) | 0.00 |
Refinement statistics were obtained with program Phenix (phenix.real_space_refinement) (ref 30) except otherwise noted.
Molprobity score was obtained from the Molprobity online server (100th percentile is the best among structures within the resolution specified) and the present structure is in the 98th percentile (N=27675, 0 – 99Å)
The assignment of Pro-1041 as a cis-Proline is supported by electron density showing a hydrogen bond between the carbonyl oxygen atom of Thr-1040 and the N atom of Gln-1353.
Supplementary Material
Acknowledgements
We are very grateful to Prof. Stephen C. Harrison for support, encouragement, suggestions and reading of the manuscript. We thank Zongli Li for cryo-EM support and acknowledge funding from NIH grant AI089618 (to Stephen Harrison). We are grateful to Min Luo and Prof. Maofu Liao of Harvard Medical School for valuable suggestions and help on cryo-EM data acquisition, and Yuhang Liu of Pfizer Inc. for insightful suggestions on image processing strategies. We thank Simon Jenni and Stephen M. Hinshaw of Harvard Medical School and Siew Pheng Lim for valuable suggestions and critiques. We thank Afzaal Shareef of Boston University for insight regarding HMPV RNA synthesis assays. JL lab was supported by Academic Research Grant Tier 3 (MOE2016-T3-1-003) from Singapore Minister of Education (MOE) and grant NRF2016NRF-CRP001-063. X.Q. was supported by Singapore MIT Alliance for Research and Technology. RF lab was supported by NIH grant AI113321.
Footnotes
Competing interests. The authors declare no competing interests
References
- 1.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med 7,719–24 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neubauer J, Ogino M, Green TJ, Ogino T Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic Acids Res. 44(1) 330–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell 162, 314–327. doi: 10.1016/j.cell.2015.06.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pflug A, Guilligay D, Reich S, Cusack S Structure of Influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–60 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Paesen GC, Collet A, Sallamand C, Debart F, Vasseur JJ, Canard B, Decroly E, Grimes JM X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat. Commun 6, 8749. doi: 10.1038/ncomms9749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu S, Ogino M, Luo M, Ogino T, Green TJ Structure and function of the N-terminal domain of the vesicular stomatitis virus RNA polymerase. J. Virol 90, 715724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deval J et al. Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2’-fluoro-4’chloromethyl-cytidine triphosphate. PLoS Pathog. 11(6):e1004995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y, Farsetta DL, Nibert ML, Harrison SC RNA synthesis in a cage-structural studies of reovirus polymerase lambda3. Cell 111, 733–745 (2002). [DOI] [PubMed] [Google Scholar]
- 9.te Velthuis AJW, Robb N, Kapanidis AN, Fodor E The role of the priming loop in influenza A virus RNA synthesis. Nat. Microbiol 1,16029. doi: 10.1038/nmicrobiol.2016.29 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Ogino M, Gupta N, Green TJ, Ogino T A dual-functional priming-capping loop of rhabdoviral RNA polymerases direct terminal de novo initiation and capping intermediate formation. Nucleic Acids Res 47, 299–309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leyrat C, Renner M, Harlos K, Grimes JM Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS One 8(11) e80371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blondot ML et al. , Structural and functional analysis of the RNA and viral phosphoprotein binding domain of respiratory syncytial virus M2–1 protein. PLoS Pathog. 8(5) e1002734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takacs AM, Barik S, Das T, Banerjee AK Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J. Virol 66, 5842–5848 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tawar RG et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Renner M, Bertinelli M, Leyrat C, Paesen G, de Oliveira L, Huiskonen J, Grimes JM Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. Elife 5:e12627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilman MSA, et al. Structure of the Respiratory Syncytial Virus Polymerase Complex. Cell pii: S0092–8674(19)30902-X. doi: 10.1016/j.cell.2019.08.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sourimant J, Rameix-Welti M-A, Gaillard A-L, Chevret D, Galloux M, Gault E, Eleouet J-F Fine mapping and characterization of the L/Polymerase binding domain of the respiratory syncytial virus phosphoprotein. J. Virol 89, 4421–4433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chenik M, Schnell M, Conzelmann KK, Blondel D Mapping the interacting domains between the Rabies virus polymerase and phosphoprotein. J. Virol 72, 1925–1930 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 19.Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ Genetic diversity between human metapneumovirus subgroups. Virology 315 (1), 1–9 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Zheng S, Palovcak E, Armache J-P, Cheng Y, Agard D MotionCor2: Anisotropic Correction of Beam-induced Motion for Improved Single-particle Electron Cryo-microscopy. Nat. Methods 14(4), 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol 183, 377–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ EMAN2: an extensible image-processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Wagner T, Merino F, Stabrin M, Moriya T, Gatsogiannis C, Raunser S SPHIRE-crYOLO: A fast and well-centering automated particle picker for cryo-EM. BioRxiv. doi: 10.1101/356584 (2018). [DOI] [Google Scholar]
- 25.Rohou A, Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheres SHW RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25(13):1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. A A47, 9 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K Features and development of Coot. Acta Cryst. D 66:486–501. doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouba T, Drncová P, and Cusack S Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol 26: 460–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appleby TC, et al. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Kleywegt GJ, Jones TA Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Cryst. D 50, 178–185 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Kalocsay M APEX Peroxidase-Catalyzed Proximity Labeling and Multiplexed Quantitative Proteomics. Methods Mol. Biol 2008:41–55 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Bond CS TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 22(2), 311–2 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD and Higgins DG Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega.Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert X, Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42(W1), W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kucukelbir A, Sigworth FJ, Tagare HD. The local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poch O, Blumberg B, Bougueleret L, Tordo N Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol 71, 1153–1162 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Van den Hoogen BG, Bestebroer TM, Osterhaus ADME, Fouchier RAM Analysis of the genomic sequence of a human metapneumovirus. Virology 295, 119–132 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Bruenn JA A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31(7), 1821–1829 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang DM, Zemla AT, Zhou CLE Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 41(3) 1464–1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason SW et al. Interactions between human respiratory syncytial virus (RSV) M2–1 and P proteins is required for reconstitution of M2–1-dependent RSV minigenome activity. J. Virol 77, 10670–10676 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asenjo A, Calvo E, Villanueva N Phosphorylation of human respiratory syncytial virus P protein at Threonine 108 controls its interaction with the M2–1 protein in the viral RNA polymerase complex. J. Gen. Virol 87, 3637–3642 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Buchholz UJ, Finke S, Conzelmann KK Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol, 73: 251–259 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates are available from the RCSB Protein Data Bank (PDB) under accession code 6U5O and the electron density map from the Electron Microscopy Data Bank (EMDB) under accession code EMD-20651. All other data generated or analyzed in this work are available from the corresponding authors upon reasonable request.