Abstract
The aim of the present study was to evaluate the expression of the chemokine ligand 18 (CCL18) gene in ovarian cancer and to investigate the effects of its overexpression or suppression on growth, invasion, and metastasis in an ovarian carcinoma cell line (SKOV3) in vitro. CCL18 mRNA expression in epithelial ovarian carcinoma (EOC), benign ovarian tumor and normal ovarian tissues was measured by fluorescence quantitative polymerase chain reaction. A CCL18 restructuring plasmid was constructed, and SKOV3 cells were transfected with the plasmid DNA in vitro. A restructuring interference vector was also transfected into CCL18-positive SKOV3 cells. The growth curves, cell cycle distribution, and invasive, migrative and adhesive capacities of SKOV3 cells following overexpression and suppression of CCL18 were evaluated by MTT assay, flow cytometry, Transwell assay, migration assay, and the fibronectin adhesion method, respectively. The positive expression rate of CCL18 in EOC was significantly higher than in benign ovarian tumor (P = 0.002) and normal ovarian tissues (P = 0.003). However, there was no statistical significance in the expression of CCL18 with regard to clinical pathology (including histological classification, pathological grade and surgical pathological stage), and the median survival times of CCL18-positive and CCL18-negative patients did not differ significantly. The invasive, migrative, and adhesive capacities of SKOV3-CCL18 cells were significantly higher than those of SKOV3 and SKOV3-vector cells (P < 0.05). However, there was no significant difference in cell proliferation between the SKOV3-CCL18 and negative control cells. The invasive, migrative, and adhesive capacities of the pSilencer4.1-CCL18-small interfering RNA127 group were significantly lower than those of non-transfected pSilencer4.1-negative and pSilencer4.1 groups (P < 0.05). In conclusion, the overexpression and silencing of CCL18 affected invasion, adhesion, and migration in EOC cells; thus CCL18 may have potential as a clinical marker for early diagnosis of malignant ovarian tumors, and as a target molecule in the treatment of ovarian cancer.
Keywords: Ovarian cancer, chemokine ligand 18 gene, fluorescence quantitative polymerase chain reaction, cell cycle, invasion, metastasis
Introduction
Epithelial ovarian carcinoma (EOC) is a common malignant tumor in females. Due to its occult onset and fast rate of tumor progression, 70% of cases are diagnosed at advanced stage [1]. A previous study confirmed that the 5-year survival rate decreases from 90% at stage I to 30-40% at stage III, with the 5-year survival rate for stage IV patients being only 10% [2]. Therefore, early diagnosis and treatment are critical to improve the survival rate of patients with EOC. Although novel diagnostic methods and markers are constantly emerging, diagnosis of EOC remains unsatisfactory for asymptomatic women and those without prior family history, as the majority of the symptoms are non-specific [3]. The sensitivity and specificity of cancer antigen-125 as an ovarian cancer marker is only 25-30% at stage I [4,5]. The serum protein originates partly from cell secretion and metabolism. When tissue undergoes pathologic changes, the altered cellular peptides, proteins, enzymes, and metabolites can be secreted into the bloodstream [6]. Chemokine ligand 18 (CCL18) was first identified in 1997 as an inflammatory molecular protein [7]. Recent reports have indicated that CCL18 serves a key role in tumor growth and development, and thus may be used as an independent prognostic marker [6,8]. A previous study by our group established a differential serum protein expression profile between subjects with malignant ovarian tumors, benign tumors and those lacking carcinoma, using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS), modified targeted liquid chromatography-MS assays with Cu2+-loaded columns (IMAC-Cu) and weak cationic exchange (WCX-2) protein chip array profiling [9]. For malignant ovarian tumor diagnosis, we evaluated 31 proteins as potential serum markers, ultimately focusing on CCL18 [9,10]. These studies suggested that high expression of CCL18 was associated with the occurrence and development of malignant ovarian tumors [9,10]. Therefore, in the present study, in vitro experiments were conducted to investigate the role of CCL18 in malignant ovarian tumors and confirm the biologic function of CCL18 in ovarian cancer cells.
Materials and methods
Clinical data
A total of 62 Chinese female patients with EOC from the Tumor Hospital affiliated to Guangxi Medical University, Nanning, China, were included in the study (malignant group). The mean age of the malignant group was 45.6 years (range 19-68 years). Tumors were classified histologically according to World Health Organization (WHO) criteria [11] as serous (n = 42) or mucinous (n = 20); and as poorly differentiated (G3) (n = 28), moderately differentiated (G2) or highly differentiated (G1) (n = 34). Tumors were staged according to the International Federation of Gynecology and Obstetrics 2014 criteria [12]; the number of patients with malignancy at stage I/II and III/IV was 23 and 39, respectively. A benign ovarian lesion group contained 40 female patients (mature teratoma, n = 30; cystadenoma, n = 10), of which the mean age was 48.5 years (range 27-73 years). A normal control group consisted of 20 females with uterine fibroids (mean age, 50.3 years; range 33-56 years); these subjects were subjected to total hysterectomy and adnexectomy and no abnormal ovarian pathology was observed. The malignant tumor group underwent primary surgery followed by platinum-based chemotherapy. All cases were followed up; 8 patients did not survive during the 60-month follow-up period and 11 patients were lost to follow up, two for relocation, 8 for mortality from other causes (1 for pneumonia, 2 for cardiac failure, 1 for suicide and 4 for car accident) and one patient for unknown reasons. The study was approved by the Ethics Committee of Guangxi Medical University. All subjects received an explanation of the aims of the study and provided written informed consent. All subjects understood that they could withdraw from the study at any time without influencing their oncological or general medical treatment.
Materials
Plasmids expressing enhanced green fluorescent protein (pEGFP-N1) and Escherichia coli (E.coli) DH5α were purchased from Clontech Laboratories, Inc. (Mountainview, CA, USA), and the SKOV3 cell line was reserved in our laboratory; the restriction enzymes XhoI, BamHI and HindIII, a first strand complementary DNA (cDNA) synthesis kit and T4 DNA ligase were purchased from Fermentas (Thermo Fisher Scientific, Inc., Waltham, MA, USA); LipofectamineTM 2000 and TRIzol reagent were purchased from Invitrogen (Thermo Fisher Scientific, Inc.); fibronectin and matrigel were purchased from the Center for Quantitative Biology of Peking University (Beijing, China); Transwell chambers were purchased from Corning, Inc. (Corning, NY, USA); a QIAquick gel extraction kit was purchased from Qiagen GmbH (Hilden, Germany); a Tiangen plasmid miniprep kit was purchased from Tiangen Biotech Co., Ltd. (Beijing, China); pSilencer4.1-CMVneo vector was purchased from Ambion, Inc. (Austin, TX, USA); and PTG-19-T vector was purchased from Shanghai Generay Biotech Co. Ltd (Shanghai, China).
Design and synthesis of CCL18 primers
According to the published gene sequence of CCL18 in Genebank (https://www.ncbi.nlm.nih.gov/genbank/; ID: NM_002988.2), Polymerase chain reaction (PCR) primers were designed to amplify the open reading fragment of the CCL18 gene using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). For reverse transcription-quantitative PCR (RT-qPCR), the following primers for the CCL18 gene (281 bp) were used: Forward, 3’-TGCCCAGCATCATGAAGG-5’ (base pairs 50-67) and reverse, 3’-TCAGGCATTCAGCTTCAGG-5’ (base pairs 330-312). Primers for GADPH (225 bp) were as follows: Forward, 3’-GAAGGTGAAGGTCGGAGT-5’ (base pairs 6-23) and reverse, 3’-GAAGATGGTGATGGGATTTC-5’ (base pairs 231-212). XhoI (restriction site CTCGAG) and BamHI (restriction site GGATCC) were used for restriction enzyme sites at the ends of the CCL18 primers sequences: Forward, 5’-AAACTCGAGCTGCCCAGCATCATGAAGG-3’ and reverse, 5’-TTTGGATCCCCTCAGGCATTCAGCTTCAG-3’. Primers were synthesized by Shanghai Generay Biotech Co. Ltd. The sequences of CCL18-small interfering (si) RNA target sites are listed in Table 1. The basic local alignment search tool (BLAST) (http://www.ncbi.nlm.gov/blast/Blast.cgi) was used to search the published database (Genebank) and to confirm the siRNA sequences. siRNA was synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The best interference fragment for CCL18 knockdown was selected by transiently transfecting each into SKOV3 cells, extracting the mRNA, and quantifying the cDNA using fluorescent qPCR.
Table 1.
siRNA sequences for chemokine ligand 18
| No. | Base position of the CCL18 sequence | siRNA sequence, 5’-3’ |
|---|---|---|
| 1 | 61 | ACAAGUUGGUACCAACAAA |
| 2 | 127 | CAUAGUUGACUAUUCUGAA |
| 3 | 224 | AGAAGUGGGUCCAGAAAUA |
| 4 | GAPDH positive control | GUAUGACAACAGCCUCAAG |
| 5 | Negative control | UUCUCCGAACGUGUCACGU |
siRNA, small interfering RNA.
Plasmid construction
Total RNA was extracted from ovarian tissue using TRIzol reagent. cDNA was synthesized using the first-strand cDNA synthesis kit according to the manufacturer’s protocol. PCR products were ionophoretically separated, purified, and recovered on a 1% low melting point agarose gel. The PCR products were connected with pEGFP-N1 following double digestion by XhoI and BamHI using T4 DNA ligase. The TTCAAGAGA sequence was used as the loop ring of plasmid pSilencer4.1-CMVneo. The oligonucleotide base pairs were designed with BamH1 (5’-GATCC-3’) and HindIII (5’-AGCTT-3’) sticky ends that expressed the sequence of CCL18-shRNA (forward, 5’-GATCCCATAGTTGACTATTCTGAATTCAAGAGATTCAGAATAGTCAACTATG CTA-3’ and reverse, 5’AGCTTAG CATAGTTGACTATTCTGAAAAGTTCTCTTTCAGAATAGTCAACTATGG-3’). The negative control was designed to have the same bases as the CCL18-shRNA but with altered sequence, as well as no homology with other genes. The insert fragment was digested and annealed to form the double chain. Plasmid pSilencer4.1-CMVNeo was double enzyme digested by BamHI and HindIII. T4 DNA ligase was used to anneal the double chain and linear pSilencer4.1-CMVneo plasmid. E. coli DH5α were transformed into competent bacteria following overnight incubation at 37°C, which was followed by the addition of 3 ml positive clones (containing 100 μg/ml ampicillin) to liquid broth (1% NaCl, 0.5% yeast extract, 1% tryptone; and adjusted to pH 7.0 with 5 M NaOH) and incubation overnight at 37°C. Plasmid was extracted using the Tiangen plasmid miniprep kit and determined by Sanger’s dideoxy method for unidirectional sequencing [13]. Homology analysis of the alignment was performed using BLAST. The sequence of pSilencer4.1-CMVneo-CCL18-siRNA127 (pSilencer4.1-CCL18-siRNA127) was compared with the designed CCL18 oligonucleotide fragment using DNAMAN 6.0 software (Lynnon Biosoft, San Ramon, CA, USA).
Determination of CCL18 mRNA in tissues
CCL18-pTG19-T and GAPDH-pTG19-T were used as templates for qPCR to establish the standard curve. Real-time qPCR amplifications were performed in an ICycler iQ real-time PCR detector (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the following cycling program: Denaturation for 5 min at 95°C, 40 cycles of 30 sec at 94°C, 30 sec at 58°C, 45 sec at 72°C (plus plate read) and 2 sec at 80°C (plus plate read). The setpoint temperature was increased by 0.4°C after cycle 2. Data was collected and analyzed at the melt curve of 55-95°C. The amplification of cDNA template from EOC, benign ovarian tumor and normal ovarian tissue samples was analyzed to obtain cycle threshold and starting quantity values according to the ΔΔCq method [14].
CCL18 overexpression and establishment of SKOV3-CCL18-siRNA127 cell lines
The level of CCL18 mRNA was determined by qPCR in all ovarian carcinoma cell lines. Total RNA was extracted using TRIzol reagent, and the RNA was reverse transcribed into cDNA for fluorescence qPCR. The liposome method [15] was used to establish SKOV3-CCL18-pEGFP-N1 overexpression. The groups included the following: (i) a CCL18-pEGFP-N1 group, transfected with plasmid containing the CCL18 gene; (ii) a pEGFP-N1 group, transfected with pEGFP-N1 empty vector; and (iii) a normal group without transfection. PSilencer4.1-CCL18-siRNA127 plasmid was transfected into the following SKOV3 cell experimental groups: (i) A CCL18-siRNA127 group, transfected with pSilencer4.1-CCL18-siRNA127 plasmid; (ii) a negative control group, transfected with pSilencer4.1-CMVneo-negative (pSilencer4.1-negative) plasmid; (iii) an empty plasmid group, transfected with pSilencer4.1-CMVneo (pSilencer4.1) empty plasmid; and (iv) a non-transfected group of SKOV3 cells plated to 80-90% confluence. Cells were seeded at a density of 3 × 105 cells per well in 6-well plates. All cells were maintained to 80-90% confluency at 37°C with 5% CO2 in a humidified incubator for 24 h. A total of 5 μl lipofectamine™ 2000 and 3.0 μ;g plasmid were each mixed with 250 μ;l RPMI 1640 medium (RPMI medium 1640 dry powder and 0.35 g HEPES dissolved in 1,000 ml double-distilled water, adjusted to pH 7.2-7.4 with NaHCO3) separately and incubated at room temperature for 5 min. The two mixtures were then combined and incubated for 20 min at room temperature. The target mixture was added to the 6-well plates and incubated at 37°C with 5% CO2 in a humidified incubator for 6 h, then the mixture was discarded and 1640 medium containing 20% fetal bovine serum (FBS; Beijing Yuanheng Shengma Biology Technology Research Institute, Peking, China) was added, and cells were further incubated at 37°C with 5% CO2 for 24 h. A fluorescence microscope was used to quantify the average number of green fluorescent cells and total cells to determine the cell transfection efficiency.
CCL18 overexpression and 3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT) assay
SKOV3 cells with stably transfected CCL18-pEGFP-N1 and pEGFP-N1 were cultured to 80% confluence and processed to create a single-cell suspension after 0.25% trypsin digestion (2 min at room temperature). Single-cell suspension was inoculated into 96-well plates with 1 × 104 cells per well. Following incubation at 37°C with 5% CO2 for 24 h, 20 μ;l MTT solution [500 mg MTT (Beijing Hengyuan Hengxin Technology Development Co., Ltd., Beijing, China) dissolved in 100 ml PBS, filtered and sterilized by a sterile filter membrane of 0.22 mm pore size] was added into each well containing the single-cell suspension and incubated for 4 h at 37°C. The supernatant was discarded, and 150 μ;l dimethyl sulfoxide was added to form a solution. The absorbance values of the solution were measured at 490 nm using a microplate reader every 24 h for 7 days.
Cell cycle detection by flow cytometry
Cultured cells were digested with 0.25% trypsin and washed with phosphate-buffered saline (PBS) twice, followed by the addition of 1 ml pre-cooled 70% ethanol. The culture was maintained at 4°C overnight. Following two washes with PBS solution, cell density was adjusted to 1 × 106 cells/ml. Subsequently, propidium iodide (PI) solution was added to the cell culture (1:1) and incubated in the dark at 4°C for 30 min. EPICS XL flow cytometry (Beckman Coulter, Inc., Brea, CA, USA) and MultiCycle 3.2 software for Windows (Beckman Coulter, Inc.) were used to analyze the DNA content and quantify the SKOV3-CCL18-pEGFP-N1 cells in G1, G2, and S phases.
Transwell migration assay
Cultured cells were digested with 0.25% tryp-sin and adjusted to 1 × 106 cells/ml with 1640 medium containing 1% FBS. A total of 100 ml of each cell suspension was added to the upper chamber of a Transwell system, and 600 ml 1640 medium with 10% FBS was added to the lower chamber. The Transwell system was incubated at 37°C and 5% CO2 for 20 h, then 10 and 60 ml MTT were added to the upper and lower chambers, respectively. After 3 h, the lower surface of the Transwell chambers was wiped with 1 cm filter paper, which was put into a 24-well plate with the suspension of the lower chambers. The upper Transwell chambers were placed into another 24-well plate, and to each well 0.5 ml DMSO was added. The dissolved solution of the both 24-well plates was transferred to a 96-well plate, and the absorbance values of the solution were measured at 450 nm using a microplate reader.
Matrigel invasion assay
The lower surface of the polycarbonate filter membrane in the Transwell system was treated with 50 ml PBS containing 5 mg fibronectin and dried overnight at room temperature. A total of 50 ml diluted matrigel (1.25 mg/ml with cold 1640 serum-free medium) was added to the upper surface of the filter membrane and incubated at 37°C for 4-5 h to form a colloid. Then, 100 ml cell suspension (1 × 106 cells/ml) was added to the upper chamber, washed with warm serum-free 1640, and 600 ml 1640 medium (with 1% FBS) was added to the lower chamber. Following incubation at 37°C and 5% CO2 for 20 h, the absorbance values of the upper and lower suspensions were measured at 450 nm.
Cell adhesion assay
A 96-well culture plate with 50 ml fibronectin (20 mg/l) in each well was dried overnight at room temperature, washed twice with PBS and mixed with 1% bovine serum albumin (Yancheng China Biotechnology Co., Ltd., Yancheng, China), followed by a 1-h incubation at 37°C. Cell density was adjusted to 0.5 × 106 cells/ml with 10% FBS, and the cells were added to the 96-well plate with 200 ml cell suspension per well, and incubated at 37°C and 5% CO2 for 60 min. The non-adhered cells were washed away with PBS and the absorbance values were measured at 450 nm.
Statistical analysis
All statistical analyses were performed using SPSS, version 17.0 (SPSS, Inc., Chicago, IL, USA). The data of normal distribution were expressed as the mean ± standard deviation. Measurement data with homogeneity of variance were compared by one-way analysis of variance, LSD method was used for pairwise comparison, and measurement data with heterogeneity of variance were compared using Wilcoxon test. Categorical data were compared using the χ2 test. Analysis of cumulative survival rate was performed using the life table method and Kaplan-Meier survival curves, whereas comparison of survival rates was conducted using the log-rank method. The related factors affecting the prognosis of malignant ovarian tumors were determined by Cox proportional hazard regression model analysis. All experiments with cell cultures were performed independently at least five times in triplicate. The P-values were based on two-sided comparisons, and P < 0.05 was considered significant.
Results
CCL18 mRNA expression in ovarian tissues
Copies of the constructed plasmid was calculated by the formula: copies/ul = con.(ng/ul) × 6.02 × 1023 × 10-9/660 × bases, DNA concentration (ng/ul) = OD260 × 50 × dilution multiple. The standard curve was made from standard plasmids with different concentration gradients, and the original copy numbers of the samples were calculated by the software. The original copy number ratio of CCL18 and GAPDH was determined as the relative original template, which was used for comparison to quantify the CCL18 gene following PCR amplification. Expression rates of CCL18 in malignant ovarian tumor, benign ovarian tumor, and normal ovarian tissues are presented in Table 2. The rate of expression of CCL18 mRNA in malignant ovarian cancer tissue was significantly higher compared with that in benign ovarian tumor (P = 0.002) and normal ovarian tissues (P = 0.003). However, the level of CCL18 expression was not significantly different between benign ovarian tumor and normal ovarian tissues (P = 0.064).
Table 2.
CCL18 mRNA expression in ovarian tissues
| Tissue | Cases, n | Relative CCL18 mRNA | ||
|---|---|---|---|---|
|
| ||||
| Rate of expression, % | Quantity of expression, mean ± standard deviation | P-value | ||
| Normal ovary | 20 | 30.0 | 0.130 ± 0.209 | 0.064a |
| Benign ovarian tumor | 40 | 28.6 | 0.046 ± 0.136 | 0.002b |
| Malignant tumor | 62 | 74.2 | 2.997 ± 6.043 | 0.003c |
CCL18 mRNA expression in ovarian tissues was compared by Kruskal-Wallis one-way analysis of variance.
Benign ovarian tumor vs. normal ovarian tissue;
malignant vs. benign ovarian tumor;
malignant tumor vs. normal ovarian tissue.
CCL18, chemokine ligand 18.
Association between CCL18 mRNA expression and clinical pathology of EOC
The independent samples t-test was used to determine the significance of expression. The level of CCL18 mRNA expression did not differ significantly with regard to histologic type (serous vs. mucinous), surgical pathological staging (phase I/II vs. phase III/IV), pathological grade (G1/2 vs. G3), residual tumor size (≤ 2 vs. > 2 cm), lymph node metastasis (present vs. absent), and ascites volume (≤ 500 vs. > 500 ml; P > 0.05; Table 3).
Table 3.
Relationship between CCL18 mRNA expression and clinical pathology
| Category | n | Relative CCL18 mRNA, mean ± standard deviation | P-value |
|---|---|---|---|
| Epithelial ovarian cancer | 62 | ||
| Serous | 42 | 3.947 ± 6.540 | 0.354 |
| Mucinous carcinoma | 20 | 2.640 ± 5.183 | |
| Surgical pathology staging | |||
| Phase I/II | 23 | 1.202 ± 2.722 | 0.105 |
| Phase III/IV | 39 | 2.715 ± 6.305 | |
| Pathologic grade | |||
| G1/G2 | 34 | 2.458 ± 3.884 | 0.085 |
| G3 | 28 | 3.966 ± 5.686 | |
| Lymph node metastasis | |||
| No | 23 | 1.974 ± 3.034 | 0.117 |
| Yes | 39 | 4.523 ± 5.650 | |
| Ascites, ml | |||
| ≤ 500 | 14 | 1.366 ± 2.393 | 0.108 |
| > 500 | 46 | 4.297 ± 6.836 | |
| Residual tumor, cm | |||
| ≤ 2 | 53 | 1.168 ± 2.679 | 0.087 |
| > 2 | 9 | 5.587 ± 7.739 |
CCL18 mRNA expression was compared by Wilcoxon test. CCL18, chemokine ligand 18.
Association between CCL18 mRNA expression and EOC prognosis
Kaplan-Meier survival curves and the log-rank method were used to determine the association between CCL18 mRNA expression and EOC prognosis (Figure 1; Table 4). The median survival time for CCL18-positive and CCL18-negative patients was 17.3 ± 3.5 and 29.5 ± 3.9 months, respectively. Although the median survival time of CCL18-positive patients was longer, there was no significant difference between the groups (P = 0.123; Figure 1).
Figure 1.

Survival curves of CCL18-positive and CCL18-negative patients. CCL18, chemokine ligand 18. Censored data stands for the 11 patients lost to follow up.
Table 4.
Life table analysis of survival in epithelial ovarian cancer patients
| Category | Probability | Relative risk | 95% confidence interval |
|---|---|---|---|
| CCL18 | 0.138 | 0.185 | 0.020-1.720 |
| Age | 0.615 | 1.021 | 0.941-1.107 |
| Histologic type | 0.113 | 0.346 | 0.093-1.286 |
| Surgical pathology staging | 0.871 | 0.001 | 0.001-0.003 |
| Pathologic grading | 0.425 | 2.426 | 0.275-21.413 |
| Ascites | 0.276 | 3.379 | 0.377-30.279 |
| Lymph node | 0.845 | 0.839 | 0.145-4.857 |
| Residual tumor size | 0.013 | 0.076 | 0.010-0.587 |
The relationship between CCL18 mRNA expression and EOC prognosis was analyzed using multivariate analysis. Expression of CCL18 mRNA together with histological type, pathologic grade, pathologic staging, lymph node metastasis, ascites volume and residual tumor size was investigated by the Cox multivariate model. According to the results, the residual tumor size regression coefficient and relative risk were 2.579 and 0.076, respectively (P = 0.013), suggesting that increased residual tumor size was independently associated with an increased risk of cancer-related mortality. Residual tumor size was the only independent factor indicated to influence the prognosis of ovarian cancer patients (Table 4).
Construction of recombinant plasmid pSilencer4.1-CCL18-siRNA127
Double-enzyme digestion with BamHI and HindIII followed by 0.8% agarose gel electrophoresis resulted in the identification of bands corresponding to PSilencer4.1-CCL18-siRNA127. However, the small fragment (55 bp) was not visible with 0.8% agarose gel electrophoresis. The presence of the expected bands corresponding to the pSilencerTM4.1-CMV Neo vector (4,944 bp, minus 55 bases between the endonuclease sites of BamHI and HindIII) suggested the successful construction of pSilencer4.1-CCL18-siRNA127 recombinant plasmid (Figure 2).
Figure 2.
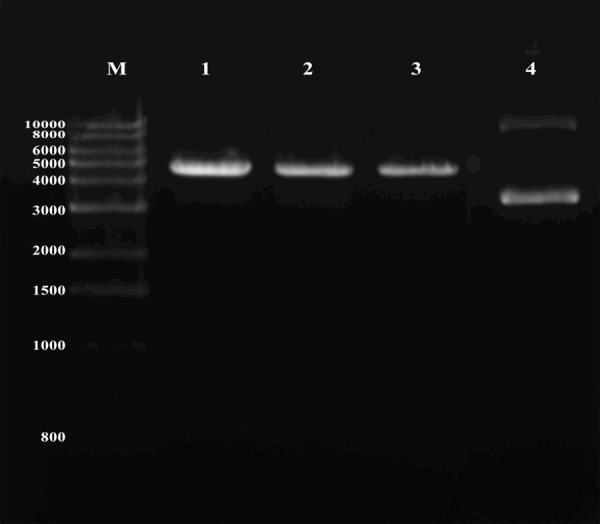
Gel electrophoresis of plasmid pSilencer4.1-CCL18-siRNA127 with BamHI and HindIII double digestion. M, Marker (bp); lanes 1-3, pSilencer4.1-CCL18-siRNA127; lane 4, pSilencer4.1. CCL18, chemokine ligand 18; siRNA127, small interfering RNA 127.
Sequencing of pSilencer4.1-CCL18-siRNA127 and pSilencer4.1-negative was performed to obtain the sequence map and compared with the original CCL18 oligonucleotide fragment using DNAMAN. The results suggested that pSilencer4.1-CCL18-siRNA127 recombinant plasmid was successfully constructed (Figure 3).
Figure 3.
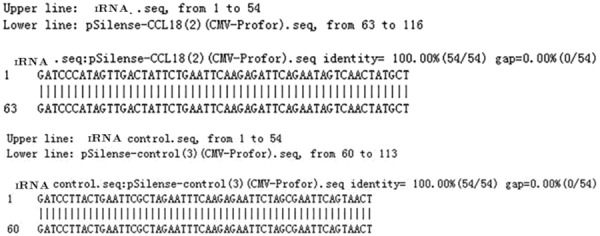
Sequence analysis of pSilencer4.1-CMVneo-CCL18 and pSilencer4.1-CMVneo-control recombinant plasmid by DNAMAN. CCL18, chemokine ligand 18.
CCL18 mRNA expression in SKOV3 cells transfected with recombinant plasmids CCL18-pEGFP-N1 and pSilencer4.1-CCL18-siRNA127
Fluorescence qPCR was used to determine the expression of CCL18 mRNA in all cell lines (Tables 5 and 6). CCL18 mRNA expression in SKOV3-CCL18-pEGFP-N1 cells was significantly increased compared with that in the SKOV3-pEGFP-N1 and SKOV3 groups (P = 0.024 and 0.015, respectively). Furthermore, CCL18 mRNA expression in SKOV3-CCL18-siRNA127 cells was significantly lower than in the SKOV3-pSilencer4.1, SKOV3-control and SKOV3 groups (P = 0.001, 0.013 and 0.001, respectively); while there was no difference between the SKOV3-pEGFP-N1 and SKOV3 groups (P = 0.580).
Table 5.
CCL18 mRNA expression in SKOV3 cells transfected with recombinant plasmid
| Group | Relative CCL18 mRNA, mean ± standard deviation | P-value |
|---|---|---|
| SKOV3 | 0.074 ± 0.1025 | 0.580a |
| SKOV3-pEGFP-N1 | 0.081 ± 0.0895 | 0.024b |
| SKOV3-CCL18-pEGFP-N1 | 0.196 ± 0.2275 | 0.015c |
CCL18 mRNA expression in SKOV3 cells was compared by one-way analysis of variance, A significant difference was found between groups (P = 0.017). LSD method was used for pairwise comparison and it demonstrated that CCL18 mRNA expression in SKOV3-CCL18-pEGFP-N1 was significant higher than in SKOV3-pEGFP-N1 and SKOV3 group (P = 0.024, P = 0.015 respectively).
SKOV3-pEGFP-N1 vs. SKOV3;
SKOV3-CCL18-pEGFP-N1 vs. SKOV3-pEGFP-N1;
SKOV3-CCL18-pEGFP-N1 vs. SKOV3.
CCL18, chemokine ligand 18.
Table 6.
Downregulation of CCL18 mRNA expression in SKOV3 cells
| Group | Relative CCL18 mRNA, mean ± standard deviation | P-value |
|---|---|---|
| SKOV3 | 0.075 ± 0.014 | 0.001a |
| SKOV3-pSilencer | 0.069 ± 0.022 | 0.001b |
| SKOV3-CCL18-siRNA127 | 0.004 ± 0.002 | |
| SKOV3-control | 0.048 ± 0.013 | 0.013c |
CCL18 mRNA expression in SKOV3 cells was compared by one-way analysis of variance. A significant difference was found between groups (P = 0.002). LSD method was used for pairwise comparison and it demonstrated that CCL18 mRNA expression was significantly lower in SKOV3-CCL18-siRNA127 than that in other groups.
SKOV3-CCL18-siRNA127 vs. SKOV3;
SKOV3-CCL18-siRNA127 vs. SKOV3-pSilencer;
SKOV3-CCL18-siRNA127 vs. SKOV3-control.
CCL18, chemokine ligand 18.
Effects of CCL18 overexpression and silencing on the growth of SKOV3 cells
MTT assays were performed to generate growth curves prior to and following transfection of SKOV3 cells (Figure 4). The growth rate of cells did not differ significantly between the SKOV3-CCL18-pEGFP-N1 and non-transfected groups (P = 0.224). In the SKOV3-CCL18-pSilencer group, the rate of cell growth appeared lower than in the negative control and empty plasmid groups; however, the differences were not statistically significant (P = 0.135 and P = 0.089, respectively). The results demonstrated that CCL18 overexpression and silencing had no marked effect on the proliferation of SKOV3 cells.
Figure 4.
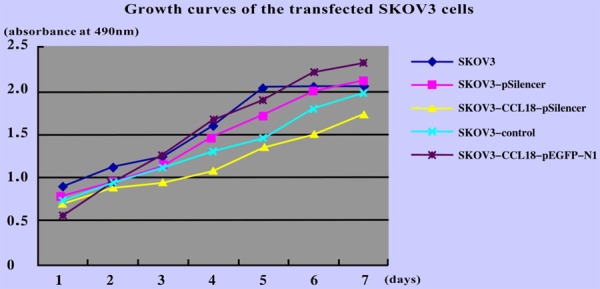
Growth curves of SKOV3 cells following transfection with CCL18 overexpression and interference vectors. CCL18, chemokine ligand 18.
Effects of CCL18 gene overexpression and silencing on SKOV3 cell cycling
The number of SKOV3-CCL18-pEGFP-N1 cells in the proliferative phases of the cell cycle (S+G2+M) was significantly higher than that of SKOV3 cells (P = 0.022); however, no significant difference was observed between SKOV3-pEGFP-N1 cells and SKOV3 cells (P = 0.378). There was a significant difference in the number of SKOV3 cells in proliferative phases (S+G2+M) in the SKOV3 group compared with that in the SKOV3-pSilencer4.1-CCL18-siRNA127 group (P = 0.015). By contrast, there was no significant difference between the SKOV3-pSilencer4.1 and SKOV3 groups (P = 0.265; Table 7). Therefore, the results indicated significant reduction in the expression of CCL18 mRNA and number of SKOV3 cells in proliferative phases (S+G2+M), and thus reduction in cell proliferation, following pSilencer4.1-CCL18-siRNA127 transfection.
Table 7.
Cell cycle phase distribution following transfection of SKOV3 cells
| Group | Cell ratios, % | P-value | |||
|---|---|---|---|---|---|
|
| |||||
| G0-G1 | S | G2-M | S+G2+M | ||
| SKOV3 | 72.9 ± 23.3 | 24.2 ± 3.7 | 2.83 ± 0.8 | 27.1 ± 4.8 | - |
| SKOV3-pEGFP-N1 | 73.0 ± 20.4 | 22.1 ± 4.9 | 4.86 ± 0.7 | 27.0 ± 4.6 | 0.378a |
| SKOV3-CCL18-pEGFP-N1 | 67.2 ± 18.7 | 27.8 ± 4.3 | 5.01 ± 0.9 | 32.8 ± 4.4 | 0.022a |
| SKOV3-pSilencer4.1-CCL18-siRNA127 | 80.3 ± 17.9 | 18.0 ± 5.9 | 1.69 ± 0.6 | 19.7 ± 4.4 | 0.015a |
| KOV3-pSilencer4.1 | 71.0 ± 20.7 | 25.1 ± 6.0 | 3.94 ± 0.7 | 29.0 ± 6.1 | 0.265a |
Values are provided as the mean percentage ± standard deviation, and compared by one-way analysis of variance.
SKOV3 group vs. other groups.
CCL18, chemokine ligand 18.
Effect of CCL18 gene overexpression and silencing on invasive, adhesive, and migrative capacities of SKOV3 cells
The invasive, migrative and adhesive capacities of SKOV3-CCL18-pEGFP-N1 cells were significantly enhanced compared with those of SKOV3 and SKOV3-pEGFP-N1 cells (P < 0.05); however, these properties did not differ significantly between SKOV3-pEGFP-N1 and SKOV3 cells (P > 0.05). The invasive, migrative, and adhesive capacities of cells were effectively inhibited in the SKOV3-pSilencer4-CCL18-siRNA127 group as compared with in the SKOV3-pSilencer4.1 and SKOV3 groups (P < 0.05; Table 8). These findings indicated that the invasive, migrative, and adhesive capacities of SKOV3 cells were effectively inhibited with decreased expression of CCL18 mRNA following pSilencer4.1-CCL18-siRNA127 transfection. This indicated an involvement of CCL18 in malignant ovarian tumor cell invasion and metastasis.
Table 8.
Effects of CCL18 gene overexpression and silencing on invasive, adhesive, and migrative capacities
| Group | Absorbance at 450 nm | ||
|---|---|---|---|
|
| |||
| Invasion | Migration | Adhesion | |
| SKOV3-CCL18-pEGFP-N1 | 0.486 ± 0.187 | 1.157 ± 0.254 | 0.387 ± 0.105 |
| SKOV3-pEGFP-N1 | 0.187 ± 0.054 | 0.377 ± 0.237 | 0.134 ± 0.117 |
| SKOV3 | 0.225 ± 0.134 | 0.356 ± 0.104 | 0.157 ± 0.033 |
| SKOV3-pSilencer4.1-CCL18-siRNA127 | 0.099 ± 0.034 | 0.168 ± 0.087 | 0.067 ± 0.043 |
| SKOV3-pSilencer4.1 | 0.195 ± 0.046 | 0.365 ± 0.097 | 0.113 ± 0.052 |
Values are provided as the mean percentage ± standard deviation, and compared by one-way analysis of variance. The invasion, migration, and adhesion capacities of SKOV3-CCL18-pEGFP-N1 group were significantly enhanced compared to SKOV3-pEGFP-N1 group (P = 0.048, 0.01, 0.034 respectively) and SKOV3 group (P = 0.036, 0.009 and 0.024 respectively). The invasion, migration, and adhesion capacities of SKOV3-pSilencer4.1-CCL18-siRNA127 were inhibited compared to SKOV3-pSilencer4.1 group (P = 0.015, 0.043, 0.035 respectively) and SKOV3 group (P = 0.010, 0.021, 0.029 respectively). However, these capacities had no obvious difference in the SKOV3-pEGFP-N1 group and SKOV3 group (P = 0.745, 0.924 and 0.802 respectively).
Discussion
CCL18 is a small (7.8 kDa) inflammatory protein. Previous studies have indicated that CCL18 overexpression may be associated with tumor growth and development, particularly when in the microenvironment of the tumor, and it is considered a useful marker for diagnosis and prognosis [16,17]. In our previous studies, we detected the protein expression profile of malignant ovarian tumor, benign ovarian lesion, and healthy ovarian tissues by SELDI-TOF-MS, IMAC-Cu and WCX-2 protein chip array profiling. For construction and analysis of a malignant ovarian tumor diagnostic model, 31 proteins were selected as potential serum markers. Among them, CCL18 was correlated with clinical characteristics [9,10]. Further study indicated that CCL18 was secreted by tumor cells and that CCL18 mRNA expression in these malignant tumor tissues is significantly increased [18].
Our present results indicated that major clinical features of EOC (including pathological staging and pathological grade) were not significantly associated with the expression of CCL18 (P > 0.05). Our group previously reported on the expression of CCL18 in 47 EOC patients; however, there was no significant difference in the expression of CCL18 among malignant tumor tissues based on findings including pathologic staging and pathologic grade [9], which is in agreement with current findings. Additionally, previous study identified that CCL18 expression in the peripheral blood of patients with gastric cancer was significantly higher than in a healthy control group and higher in clinical stage I-II patients than in stage III-IV patients [19]. In patients with gastric cancer (with no extranodal metastasis or lymph node metastasis), the positive rate of CCL18 expression was significantly higher than in patients with advanced gastric cancer and no lymph node metastasis [20]. The current study analyzed the clinicopathologic characteristics and prognosis of patients with malignant ovarian tumors by the Cox proportional hazard regression model. Results from the analysis demonstrated that residual tumor size was an important independent factor that influences the prognosis of ovarian cancer patients (P < 0.05); the larger the residual tumor, the poorer the prognosis (with a low 5-year survival rate; data not shown). Pathologic grade, clinical stage, lymph node metastasis and positive expression of CCL18 did not appear to affect the prognosis of ovarian cancer patients. Results from an analysis of 89 gastric cancer patients in a study by Leung et al. [21] demonstrated that high expression of CCL18 was significantly correlated with overall survival (OS) and disease-free survival (DFS). Furthermore, results from multivariate analysis of OS and DFS showed that CCL18 and tumor stage were independent prognostic factors, which is in disagreement with the present results. We speculate that although CCL18 may be used as a marker for early diagnosis of both gastric and ovarian cancer [8,21], the protein expression differs in different cell sources. From the present study, it was concluded that CCL18 is secreted by tumor cells, and the rate of mRNA expression in malignant tumor tissues was significantly increased.
CCL18 is a member of the superfamily of chemokines, which are mainly secreted by immature dendritic cells (DC) and macrophages [22]. Additionally, interleukin (IL)-10 was identified to promote monocyte-derived DCs to increase expression of CCL18, and promote immature DC and T cells to participate in primary immune responses [23,24]. IL-10 may serve as an inhibitor of major histocompatibility complex (MHC) class II molecules, as an increase of IL-10 could inhibit mononuclear macrophage production [25]. Studies have also found high-level expression of CCL18 in patients with early-stage tumors, highlighting the involvement of white blood cells [26]. In addition, high expression of chemokines and leukocyte-mediated chemokine expression result in tissue damage. Consequently, the protective effect of white blood cells can be reduced, resulting in the growth and development of cancer cells [27]. The current study indicated that CCL18 overexpression did not affect the growth curve of cancer cells; however, the ratio of cells in proliferative phases (S+G2+M) was significantly increased.
In investigating the effects of CCL18 overexpression and silencing on the biologic behavior of ovarian cancer cells in vitro, there was no statistical significance in the growth rates of CCL18-pEGFP-N1, pEGFP-N1 and normal SKOV3 cells. Nonetheless, cell ratio in the S+G2+M phases for the CCL18-pEGFP-N1, PEGFP-N1, and non-transfected groups was 32.8, 27.0 and 27.1%, respectively. The results demonstrated that CCL18 overexpression may somewhat promote DNA replication in ovarian cancer cells, evident with the increased cell populations in the proliferation phases of the cell cycle associated with aneuploidy. The growth curve of SKOV3 cells exhibited a delay following pSilencer4.1-CCL18 transfection; however, the difference was not significant among the SKOV3, SKOV3-control and SKOV3-pSilencer4.1 groups. The proportion of SKOV3-pSilencer4.1-CCL18-siRNA127 cells in S+G2+M phases was 19.7%, which was comparatively less than that in the non-transfected, negative control and empty vector-transfected groups. Chromosomal abnormalities are a characteristic feature of malignant tumor cells, including euploidy and aneuploidy [28]. Since CCL18 had little apparent effect on tumor cell growth rate, it may be speculated that SKOV3 DNA replication increases following CCL18 overexpression, resulting in ploidy changes and possibly promoting carcinogenesis. Indeed, CCL18 has been proposed to affect the replicative ability of tumor cells by altering the number of tumor cell chromosomes and promoting cell transformation [29,30].
A Transwell assay was used to demonstrate the association between CCL18 expression and tumor invasion and metastasis. Comparison of the invasive and migrative capacities of SKOV3 cells pre- and post-transfection demonstrated that CCL18 overexpression accelerated tumor cell metastasis. Reduced cell adhesion allows tumor cells to leave the primary site. This reduction in adhesion is correlated with tumor invasion and metastasis [31]. In the current study, the adhesive capacity of the CCL18-pEGFP-N1 group was compared with that of the control group. There was a significant increase in adhesive capacity following CCL18 overexpression. This may allow cancer cells to adhere to the extracellular matrix; and thereby explains the reduced invasive, migrative, and adhesive abilities of SKOV3-pSilencer4.1-CCL18-siRNA127 cells. Therefore, CCL18 was verified seemingly as the first inflammatory chemokine capable of increasing adhesion and promoting the processes of invasion and metastasis. Previous studies have shown that once tumor cells leave the primary tumor site, CCL18 may increase cell invasion capacity, promote adhesion between the cells in the extracellular matrix and the vascular wall, and enhance the migrative ability of the tumor cells [10,17,32]. Multiple regression analyses have further demonstrated that the expression of CCL18 was positively associated with tumor invasion [21]. These findings are in agreement with the present results. Therefore, the expression of CCL18 may enhance the ability of cancer cells to infiltrate into the surrounding tissue. CCL18 is secreted by activated macrophages, mainly trigger naive T lymphocytes and immature dendritic cells, and stimulate the CD4 T lymphocytes into regulatory T cells (Treg). Treg mainly secrete IL-10, which is an MHC II molecular inhibitor; that is, an increase of IL-10 can inhibit the generation of mononuclear macrophage [33]. Altogether, findings indicate that the production of malignant tumor cells may be caused by gene mutation, followed by abnormal increase of CCL18, which may activate the immune chain reaction above. Decrease in MHC-II may reduce immunity and lymphocyte assault on tumor cells, thereby allowing the cells to escape the immune response and metastasize.
In conclusion, the present findings suggest that CCL18 may be developed as a clinical marker for early diagnosis of malignant ovarian tumors, particularly EOC, and has the potential to be a target molecule for ovarian cancer treatment. Further research is required to establish the association between CCL18 and malignant ovarian tumors in animal models and clinical trials.
Acknowledgements
The present work was supported by the Natural Science Foundation of Guangxi (grant no. 2014jjAA40637) and the Key Health Science Foundation of Guangxi (grant no. 14124004-1-24).
Disclosure of conflict of interest
None.
References
- 1.Shaaban A, Rezvani M. Ovarian cancer: detection and radiologic staging. Clin Obstet Gynecol. 2009;52:73–93. doi: 10.1097/GRF.0b013e3181961625. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zhang SW, Zou XN, Zhao P. Cancer incidence and mortality in China, 2006. Chin J Cancer Res. 2011;23:3–9. doi: 10.1007/s11670-011-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JY, Kim HS, Suh DH, Kim MK, Chung HH, Song YS. Ovarian cancer biomarker discovery based on genomic approaches. J Cancer Prev. 2013;18:298–312. doi: 10.15430/JCP.2013.18.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia AA, Blessing JA, Lenz HJ, Darcy KM, Mannel RS, Miller DS, Husseinzadeh N. Phase II clinical trial of capecitabine in ovarian carcinoma recurrent 6-12 months after completion of primary chemotherapy with exploratory TS, DPD, and TP correlates: a Gynecologic Oncology Group study. Gynecol Oncol. 2005;96:810–817. doi: 10.1016/j.ygyno.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Zanetta G, Rota S, Lissoni A, Meni A, Brancatelli G, Buda A. Ultrasound, physical examination, and CA 125 measurement for the detection of recurrence after conservative surgery for early borderline ovarian tumors. Gynecol Oncol. 2001;81:63–66. doi: 10.1006/gyno.2000.6099. [DOI] [PubMed] [Google Scholar]
- 6.Sakane R, Tsubamoto H, Sakata K, Inoue K, Ogino M, Shibahara H, Hao H, Hirota S. Expression of chemokine ligand 18 in stage IA low-grade endometrial cancer. Anticancer Res. 2014;34:5331–5336. [PubMed] [Google Scholar]
- 7.Adema GJ, Hartgers F, Verstraten R, Marland G, Menon S, Foster J, Xu Y, Nooyen P, McClanahan T, Bacon KB, Figdor CG. A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature. 1997;387:713–717. doi: 10.1038/42716. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Liang YX, Zhu JG, Fu X, Chen YF, Mo RJ, Zhou L, Fu H, Bi XC, He HC, Yang SB, Wu YD, Jiang FN, Zhong WD. CC chemokine ligand 18 correlates with malignant progression of prostate cancer. Biomed Res Int. 2014;2014:230183. doi: 10.1155/2014/230183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Li L, Li DR, Zhang W, Wei X, Zhang JQ, Tang Y. Identification of serum biomarkers for ovarian cancer using protein chips and time of flight mass spectrometry technology. Zhonghua Fu Chan Ke Za Zhi. 2006;41:544–548. [PubMed] [Google Scholar]
- 10.Wang Q, Li D, Zhang W, Tang B, Li QQ, Li L. Evaluation of proteomics-identified CCL18 and CXCL1 as circulating tumor markers for differential diagnosis between ovarian carcinomas and benign pelvic masses. Int J Biol Markers. 2011;26:262–273. doi: 10.5301/JBM.2011.8616. [DOI] [PubMed] [Google Scholar]
- 11.Meinhold-Heerlein I, Fotopoulou C, Harter P, Kurzeder C, Mustea A, Wimberger P, Hauptmann S, Sehouli J. The new WHO classification of ovarian, fallopian tube, and primary peritoneal cancer and its clinical implications. Arch Gynecol Obstet. 2016;293:695–700. doi: 10.1007/s00404-016-4035-8. [DOI] [PubMed] [Google Scholar]
- 12.Prat J FIGO Committee on Gynecologic Oncology. Abridged republication of FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum. Cancer. 2015;121:3452–3454. doi: 10.1002/cncr.29524. [DOI] [PubMed] [Google Scholar]
- 13.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofectin: a highly efficient lipid-mediated DNA transfection procedure. Proc Natl Acad Sci U S A. 1987;84:7413–7. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schutyser E, Struyf S, Proost P, Opdenakker G, Laureys G, Verhasselt B, Peperstraete L, Van de Putte I, Saccani A, Allavena P, Mantovani A, Van Damme J. Identification of biologically active chemokine isoforms from ascitic fluid and elevated levels of CCL18/pulmonary and activation-regulated chemokine in ovarian carcinoma. J Biol Chem. 2002;277:24584–24593. doi: 10.1074/jbc.M112275200. [DOI] [PubMed] [Google Scholar]
- 17.Zohny SF, Fayed ST. Clinical utility of circulating matrix metalloproteinase-7 (MMP-7), CC chemokine ligand 18 (CCL18) and CC chemokine ligand 11 (CCL11) as markers for diagnosis of epithelial ovarian cancer. Med Oncol. 2010;27:1246–1253. doi: 10.1007/s12032-009-9366-x. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Tang Y, Yu H, Yin Q, Li M, Shi L, Zhang W, Li D, Li L. CCL18 from tumor-cells promotes epithelial ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog. 2016;55:1688–1699. doi: 10.1002/mc.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Liu X, Wang Y. Predictive value of preoperative serum CCL2, CCL18, and VEGF for the patients with gastric cancer. BMC Clin Pathol. 2013;13:15. doi: 10.1186/1472-6890-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JH, Wang YN, Ou ZL, Shi YQ. Preoperative detection of CCL18 in gastric cancer patients and its significance. China Oncology. 2006;16:835–837. [Google Scholar]
- 21.Leung SY, Yuen ST, Chu KM, Mathy JA, Li R, Chan AS, Law S, Wong J, Chen X, So S. Expression profiling identifies chemokine (C-C motif) ligand 18 as an independent prognostic indicator in gastric cancer. Gastroenterology. 2004;127:457–469. doi: 10.1053/j.gastro.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 22.Tecimer C, Doering DL, Goldsmith LJ, Meyer JS, Abdulhay G, Wittliff JL. Clinical relevance of urokinase-type plasminogen activator, its recep tor and inhibitor type 1 in ovarian cancer. Int J Gynecol Cancer. 2000;10:372–381. doi: 10.1046/j.1525-1438.2000.010005372.x. [DOI] [PubMed] [Google Scholar]
- 23.Vulcano M, Struyf S, Scapini P, Cassatella M, Bernasconi S, Bonecchi R, Calleri A, Penna G, Adorini L, Luini W, Mantovani A, Van Damme J, Sozzani S. Unique regulation of CCL18 production by maturing dendritic cells. J Immunol. 2003;170:3843–3849. doi: 10.4049/jimmunol.170.7.3843. [DOI] [PubMed] [Google Scholar]
- 24.Yuan R, Chen Y, He X, Wu X, Ke J, Zou Y, Cai Z, Zeng Y, Wang L, Wang J, Fan X, Wu X, Lan P. CCL18 as an independent favorable prognostic biomarker in patients with colorectal cancer. J Surg Res. 2013;183:163–169. doi: 10.1016/j.jss.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Schutyser E, Richmond A, Van Damme J. Involvement of CC Chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Günther C, Zimmermann N, Berndt N, Grosser M, Stein A, Koch A, Meurer M. Up-regulation of the chemokine CCL18 by macrophages is a potential immunomodulatory pathway in cutaneous T-cell lymphoma. Am J Pathol. 2011;179:1434–1442. doi: 10.1016/j.ajpath.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L, Yao H, Su F, Anderson KS, Liu Q, Ewen ME, Yao X, Song E. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JY, Kim HS, Suh DH, Kim MK, Chung HH, Song YS. Ovarian cancer biomarker discovery based on genomic approaches. J Cancer Prev. 2013;18:298–312. doi: 10.15430/JCP.2013.18.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urquidi V, Kim J, Chang M, Dai Y, Rosser CJ, Goodison S. CCL18 in a multiplex urine-based assay for the detection of bladder cancer. PLoS One. 2012;7:e37797–e37803. doi: 10.1371/journal.pone.0037797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajkumar T, Sabitha K, Vijayalakshmi N, Shirley S, Bose MV, Gopal G, Selvaluxmy G. Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer. 2011;22:80–92. doi: 10.1186/1471-2407-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Zhang T, Ma Z, Wang Y, Cheng Z, Xu H, Li W, Wang X. The interaction of coagulation factor XII and monocyte/macrophages mediating peritoneal metastasis of epithelial ovarian cancer. Gynecol Oncol. 2010;117:460–466. doi: 10.1016/j.ygyno.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen JS, Fuentes-Duculan J, Suárez-Fariñas M, Pierson KC, Pitts-Kiefer A, Fan L, Belkin DA, Wang CQ, Bhuvanendran S, Johnson-Huang LM, Bluth MJ, Krueger JG, Lowes MA, Carucci JA. Tumor-associated macrophages in the cutaneous SCC microenvironment are heterogeneously activated. J Invest Dermatol. 2011;131:1322–1330. doi: 10.103/jid.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]


