Abstract
Human cytomegalovirus (HCMV), a ubiquitous pathogen, can cause severe illness in immunocompromised individuals. Typically, glioma is one of the most common malignant primary brain tumors and originates in the central nervous system. The IE86 gene of HCMV exerts a major role in regulating virus replication. By using coimmunoprecipitation combined with mass spectrometry, the components of the IE86 complex were identified, and the heterogeneous ribonucleoprotein A2/B1 (hnRNP A2/B1) was recognized as one of the IE86 complex components. hnRNP A2/B1 is highly expressed in U251 cells, and the data suggest that IE86 can promote hnRNP A2/B1 expression. Furthermore, the knockdown of hnRNP A2/B1 significantly attenuates IE86-mediated apoptosis and cell proliferation. Importantly, IE86 can also inhibit the alternative splicing of Bcl-x by decreasing the Bcl-xS/Bcl-xL ratio, which is closely related to apoptosis. Meanwhile, the knockdown of hnRNP A2/B1 can mitigate the inhibitory effect of IE86 on the alternative splicing of Bcl-x. In conclusion, the inhibition of apoptosis and enhancement of cell proliferation by IE86 may be related to the hnRNP A2/B1-mediated alternative splicing of Bcl-x.
Keywords: IE86, Bcl-x, hnRNP A2/B1
Introduction
HCMV, namely, human cytomegalovirus, is a component of the subfamily of herpesvirus β, which is the largest virus among the human herpesviruses. The HCMV genome is composed of double-stranded linear DNA that is approximately 235 kb long. There are 165 proteins encoded in the whole genome [1]. HCMV infection leads to the sequential expression of the viral genes, which can be classified into late (L), early (E) and immediately early (IE) genes. There is an immediate synthesis of HCMV IE2 after injection. The corresponding gene product serves as a classical transcriptional regulatory protein [2]. The IE2 gene encodes IE86 and plays a critical function during the process of HCMV infection of cells, without which HCMV infection will not generate infectious progeny virus particles [3]. The expression of genes can be affected by IE86 through the recruitment of some transcriptional activators.
There are two homologous proteins composing heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1). These two kinds of proteins are classified into the hnRNP family [4]. The hnRNPs serve as members of a large family of RNA-binding proteins that are associated with preneonatal mRNA transcripts within eukaryotic cells. They have various functions during the processes of stability, translation, localization, mRNA output, pre-mRNA splicing and RNA processing [5]. There is an overexpression of hnRNP A2/B1 in different tumor cells, such as lung carcinoma [10], and in gastric [9], breast [8], liver [7], and pancreatic cells [6]. Recently, it has been found that hnRNP A2/B1 acts as a selective splicing regulator of multiple tumor oncogenes and suppressors, for example, Bcl-x [11]. HnRNP A2/B1 serves as a kind of antiapoptotic protein, which is classified into the well-known Bcl-2 category. Bcl-x serves as an antiapoptotic component of the bcl-2 gene family and plays an important role in the regulation of mammalian cell apoptosis. There are two protein isotypes, bcl-xS and bcl-xL, with opposite functions that are produced by the alternative splicing of bcl-x. There is growing evidence proving that hnRNP A2/B1 inhibits cell growth [12] while apoptosis is enhanced [13]. There is an increasing number of studies proving that HCMV exists in prostate cancer, colon cancer, glioma, and other tumors [14]. In recent years, an association has been found between IE86 and apoptosis and the proliferation of tumor cells [15].
Immunoprecipitation was applied to identify the composition of the IE86 complex and was coupled with liquid chromatography-mass spectrometry. It was determined that hnRNP A2/B1 serves as one of the components of the IE86 immune complex. Thus, investigating the potential function of the hnRNP A2/B1-mediated signaling pathway within glioma cells transfected with IE86 is very important. According to this study, the function of IE86 in the apoptosis and proliferation of U251 cells was further characterized, followed by the potential function of hnRNP A2/B1 and the corresponding systems during these processes.
Materials and methods
Cell culture and mouse model
The Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China) was the source of the human glioblastoma cell line U251. Dulbecco’s High Glucose Modified Eagle Medium (HyClone) supplemented with 10% FBS was used for maintenance of the U251 cells. All the cells were grown under adherent cultivation conditions at 37°C using 5% CO2. All of the animal experiments were carried out based on the China animal welfare law and approved by the local authorities. Cyagen Biosciences Inc. customized the IE86 transgenic mice, which were kept in pathogen-free conditions.
Plasmids and shRNA
Sino Biological Inc. (Beijing, China) was the source of the peGFP-N3-IE86 and peGFP-N3 vectors. When the cells reached 50% confluence, they were transfected with 2.5 μg of DNA using the transfection reagent Lipofectamine 3000 (Invitrogen, USA) based on the manufacturer’s protocol. The first cultivation of the transfected cells occurred in an antibiotic-free medium for 6 h. Afterwards, the transfected cells were moved to a fresh medium and cultured for 48 h, prior to further processing.
The cells were cultivated in MEM for the knockdown of hnRNP A2/B1 expression. Then a small hairpin RNA (shRNA) vector was transfected via Lipofectamine 2000 following the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). The stable sublines were transfected with the hnRNP A2/B1 shRNA vector (sh1A2/B1 sense primer: 5’-CCGGCAGAAATACCATACCATCAATCTCGAGATTGATGGTATGGTATTTCTGTTTTTG-3’, sh1A2/B1 antisense primer: 5’-AATTCAAAAACAGAAATACCATACCATCAATCTCGAGATTGATGGTATGGTATTTCTG-3’; sh2A2/B1 sense primer: 5’-CCGGAGAAGCTGTTTGTTGGCGGAACTCGAGTTCCGCCAACAAACAGCTTCTTTTTTG-3’, sh2A2/B1 antisense primer: 5’-AATTCAAAAAAGAAGCTGTTTGTTGGCGGAACTCGAGTTCCGCCAACAAACAGCTTCT-3’; sh3A2/B1 sense primer: 5’-CCGGTGACAACTATGGAGGAGGAAACTCGAGTTTCCTCCTCCATAGTTGTCATTTTTG-3’, sh3A2/B1 antisense primer: 5’-AATTCAAAAATGACAACTATGGAGGAGGAAACTCGAGTTTCCTCCTCCATAGTTGTCA-3’) were identified as U251-sh1A2/B1, U251-sh2A2/B1, and U251-sh3A2/B1, respectively.
Total protein extraction
A RIPA buffer (Solarbio) was used for extraction of the total proteins, which was supplemented with 10 μL of PMSF (Solarbio) per milliliter, at 4°C for 20 min. Centrifugation was carried out for the collection of the lysates at 13,000 × g for 15 min.
Coimmunoprecipitation
Five micrograms of murine IE86 antibody were added to the 500 μL of total cell protein. Afterwards, the mixture was incubated on a shaker at 4°C for 4 h to establish an antigen-antibody complex. A total of 25 μL of protein G immunomagnetic beads was added for 1 h at 4°C. Later, the beads were placed on the magnetic stand. The immunomagnetic beads absorbed the antigen-antibody complex. Afterwards, a 1 × SDS loading buffer was added. Then, the antigen-antibody complex was eluted from the immunomagnetic beads. Boiling was performed for 5 min to obtain the protein complex. Mass spectrometry was performed on the IE86 protein complex after coomassie blue staining and SDS-PAGE gel electrophoresis.
Liquid chromatography-mass spectrometry analysis and database search
Hangzhou Jingjie Biotechnology Co., Ltd. completed the mincing of the Coomassie-stained strips. Next, the strips were digested overnight at 37°C. The enzymatically decomposed peptide was dissolved in mobile phase A (aqueous solution with 20 g/L acetonitrile and 1 g/L formic acid) and separated via an EASY-nLC1000 ultraefficient liquid phase mechanism. The peptides were injected and separated using an NSI ion source for ionization, and then an analysis was conducted via the ThermoScientificTM QExactiveTM mass spectrometry system.
The source voltage was set to 2.0 kV, the primary mass spectrometer scan range was set between 350 and 1,800 m/z, the scan resolution was set to 70,000, and the Orbitrap scan resolution was set to 17,500. A data-dependent scanner mode was used for the data collection. Secondary mass spectrometry was also conducted in sequence. Proteome Discoverer 1.3 was used for the searching of the secondary mass spectrometry data.
RNA extraction and reverse transcription PCR
TRIzol Reagent (Invitrogen) was used for the extraction of the total RNA. Afterwards, a spectrophotometer at 260 and 280 nm (A260/280) was used to measure the concentration of these RNA samples. Reverse transcription of total RNA was performed via the application of a Prime-Script RT Reagent Kit (TaKaRa, Dalian, China). hnRNP A2/B1 expression was measured via a SYBR® Green PCR Kit (Qiagen, Germany). The primers used are listed below: IE86 forward 5’-CCGCAAGAAGAAGAGCAAACG-3’ and reverse 5’-CACCTGGTGCATACTGGGAAT-3’. Bcl-x was detected via application of the forward primer 5’-GGGTCTAGAAGTGGATGGTCAGTGTCTGGT-3’ and the reverse primer 5’-GGGGAATTCTTGGACAATGGACTGGTTGA-3’, GAPDH via forward 5’-GAAGGTGAAGGTCGGAGTC-3’ and reverse 5’-GAAGATGGTGATGGGATTTC-3’, and hnRNP A2/B1 via forward 5’-TCATGGCTCGCCTTCCTCTCAG-3’ and reverse 5’-TGTGGCTTCCGAACGCAATGG-3’. qPCR conditions consisted of 95°C for 3 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Bcl-xL (779 bp) and Bcl-xS (590 bp) were amplified using Bcl-x primers. The PCR amplification was performed as follows: after the first denaturation at 94°C for 2 min, there were 30 cycles at 94°C for 30 s, 60°C (IE86)/56°C (Bcl-x)/55°C (GAPDH) for 30 s, and 72°C for 60 s. The reaction was stopped by a final extension step at 72°C for 5 min. The PCR products were separated through gel electrophoresis in a 1.2% agarose gel containing ethidium bromide. Afterwards, PCR products were visualized with ultraviolet (UV) light. GAPDH was used for the normalization of the mRNA expression of hnRNP A2/B1 and IE86. These experiments were conducted using a iQ5 Real-Time PCR Detection system (Bio-Rad, USA), the results of which were analyzed using the 2-ΔΔCt method.
CCK8 assay
A Cell Counting Kit (7 Sea Biotech, Shanghai, China) was used to detect cell proliferation. The growth of cells occurred in 96-well plates with 1 × 104 cells per well. Then, the cells were incubated at 37°C using CO2 until the cell confluence rate increased up to 70%. After transfection with shRNA and plasmid for 48 h, the cells were incubated for 24, 48, 72, and 96 h. A total of 10 μL of CCK8 solution was used for every well. The absorbance at 450 nm was measured using a SUNRISE Microplate Reader (Tecan, Switzerland).
Flow cytometry
After transfection for 48 h, the cells were washed, centrifuged and harvested in phosphate-buffered saline three times. A total of 100 μL of 5% MEM was added to the resuspended cells in a 1.5 mL centrifuge tube. Incubation occurred for each tube at room temperature in the dark for 20 min using Guava Nexin Reagent (Millipore Guava Technologies, Billerica, MA, USA). An analysis was conducted on the samples using a Guava EasyCyte Mini Flow cytometry instrument (Millipore Guava). Every sample analysis was repeated three times.
Western blot
The proteins were separated in a 12% SDS-PAGE gel. Then, they were transferred onto polyvinylidene fluoride membranes. After blocking with 5% nonfat milk for 1 h at room temperature, the membranes were incubated with the corresponding primary antibodies at 4°C overnight. Afterwards, the membranes were washed and incubated with secondary antibodies at room temperature for 1 h. Various antibodies were used, including horseradish peroxidase-conjugated goat anti-rabbit IgG (Abgent, USA), rabbit anti-β-actin (Bioss, Beijing, China), rabbit anti-hnRNP A2/B1 (Abcam, UK) and rabbit anti-IE86, rabbit anti-caspase 3 (Abcam, UK). ECL reagents (Thermo Fisher, USA) were used for the development of these membranes, which were visualized using a BIO-PRINT ST4 gel imaging system (Vilber Lourmat, Marne-la-Vallée, France). ImageJ software was used to analyze the gray intensity of the bands.
Statistical analysis
All the data are presented as the mean ± SD for three independent studies. A one-way analysis of variance (ANOVA) and a two-tailed Student’s t-test were used for the statistical analysis. A p-value smaller than 0.05 was considered statistically significant.
Results
Composition of the IE86 immune complex identified by mass spectrometry
In this study, IE86 was first successfully expressed in U251 cells Figure 1A and 1B, and Western blot assays confirmed that the IE86 protein was successfully expressed in the cells; subsequently, total protein was extracted from the transfected cells for the subsequent coimmunoprecipitation experiments. The IE86 protein antibody was immunoprecipitated with the total protein extracted from U251 cells, while the control group was immunoprecipitated with murine IgG. Afterwards, the complex was denatured and subjected to SDS-PAGE followed by coomassie blue staining, as presented in Figure 1C. At the same time, the interaction of IE86 with hnRNP A2/B1 is also shown in Figure 1D. Thereafter, the coomassie-stained strips were excised and subjected to mass spectrometry after in-gel digestion. Proteins identified by this approach are listed in Table 1. A total of 8 splicing-associated proteins that interacted with IE86 were successfully identified through immunoprecipitation combined with mass spectrometry, including hnRNPA1, hnRNP A2/B1, hnRNPC, hnRNPM, hnRNPK and hnRNPH1. The splicing factor, hnRNPM, has been proven to promote breast cancer metastasis by activating a switch of alternative splicing during the epithelial-mesenchymal transition (EMT) [16]. Moreover, hnRNP A2/B1 is a member of the hnRNP family and plays an important role in the posttranscriptional regulation of mRNA. In addition, hnRNP A2/B1 is also an essential oncogenic gene that is overexpressed in many tumor types and is closely related to tumor formation [17,18].
Figure 1.
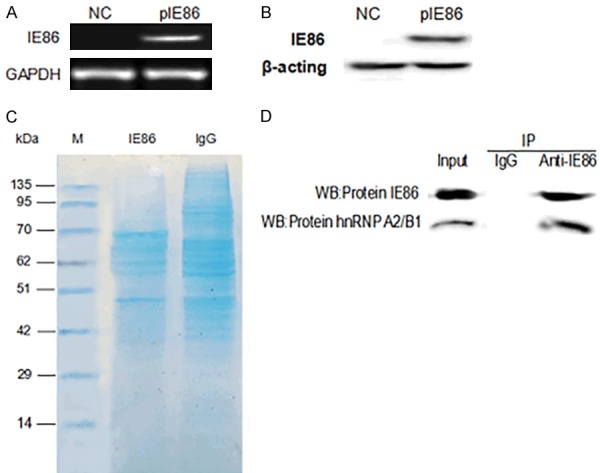
Identification of cellular factors that bind to IE86. (A) IE86 levels were measured by reverse transcription PCR (RT-PCR) and Western blotting (B) after U251 cells were transfected with peGFP-N3 or peGFP-N3-IE86 for 48 h. (C) Results of SDS-PAGE gel electrophoresis and coomassie blue staining. (D) Co-IP of IE86 and hnRNP A2/B1.
Table 1.
Protein Interacting With IE86
| Gene Name | Protein Name | Species | Protein score |
|---|---|---|---|
| HnRNP A2/B1 | Heterogeneous nuclearribonucleoproteins A2/B1 | Human | 367.75 |
| HNRNPA1 | Heterogeneous nuclearribonucleoprotein A1 | Human | 324.27 |
| HNRNPC | Heterogeneous nuclearribonucleoproteins C1/C2 | Human | 187.97 |
| HNRNPM | Heterogeneous nuclearribonucleoprotein M | Human | 148.54 |
| SYNCRIP | Heterogeneous nuclearribonucleoprotein Q | Human | 81.74 |
| HNRNPK | Heterogeneous nuclearribonucleoprotein K | Human | 80.26 |
| PTBP1 | Polypyrimidine tract-bindingprotein 1 | Human | 44.24 |
| HNRNPH1 | Heterogeneous nuclearribonucleoprotein H | Human | 40.1 |
IE86 promoted hnRNP A2/B1 expression
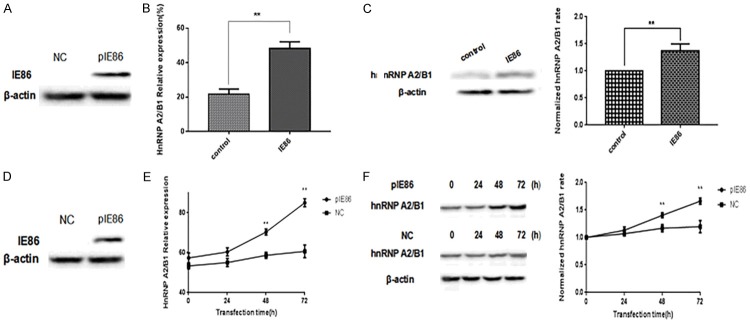
Using proteomics techniques, the splicing factor hnRNP A2/B1 was identified as one of the components of the IE86 complex, and it is the strongest combination with IE86, which has been recognized to exert a pivotal role in the growth, survival, apoptosis, and invasion of glioma cells. Thus, it is necessary to understand the effect of IE86 on hnRNP A2/B1. In this study, IE86 was first successfully expressed in IE86 transgenic mice (Figure 2A), and the effect of IE86 on hnRNP A2/B1 in IE86 transgenic mice was studied. Two groups (five mice per group) of IE86 transgenic-positive and negative mice were examined to determine any differences in their hnRNP A2/B1 expression levels. Specifically, the two groups of mice were fed the same feeds and were raised under the same environmental conditions. At the 10th week, the mice were sacrificed, and RNA and total protein were extracted from their brain tissues, the hnRNP A2/B1 mRNA expression levels were detected by qPCR, and the hnRNP A2/B1 protein expression levels were examined by Western blot. The data showed that IE86 could promote the mRNA and protein expression levels of hnRNP A2/B1 in IE86 transgenic mice (Figure 2B and 2C). Subsequently, U251 cells were transfected with peGFP-N3-IE86 (Figure 2D). Then, the mRNA and protein expression levels of hnRNP A2/B1 were measured 24, 48, and 72 h after transfection with peGFP-N3-IE86. The results indicated that IE86 could also promote hnRNP A2/B1 expression in glioma cells (Figure 2E and 2F).
Figure 2.
IE86 increases hnRNP A2/B1 mRNA and protein expression levels. A. MouseIE86 levels were measured by Western blot. B. IE86 transgenic mice and wild-type mice were given the same feeding and environmental conditions. They were sacrificed at the tenth week to extract the brain tissue RNA and total protein; the relative mRNA expression of hnRNP A2/B1 was detected by qPCR. Results are representative of three independent experiments. C. Mouse hnRNP A2/B1 levels were measured by Western blotting. The blots are representative of three independent experiments. D. The IE86 levels in the U251 cells were measured by Western blot. E. U251 cells were transfected with peGFP-N3-IE86 as indicated for 24, 48, and 72 h, and the hnRNP A2/B1 levels were determined by qPCR. NC were cells not transfected with peGFP-N3-IE86. Results are representative of three independent experiments. F. Cell hnRNP A2/B1 protein levels were measured by Western blot. ** P < 0.01.
IE86 inhibited the splicing of Bcl-x via promoting hnRNP A2/B1 expression
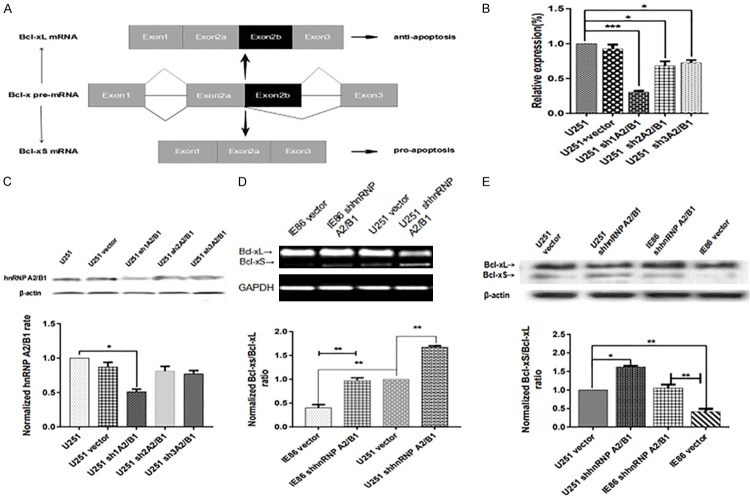
HnRNP A2/B1 has also been shown to play a critical role in regulating apoptosis by participating in the alternative splicing of Bcl-x, which will produce the antiapoptotic Bcl-xL or the proapoptotic Bcl-xS [19,21]. Thus, the influence of IE86 on the alternative splicing of Bcl-x, as well as the potential role of hnRNP A2/B1, were investigated in this study. The alternative splicing of the Bcl-x gene could generate Bcl-xL and Bcl-xS variants, which is achieved using the alternative 5’ splice site within exon 2 (Figure 3A).
Figure 3.
IE86 inhibits bcl-x splicing by promoting hnRNP A2/B1 expression. (A) Schematic splicing of Bcl-x pre-mRNA generating Bcl-xL and Bcl-xS mRNA via alternative 5’ splice site selection. (B) qPCR. U251 cells with stable hnRNP A2/B1 knockdown were grown in cell culture dishes and subjected to qPCR to determine hnRNP A2/B1 mRNA expression. The results are representative of three independent experiments, compared with the untransfected cell groups. (C) Western blot. The same cells were subjected to Western blot, compared with the untransfected cell groups. (D) After transfection with peGFP-N3-IE86 for 24 h, both U251 cells and cells transfected with peGFP-N3-IE86 were treated with sh1A2/B1. Afterwards, the Bcl-xL and Bcl-xS levels were determined by RT-PCR and (E) Western blot. *P < 0.05, **P < 0.01 and ***P < 0.001.
Compared with the hnRNP A2/B1 expression in U251 cells, the hnRNP A2/B1 mRNA expression in the U251 cells stably transfected with hnRNP A2/B1 shRNA was markedly downregulated (Figure 3B and 3C). Because the plasmid transfection efficiency was different, the sh1A2/B1-transfected U251 cells were finally selected as the target cells to research the effect of the hnRNP A2/B1 knockdown on the cells. The U251 sh3A2/B1 and U251 sh2A2/B1 transfected cells (hnRNP A2/B1 mRNA expression decreased < 31%, the protein expression decreased 17%) were excluded from this research. No significant differences were observed between the control U251 and the vector-transfected U251 cells. Afterwards, the role of hnRNP A2/B1 in the IE86-mediated alternative splicing of Bcl-x was investigated. As shown in Figure 3D and 3E, hnRNP A2/B1 partially enhanced the inhibition of IE86 on Bcl-x splicing (P < 0.01). In addition, the knockdown of hnRNP A2/B1 increased the Bcl-xS/Bcl-xL ratio.
IE86 affected the apoptosis and proliferation of U251 cells through the hnRNP A2/B1-mediated pathway
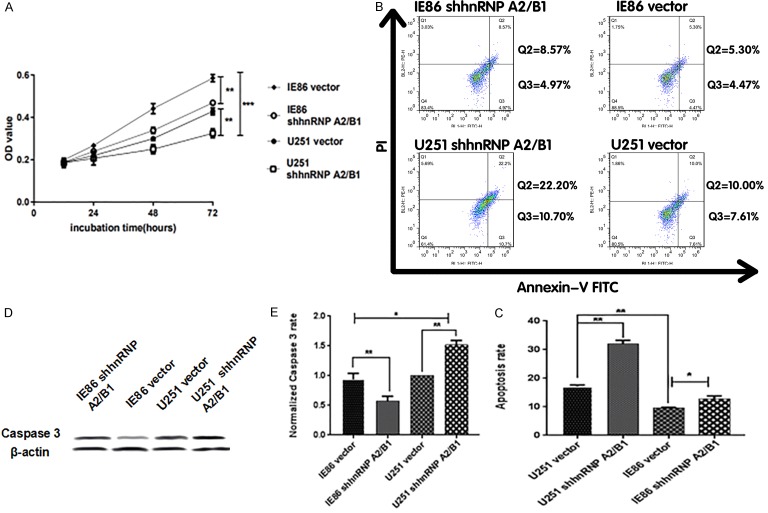
Subsequently, the influence of hnRNP A2/B1 knockdown on cell proliferation was also examined. To research the role of hnRNP A2/B1 in IE86-promoted cell proliferation, U251 cells were first transfected with a vector control or with shhnRNP A2/B1. Specifically, compared with the growth rate of the U251-vector cells, the growth rate of the IE86-vector cells increased to 75%; meanwhile, compared with the growth rate of the U251-vector cells, the growth rate of the U251-shhnRNP A2/B1 cells decreased to 42%. Compared with the growth rate of the IE86-vector cells, the growth rate of the IE86-shhnRNP A2/B1 cells decreased to 29% (Figure 4A). These results suggest that IE86 can promote cell proliferation by enhancing hnRNP A2/B1 expression. Finally, the influence of hnRNP A2/B1 knockdown on apoptosis was also examined. Cell apoptosis detected through cytometry showed that the apoptosis of IE86-vector cells was noticeably reduced compared with the apoptosis of the U251-vector cells; the apoptosis of the U251-shhnRNP A2/B1 cells was remarkably elevated compared with the apoptosis of the U251-vector cells; and the apoptosis of IE86-shhnRNP A2/B1 cells was also increased relative to the IE86-vector cells (Figure 4B and 4C). In addition, flow cytometry demonstrated that IE86 can inhibit apoptosis by promoting hnRNP A2/B1 expression. Figure 4D and 4E show that IE86 can inhibit caspase 3 by promoting hnRNP A2/B1 expression.
Figure 4.
IE86 promotes cell proliferation and inhibits apoptosis through the hnRNP A2/B1-mediated pathway. (A) Effects of hnRNP A2/B1 knockdown on the regulation of cell proliferation in U251 cells. Cells transfected with peGFP-N3-IE86 were examined using a CCK8 assay. (B) Effects of hnRNP A2/B1 knockdown on the apoptosis of U251 cells. Cells transfected with peGFP-N3-IE86 were examined by flow cytometry. (C) Statistical analysis of (B). (D) The expression of caspase 3 of different groups was determined by Western blotting. The blots are representative of three independent experiments. (E) Statistical analysis of (D) *P < 0.05, **P < 0.01.
Discussion
Human cytomegalovirus (HCMV) is a widely distributed, species-specific virus that not only can establish infections in most people, especially patients with low immunity due to organ transplantation or AIDS, but also can cause congenital malformations in an infant’s nervous system, which can place a heavy burden on society and families [22]. The IE genes are the first group of genes expressed after the virus enters the cell and can encode a variety of functional proteins [24]. Among them, IE86 is one of the most abundantly expressed proteins [28]. IE86 is first expressed by HCMV after infection and plays a crucial regulatory role in host cells. Previous studies have confirmed that IE86 inhibits apoptosis and promotes cell proliferation [25]. HnRNP A2/B1 is a well-known splicing factor that was successfully identified as one of the components of the immune complex of IE86.
In recent years, increasing attention has been focused on hnRNP A2/B1 because of its overexpression in various tumors, such as human glioma tissues [26]. Additionally, emerging evidence shows that the levels of hnRNP A2/B1 are strongly associated with a poor prognosis and more aggressive tumor phenotypes. Within human glioblastoma, it has been found that hnRNP A2/B1 is a carcinogenic driver that is closely associated with tumor cell apoptosis, growth and survival [18,26]. Thus, a potential approach to therapeutic intervention in gliomas can be provided by targeting hnRNP A2/B1. It has been found in this study that the expression level of hnRNP A2/B1 can be promoted by IE86. Importantly, the effect of IE86 on proliferation and apoptosis can be reversed dramatically by the knockdown of hnRNP A2/B1. This result implies that hnRNP A2/B1 is of great importance for IE86-mediated cellular activity. Splicing is of great importance for the creation of various protein isoforms having opposite functions by splicing in particular sites in the pre-mRNA of the target gene [27]. HnRNP A2/B1 serves as a well-characterized RNA-binding splicing factor that has been proven to regulate cell invasiveness, survival and proliferation through the replacement of various pre-mRNA targets [5]. Bcl-x serves as an example, which can be spliced to proapoptotic Bcl-xS and antiapoptotic Bcl-xL. Various viruses have developed important regulatory proteins to prohibit apoptosis during long-term evolution. This effect provides important conditions for viral replication and infection [28]. Within tumor cells, it has been found that hnRNP A2/B1 serves as the basic system, and hnRNP A2/B1 and Bcl-x pre-mRNA bind with BC200 (an ER-regulated lncRNA) to generate the BC200-Bcl-x-hnRNP A2/B1 complex, which results in the promotion of Bcl-xL expression [20,21] and the inhibition of Bcl-xS expression. Meanwhile, it was shown that the ratio of Bcl-xS/Bcl-xL at both the mRNA and protein levels is attenuated by IE86, suggesting that the alternative splicing of Bcl-x has been inhibited. Moreover, the inhibitory effect of IE86 on the alternative splicing of Bcl-x in U251 cells was attenuated by hnRNP A2/B1 knockdown Figure 5.
Figure 5.
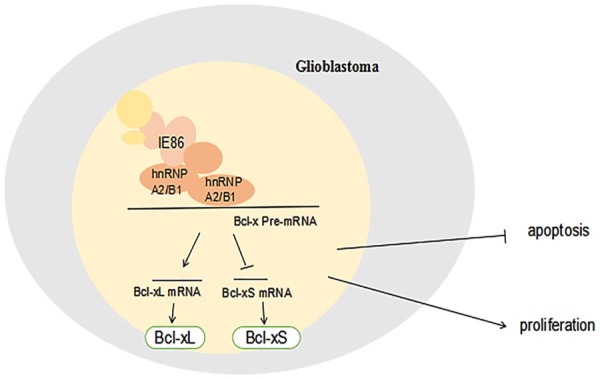
Model describing the function of hnRNP A2/B1 during IE86-infected cells.
In conclusion, the results obtained in this study indicate that IE86 promotes U251 cell proliferation and inhibits apoptosis by promoting the hnRNP A2/B1-mediated signaling pathway. On the one hand, IE86 inhibits the alternative splicing of Bcl-x and decreases the ratio of Bcl-xS/Bcl-xL by promoting the expression of hnRNP A2/B1, which may be related to the apoptosis pathway associated with IE86 inhibition. On the other hand, IE86 promotes cell proliferation by promoting an increase in the expression of hnRNP A2/B1. Overall, the study confirms the role of hnRNP A2/B1 on the effects of IE86 on tumor cell activity, and the results are crucial for inhibiting tumor growth and expanding new methods for tumor therapy and prevention.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81601754) and the Key Research and Development Project of Shandong Province (grant no. 2018GSF118141).
Disclosure of conflict of interest
None.
References
- 1.Bhattacharjee B, Renzette N, Kowalik TF. Genetic analysis of cytomegalovirus in malignant gliomas. J Virol. 2012;86:6815–24. doi: 10.1128/JVI.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mocarski ES. In: In the human herpesviruses. Roizman B, Whitley RJ, Lopez C, editors. Raven Pr N Yrk; 1993. pp. 173–226. [Google Scholar]
- 3.Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growthcontrol. Gene. 2002;290:19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, Murray GI. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim Biophys Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 6.Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183:215–220. doi: 10.1016/s0304-3835(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 7.Lee CL, Hsiao HH, Lin CW, Wu SP, Huang SY, Wu CY, Wang AH, Khoo KH. Strategic shotgun proteomics approach for efficient construction of an expression map of targeted protein families in hepatoma cell lines. Proteomics. 2003;3:2472–2486. doi: 10.1002/pmic.200300586. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Allred DC, Avis I, Martínez A, Vos MD, Smith L, Treston AM, Mulshine JL. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res Treat. 2001;66:217–224. doi: 10.1023/a:1010631915831. [DOI] [PubMed] [Google Scholar]
- 9.Lee CH, Lum JH, Cheung BP, Wong MS, Butt YK, Tam MF, Chan WY, Chow C, Hui PK, Kwok FS, Lo SC, Fan DM. Identification of the heterogeneous nuclear ribonucleoprotein A2/B1 as the antigen for the gastrointestinal cancer specific monoclonal antibody MG7. Proteomics. 2005;5:1160–1166. doi: 10.1002/pmic.200401159. [DOI] [PubMed] [Google Scholar]
- 10.Dowling P, Pollard D, Larkin A, Henry M, Meleady P, Gately K, O’Byrne K, Barr MP, Lynch V, Ballot J, Crown J, Moriarty M, O’Brien E, Morgan R, Clynes M. Abnormal levels of heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) in tumour tissue and blood samples from patients diagnosed with lung cancer. Mol Biosyst. 2015;11:743–752. doi: 10.1039/c4mb00384e. [DOI] [PubMed] [Google Scholar]
- 11.Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, Watabe K, Lu Z, Mo YY. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Chen S, Wang F, Zhao H, Xie Z, Xu Z, Zhang Q, Liang P, Zhai X, Cheng Y. Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol Neurobiol. 2016;53:1132–44. doi: 10.1007/s12035-014-9080-3. [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Ran L, Liu Y, Zhong SH, Zhou PP, Liao MX, Fang W. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cellcycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol Rep. 2018;39:939–950. doi: 10.3892/or.2018.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster H, Ulasov IV, Cobbs CS. Human cytomegalovirus-mediated immunomodulation: effects on glioblastoma progression. Biochim Biophys Acta Rev Cancer. 2017;1868:273–276. doi: 10.1016/j.bbcan.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Landi D, Hegde M, Ahmed N. Human cytomegalovirus antigens in malignant gliomas as targets for adoptive cellular therapy. Front Oncol. 2014;4:338. doi: 10.3389/fonc.2014.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, Siziopikou KP, Peng J, Xiao X, Cheng C. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specifi cities of the drosophila hnRNP A/B family members. Mol Cell. 2009;33:438–49. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakacs A, Coppola L, Karni R. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464–72. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, Watabe K, Lu Z, Mo YY. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, Liu XD, Wang SG, Bie P, Jiang P, Dong JH, Li XW. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis. 2011;32:1419–1426. doi: 10.1093/carcin/bgr088. [DOI] [PubMed] [Google Scholar]
- 21.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halwachs-Baumann G, Genser B, Danda M, Engele H, Rosegger H, Fölsch B, Maurer U, Lackner H, Truschnig-Wilders M. Screening and diagnosis of congenital cytomegalovirus infection: a 52y study. Scand J Infect Dis. 2000;32:1372–142. doi: 10.1080/003655400750045222. [DOI] [PubMed] [Google Scholar]
- 23.Murphy EA, Streblow DN, Nelson JA, Stinski MF. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castillo JP, Kowalik TF. Human cytomegalovirus im2 mediate early proteins and cell growth control. Gene. 2002;290:19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K, Zou JP, Takeda K, Ferrans VJ, Sandford GR, Johnson TM, Finkel T, Epstein SE. Effects of human cytomegalovirus immediate2early proteins on p532-mediated apoptosis in coronary artery smooth muscle cells. Circulation. 1999;99:1656–1659. doi: 10.1161/01.cir.99.13.1656. [DOI] [PubMed] [Google Scholar]
- 26.Deng J, Chen S, Wang F, Zhao H, Xie Z, Xu Z, Zhang Q, Liang P, Zhai X, Cheng Y. Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol Neurobiol. 2016;53:1132–44. doi: 10.1007/s12035-014-9080-3. [DOI] [PubMed] [Google Scholar]
- 27.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Shenk TE. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]