Abstract
Background: Acute respiratory distress syndrome (ARDS) is a common clinical syndrome with high a mortality rate, which is associated with diffuse alveolar injury and capillary endothelial damage. In recent years, numerous studies have been performed to explore the roles of long non-coding RNAs (lncRNAs) in various diseases in which lncRNA serves as a microRNA (miRNA) sponge to regulate targeted gene expression. However, whether lncRNAs participate in ARDS progression remains unclear. Materials/Methods: The dual-luciferase reporter assay was employed to identify the interaction between lncRNA XIST and miR-204, as well as the correlation between miR-204 and interferon regulatory factor 2 (IRF2). Then, PaO2/FiO2 was determined in lipopolysaccharide (LPS)-induced ARDS. In addition, the concentrations of cytokines, including IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R were analyzed by ELISA. lncRNA XIST, miR-204, and IRF2 levels were determined by qRT-PCR assay, and the IRF2 expression was evaluated by western blot. Furthermore, levels of inflammation and conditions of alveoli were evaluated by hematoxylin (H&E)-staining in LPS-induced ARDS. Results: Our findings indicated that lncRNA XIST served as a sponge for miR-204. miR-204 directly regulated IRF2, andlncRNA XIST upregulated IRF2 expression by targeting miR-204. LncRNA XIST and miR-204 inhibitors significantly decreased the PaO2/FiO2 ratio, whereas miR-204 and silencing of IRF2 significantly increased the PaO2/FiO2 ratio in LPS-induced ARDS. In addition, lncRNA XIST and miR-204 inhibitors significantly increased levels of IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R, whereas miR-204 and silencing of IRF2 dramatically decreased related cytokines in LPS-induced ARDS. Furthermore, we demonstrated that lncRNA XIST and miR-204 inhibitors aggravated inflammatory cell infiltration, alveolitis, and the degree of fibrosis, whereas miR-204 and silencing of IRF2 alleviated inflammation and conditions of the alveoli. Conclusion: In this study, we verified that lncRNA XIST serves as a sponge for miR-204 to aggravate LPS-induced ARDS in mice by upregulating IRF2.
Keywords: Acute respiratory distress syndrome, interferon regulatory factor 2, lncRNA XIST, microRNA-204
Introduction
Acute respiratory distress syndrome (ARDS) is a common critical disease that may occur due to direct injury, such as contusion, inhalation injury, pneumonia, aspiration, mechanic ventilation or indirect injury, including sepsis, blood transfusion, pancreatitis, and traumatic brain injury [1]. It has been well-established that ARDS is characterized by both endothelial and epithelial injury as a consequence of inflammatory reactions [2]. Previous studies have shown that cytokines and chemokines play essential roles in initiating and mediating pulmonary inflammation [3]. At present, ARDS is still a common cause of morbidity and mortality among patients in the intensive care unit (ICU), and accounts for ~5-20% of all mechanically ventilated patients [4,5]. Although the use of lung protective ventilation strategies has improved the management of ARDS, the mortality rate of severe ARDS is still unacceptably high [6]. Currently, no effective biomarkers and drug targets exist that can be applied to ARDS patients [7]. Therefore, it is of utmost importance to identify new and effective biomarkers for the prognosis and prediction of ARDS.
Long non-coding RNAs (lncRNAs) of more than 200 nucleotides in length and microRNAs (miRNAs) of less than 22 nucleotides are the two most important members of non-coding RNAs (ncRNAs) without protein-coding potential [8]. In previous studies, it has been demonstrated that lncRNAs and miRNAs participated in various processes, including cell proliferation, tissue differentiation, metabolic regulation, cell cycle, apoptosis, and metastasis [8,9]. Dysregulation of lncRNAs and miRNAs may lead to a series of dysfunctions and diseases [9]. Recently, emerging evidence has shown that cross-modulation exists between lncRNA and miRNA [10,11]. LncRNAs act as a competing endogenous RNA (ceRNA) or a RNA sponge to affect the expression and function of miRNAs [12-14]. In addition, miRNAs have been shown to be involved in the regulation of gene expression by targeting the 3’-untranslated region (3’-UTR) of target genes [15]. The interaction of lncRNA and miRNA regulates the occurrence and development of miscellaneous diseases by directly or indirectly affecting the expression levels of related genes [16]. LncRNA XIST, which is located on the X chromosome, has been studied in various diseases, including osteoarthritis chondrocytes [17], pancreatic carcinoma [18], lung cancer [19], myocardial infarction [20], and cervical cancer [21]. In addition, it has previously been reported that knockdown of lncRNA XIST could relieve endothelial cell injury that was mediated by oxidative low-density lipoprotein through the miR-320/NOD2 axis [22]. However, the underlying mechanism involved, and the role of lncRNA XIST in ARDS are still unknown.
In the present study, we explored whether lncRNA XIST can serve as a sponge of miR-204 to regulate miR-204 expression, and whether miR-204 can directly regulate IRF2 by binding to the 3’-UTR. We next evaluated the influence of lncRNA XIST, miR-204, and IRF2 on the PaO2/FiO2 ratio in LPS-induced ARDS in mice. In addition, we assessed the effects of lncRNA XIST, miR-204, and IRF2 on levels of these cytokines: IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R. Furthermore, we showed changes in inflammation and conditions of the alveoli that are mediated by lncRNA XIST, miR-204, and IRF2 in LPS-induced ARDS.
Materials and methods
Animals
A total of 72 healthy male mice (age, 6-8 weeks; weight, 20-30 g; body temperature, 36.5-38°C; specific pathogen free (SPF)) were provided by the Animal Experiment Center of Institute of Radiation Medicine of the Chinese Academy of Medical Sciences (Tianjin, China), and were housed in a controlled environment (food and water available ad libitum, 21 ± 1°C, humidity 60%, lights on from 7:00 AM-7:00 PM). This study was approved by the Ethics Committee of the Ningbo No. 6 Hospital (Ningbo, China), and all procedures were performed in accordance with the National Institutes of Health guidelines.
Vector construction
Full-length cDNA of lncRNA XIST was amplified by RT-PCR by using PrimerSTAR Max DNA Polymerase Mix (Takara, Shanghai, China) with specific primers. Next, PCR products were inserted into the pcDNA3.0 vector (Invitrogen, Carlsbad, CA, USA).
Oligonucleotide transfection
The control, miR-204 inhibitors, and miR-204 mimics were synthesized by Gene Pharma (Shanghai, China). All transfections were performed using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) according the manufacturer’s protocol.
Establishing model mice and grouping
As previously described [23], an ARDS model was established in mice by atomizing inhalation of LPS in the airways. In brief, mice were anesthetized (ketamine 100 mg/kg and xylazine 10 mg/kg) using a 100 μl micropipette, and then were treated with LPS (10 μg/mouse) in 50 μl of PBS by orotracheal instillation. Subsequently, ARDS mice were divided into groups as follows: a negative control (NC) group, lncRNA XIST group, miR-204 inhibitors group, miR-204 mimics group, and IRF2 siRNA (si-IRF2) group, respectively.
Cell culture
For cell culture studies, 293T cells were used that were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, CA, USA), containing 10% fetal bovine serum (FBS) in a humidified incubator containing 5% CO2.
RNA extraction and quantitative real-time PCR assay
For quantitative real-time PCR (qRT-PCR), total RNA was extracted by using Trizol (Invitrogen, Carlsbad, CA, USA), and cDNA was obtained by using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA, USA) according to the manufacturer’s instructions). The expression of lncRNA XIST, miR-204, and IRF2 was analyzed by using SYBRs GREEN PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The data obtained were assessed on an ABI7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA).
Relative mRNA expression levels were analyzed using the 2-ΔΔCt method [24]. Primers used are shown in Table 1.
Table 1.
Primer sequences for qRT-PCR analysis
| Gene | Primer sequences |
|---|---|
| GAPDH | Forward: 5’-TATGATGATATCAAGAGGGTAGT-3’ |
| Reverse: 5’-TGTATCCAAACTCATTGTCATAC-3’ | |
| U6 | Forward: 5’-CTCGCTTCGGCAGCACATA-3’ |
| Reverse: 5’-AACGATTCACGAATTTGCGT-3’ | |
| LncRNA | Forward: 5’-AGGGTGTGTGTGCATATGGA -3’ |
| XIST | Reverse: 5’-CCGCCATCTTTTCCTGTACG -3’ |
| miR-204 | Forward: 5’-ACACTCCAGCTGGGTTCCCTTTGTCATCCT-3’ |
| Reverse: 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGCATAG-3’ | |
| IRF2 | Forward: 5’-AGTGTGGCCAGTGATGAAGA-3’ |
| Reverse: 5’-GAGCTGTTGTAAGGCATCGG-3’ |
Western blot analysis
Total proteins were obtained by using RIPA buffer (Beyotime, Shanghai, China) containing a protease inhibitor cocktail (P8340; Sigma, St. Louis, MO, USA). Proteins (30 μg) were separated on 10% SDS-PAGE gels, then transferred to a polyvinyl difluoride (PVDF) membrane (PerkinElmer, Boston, MA, USA). Next, membranes were blocked using 5% skimmed milk (dissolved in Tris-buffered saline, containing 0.05% Tween-20 (TBST), BD Biosciences) for 90 min at 27°C, then membranes were incubated overnight at 4°C with the following primary antibodies: anti-GAPDH antibody (dilution 1:2000; Abcam, Cambridge, UK; ab8245), anti-IRF2 antibody (dilution 1:1000; Abcam, Cambridge, MA, USA; ab86018). Then, PVDF membranes were incubated with horseradish peroxidase conjugated secondary antibody (Santa Cruz Biotechnology, CA, USA) for 60 min in 27°C. Protein bands were visualized by using the enhanced chemiluminescence substrate kit in an enhanced chemiluminescence detection system (Amersham Biosciences Inc., Piscataway, NJ, USA).
ELISA
After treatment, blood was collected from each mouse and centrifuged (14,000 rpm/min, 4 mins) to prepare serum for ELISA. Levels of proinflammatory factors were determined by using commercial ELISA kits following the manufacturer’s guidelines. IFN-γ ELISA Kit (Abcam, Cambridge, MA, USA; ab100689), IL-6 ELISA Kit (Nanjing Jiancheng Bioengineering Institute; cat no. H007), IL-17, ELISA Kit (Abcam, Cambridge, UK; ab79056), TNF-α ELISA kit (CUSABIO, CSB-E11987r), IL-1β ELISA Kit (Nanjing Jiancheng Bioengineering Institute; cat no. H002), and IL-6R ELISA Kit (Qiaoyu Biotechnology Co., Ltd., China; QY-PF9440).
Hematoxylin and eosin staining
Lungs from mice in each group were harvested and lung tissue was fixed in 4% paraformaldehyde for 12 h at room temperature and dehydrated with different concentrations of alcohol. Then, tissue was embedded in paraffin, 5-μm sections were cut, and stained with hematoxylin and eosin (H&E) (H8070, Solarbio, China). Images were obtained by using a microscope (Nikon, Eclipse Ci, Japan) at 400 × magnification.
Dual-luciferase reporter assay
To generate reporter plasmids, the binding sites of lncRNA XIST including wild type-lncRNA XIST, mutant lncRNA XIST, and IRF2 including wild type-IRF2 and mutant IRF2 were constructed into the promoter vector (Realgene, Nanjing, China). Briefly, 293T cells (5 × 104 cells/well) were seeded into 24-well plates and transfected with corresponding reporter plasmids using Lipofectamine 2000 (Invitrogen, Shanghai, China). After 48 h, the fluorescence intensity of 293T cells was determined by Dual Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed on SPSS software version 15.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Statistically significant differences were calculated by using one-way ANOVA followed by post-hoc Tukey’s analysis of variance. P < 0.05 was considered significant.
Results
LncRNA XIST serves as a sponge for miR-204
TargetScan, an online tool for predicting lncRNAs and their interacting miRNAs, was used to screen the possible miRNAs that may be targeted by lncRNA XIST. We found a putative complementary site between lncRNA XIST and miR-204. The binding site is presented in Figure 1. In addition, we constructed a mutant sequence of lncRNA XIST. In brief, 293T cells were co-transfected with either wild-type lncRNA XIST or mutant lncRNA XIST and miR-204 mimics or control, respectively. Our data indicated a decrease in luciferase intensity between wild-type lncRNA XIST and miR-204, whereas no changes were observed in luciferase intensity between mutant lncRNA XIST and miR-204 (P < 0.05, Figure 1).
Figure 1.
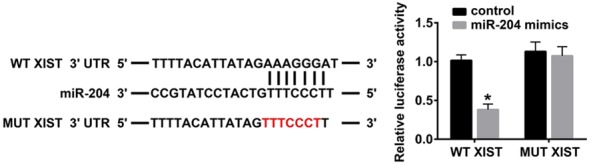
LncRNA XIST serves as a sponge for miR-204. Sequence alignment of lncRNA XIST with miR-204. MUT XIST: mutations in the lncRNA XIST sequence to create the mutant luciferase reporter construct. The activity of a luciferase reporter containing wild-type lncRNA XIST 3’UTR in 293T cells after transfection with a negative control construct or miR-204 mimics (*P < 0.05).
IRF2 is a target gene of miR-204
According to the data obtained by TargetScan, miRDB, and microrna.org, we predicted that there was a binding site between miR-204 and IRF2. The relative luciferase intensities of IRF2 wild-type or mutated 3’UTRs and miR-204 or control oligonucleotides were assessed for 24 h, then we determined the luciferase intensity by using Dual-Luciferase Reporter Assay. Our data indicated that miR-204 significantly reduced the luciferase intensity of wild-type IRF2, whereas miR-204 had no effect on the mutated IRF (P < 0.05, Figure 2). Therefore, we hypothesized that miR-204 negatively regulated IRF2.
Figure 2.
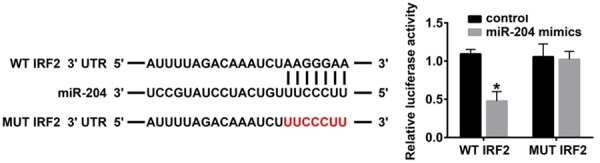
IRF2 is a target gene of miR-204. TargetScan, miRDB and microrna.org were used to predict the target gene of miR-204, and IRF2 was a candidate. Wild type (WT) and Mut 3’-UTR sequences of IRF2 are shown. The relative luciferase intensities of IRF2 3’UTR (WT and mutant constructs) were analyzed after co-transfection of firefly luciferase constructs containing the IRF2 wild-type or mutated 3’UTRs and miR-204 or control oligonucleotides for 24 h in 293T cells (*P < 0.05).
The effects of lncRNA XIST and miR-204 on PaO2/FiO2 in LPS-induced ARDS
In this study, LPS was used to induce ARDS in mice. In brief, mice were divided into the NC group, lncRNA XIST group, miR-204 inhibitors group, miR-204 mimics group, and IRF2 siRNA (si-IRF2) group, respectively. We demonstrated that the PaO2/FiO2 ratio was significantly decreased in the lncRNA XIST group and the miR-204 inhibitors group when compared to the NC group. In addition, the PaO2/FiO2 ratio was significantly increased in the miR-204 group and the si-IRF2 group when compared with the NC group (P < 0.05, Figure 3).
Figure 3.
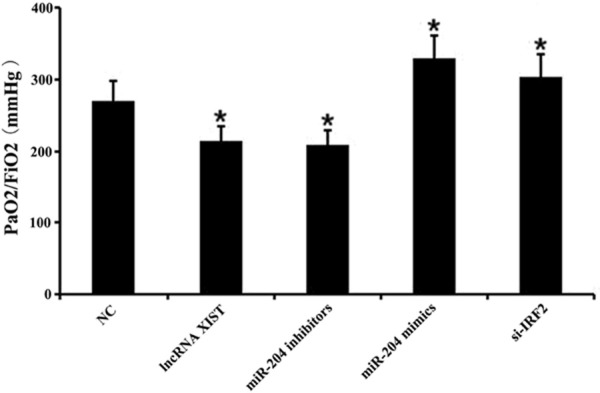
The effects of lncRNA XIST and miR-204 on PaO2/FiO2 in lipopolysaccharide-induced acute respiratory distress syndrome. An acute respiratory distress syndrome (ARDS) model in mice was established in which mice were administered with lipopolysaccharide (LPS). Mice were divided into a negative control (NC) group, lncRNA XIST group, miR-204 inhibitors group, miR-204 mimics group, and IRF2 siRNA (si-IRF2) group, respectively. The PaO2/FiO2 ratio was measured in each group (*P < 0.05 vs. NC group).
LncRNA XIST and miR-204 regulated cytokines in LPS-induced ARDS
Next, we evaluated the roles of lncRNA XIST, miR-204, and IRF2 on cytokines in LPS-induced ARDS. Our findings indicated that IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R were increased in the lncRNA XIST group and miR-204 inhibitors group relative to the NC group. Furthermore, IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R were dramatically decreased in the miR-204 group and si-IRF2 group when compared with the NC group (P < 0.05, Figure 4A). In addition, lncRNA XIST, miR-204, and IRF2 expression were analyzed. Results showed that lncRNA XIST expression was significantly upregulated in the lncRNA XIST group and miR-204 inhibitors group when compared to the NC group; however, expression was significantly downregulated in the miR-204 group and si-IRF2 group when compared with the NC group (P < 0.05, Figure 4B). The expression of miR-204 was observably downregulated in the lncRNA XIST group and the miR-204 inhibitors group when compared to the NC group, and significantly upregulated in the miR-204 group and si-IRF2 group when compared with the NC group (P < 0.05, Figure 4B). We also found that IRF2 expression was dramatically increased in the lncRNA XIST group and the miR-204 inhibitors group when compared to the NC group, and significantly decreased in the miR-204 group and si-IRF2 group when compared with the NC group (P < 0.05, Figure 4B and 4C).
Figure 4.
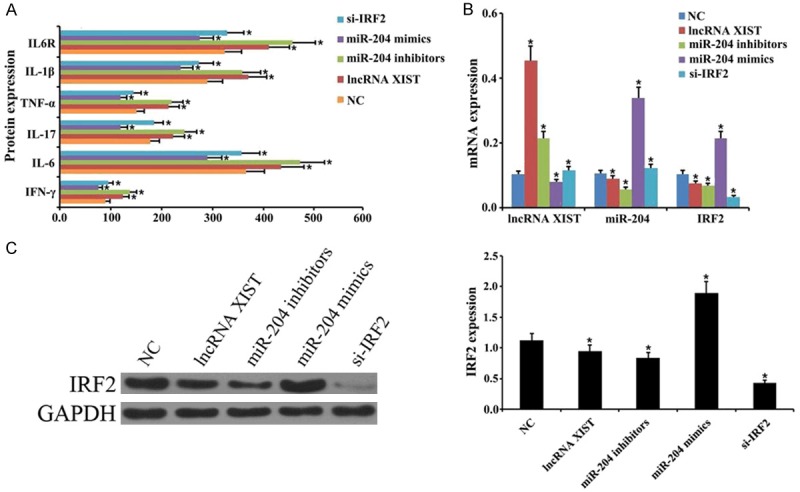
lncRNA XIST and miR-204 regulate cytokines in lipopolysaccharide-induced acute respiratory distress syndrome. An ARDS model was established in mice that were induced with lipopolysaccharide (LPS). The mice were divided into a negative control (NC) group, lncRNA XIST group, miR-204 inhibitors group, miR-204 mimics group, and IRF2 siRNA (si-IRF2) group, respectively. A. The levels of IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R as measured by ELISA (*P < 0.05 vs. NC group). B. Dot plots of lncRNA XIST, miR-204, and IRF2 levels as determined by qRT-PCR assay (*P < 0.05 vs. NC group). C. Protein expression level of IRF2 as measured by western blot analysis. GAPDH was used as an internal reference. Quantitative proteins were analyzed based on the protein gray values (*P < 0.05 vs. NC group).
LncRNA XIST and miR-204 change inflammatory conditions in LPS-induced ARDS
H&E-staining showed a very small amount of inflammatory cell infiltrate in the NC group, whereas inflammatory cell infiltrate was significant in the lncRNA XIST group and miR-204 inhibitors group when compared to the model group. Moreover, inflammatory cell infiltration was dramatically alleviated in the miR-204 group and si-IRF2 group when compared to the model group (Figure 5).
Figure 5.
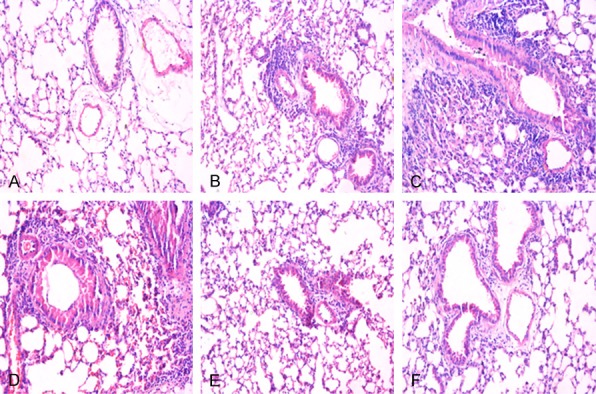
Inflammatory conditions were altered by lncRNA XIST and miR-204 in lipopolysaccharide-induced acute respiratory distress syndrome. The changes in inflammatory conditions were assessed by hematoxylin and eosin (H&E)-staining. A: Negative control (NC) group, B: Model group, C: lncRNA XIST group, D: miR-204 inhibitors group, E: miR-204 mimics group, F: IRF2 siRNA (si-IRF2) group. Magnification, × 200.
The alveoli were changed by lncRNA XIST and miR-204 in LPS-induced ARDS
In this study, lung pathology in each group was determined by H&E-staining in each group. As shown in Figure 6, lung tissue was normal in the control group, whereas when compared to the model group, alveolitis and the degree of fibrosis were increased in the lncRNA XIST group and miR-204 inhibitors group. Moreover, alveolitis and the degree of fibrosis were decreased in the miR-204 group and si-IRF2 group when compared to the model group (Figure 6).
Figure 6.
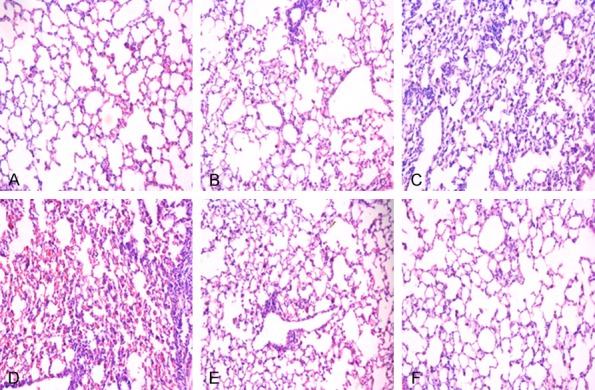
LncRNA XIST and miR-204-induced changes in alveoli in lipopolysaccharide-induced ARDS. Alveoli were stained by hematoxylin and eosin (H&E) staining. A: Negative control (NC) group, B: Model group, C: lncRNA XIST group, D: miR-204 inhibitors group, E: miR-204 mimics group, F: IRF2 siRNA (si-IRF2) group. Magnification, × 200.
LncRNA XIST upregulated IRF2 expression by miR-204
To further explore the regulatory effects of lncRNA XIST and miR-204 on IRF2 expression, ARDS model were co-transfected with lncRNA XIST and miR-204 mimics or miR-204 inhibitors and si-IRF2, and RT-qPCR and western blot analysis was performed. The results revealed that overexpression of lncRNA XIST significantly upregulated IRF2 expression, while miR-204 mimics dramatically attenuated this upregulation induced by lncRNA XIST (P < 0.01, P < 0.001, Figure 7A). In addition, the results indicated that overexpression of miR-204 inhibitors significantly upregulated IRF2 expression, while knockdown of IRF2 significantly weakened the upregulation induced by miR-204 inhibitors (P < 0.01, P < 0.001, Figure 7B). Therefore, we suggest that lncRNA XIST may upregulate IRF2 expression by targeting miR-204.
Figure 7.
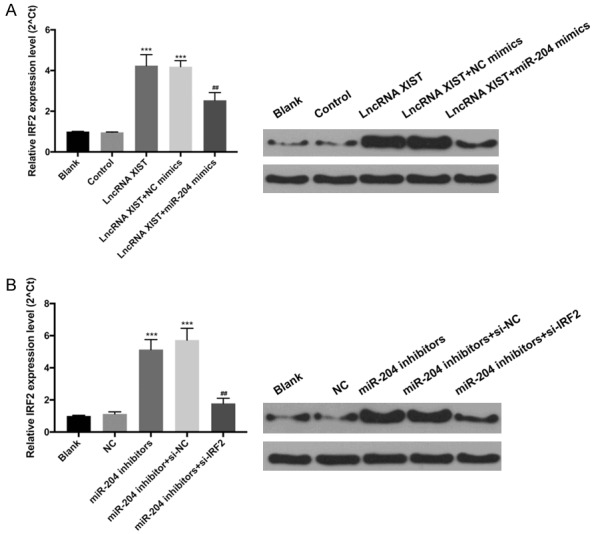
LncRNA XIST upregulated IRF2 expression by miR-204. A. The ARDS model mice were divided into Blank, control, lncRNA XIST, lncRNA XIST+NC mimics, and lncRNA XIST+miR-204 mimics group, respectively. The expression level of IRF2 was analyzed by RT-qPCR and western blot. ***P < 0.001 vs. control group, ##P < 0.01 vs. lncRNA XIST+NC mimics group. B. The ARDS model mice were divided into Blank, NC, miR-204 inhibitors, miR-204 inhibitor+si-NC and miR-204 inhibitors+si-IRF2 groups, respectively. RT-qPCR and western blot were performed to evaluate IRF2. ***P < 0.001 vs. control group, ##P < 0.01 vs. miR-204 inhibitor+si-NC group.
Discussion
In a previous study, the ceRNA hypothesis was believed to be a new regulatory mechanism between coding RNA and non-coding RNA [25]. LncRNAs play essential regulatory roles in the regulation of gene expression and contribute to the occurrence and development course of diseases. Previous studies have shown that lncRNAs could interact with target miRNAs by miRNA-binding sites (MREs) to regulate gene expression [26-28]. The crosstalk of the ceRNA network has been studied in a variety of diseases [29]. Recently, the mechanism and effects of lncRNA XIST were investigated in different diseases. For example, lncRNA XIST interacts with miR-211 to regulate the proliferation and apoptosis of osteoarthritis chondrocytes by CXCR4 and MAPK signaling [17], and the lncRNA XIST/miR-29c axis plays an essential role in the chemoresistance of glioma by the DNA mismatch repair pathway [30]. Moreover, lncRNA XIST served as a molecular sponge of miR-200a, promoted cervical cancer progression by Fus [21], and regulated MAPK1 expression in hepatocellular carcinoma by targeting miR-194-5p [31]. In our study, we further confirmed that lncRNA XIST serves as a sponge for miR-204.
miRNAs can lead to translation repression or RNA degradation by binding to the 3’UTR of target mRNA [32]. In various studies, it has been confirmed that miRNAs play a role in numerous physiologic processes, including differentiation, proliferation, metabolism, hemostasis, apoptosis, and metastasis [33-35]. In a previous study, it was shown that miR-204 was downregulated in cardiopulmonary bypass-induced acute lung injury [36]. Furthermore, it has previously been demonstrated that miR-204, which acts as a tumor suppressor, could inhibit CYP27A1 expression in glioblastoma [37]. Moreover, miR-204 motivates fracture healing by accelerating cell viability of osteoblasts [38], and miR-204 functions as a crucial therapeutic target of acute myeloid leukemia by promoting BIRC6-mediated apoptosis [39]. miR-204 inhibits glioblastoma progression by targeting ATF2 [40], and inhibits migration and invasion of non-small-cell lung carcinoma by targeting JAK2 [41]. In addition, miR-204 suppressed epithelial-mesenchymal transition ischemia-reperfusion-induced acute kidney injury by targeting SP1 [42]. In the present study, we further identified that IRF2 was a target gene of miR-204. IRF2 is a member of the transcription factor family, and plays a role in the regulation of IFN-induced immune responses [43], which has significant effects on biologic processes [44,45].
ARDS involves acute respiratory failure that is characterized by progressive difficulty of breathing and refractory hypoxemia [46]. Cytokines and inflammatory mediators play important roles in the occurrence and development of ARDS patients with local inflammatory and systemic inflammatory responses [47]. Although mechanical ventilation and protective lung ventilation strategies can be used to improve the rescue success rate of ARDS patients, no effective method is yet available to prevent the inflammatory lung injury that is often seen in ARDS [48]. At present, 200 mmHg < PaO2/FiO2 ratio < 300 mmHg has become a vital indicator for ARDS [49]. In our study, we demonstrated that lncRNA XIST or miR-204 inhibitors significantly decreased the PaO2/FiO2 ratio, while the PaO2/FiO2 ratio was significantly increased by miR-204 mimics or IRF2 knock-down. In addition, we demonstrated that lncRNA XIST increased inflammatory cytokines, while miR-204 or silencing of IRF2 decreased inflammatory cytokines. Thus, our data suggested that lncRNA XIST promoted ARDS progression, whereas miR-204 or silencing of IRF2 inhibited ARDS progression. Furthermore, we confirmed that lncRNA XIST or miR-204 inhibitors promote inflammatory cell infiltration in LPS-induced ARDS, and that miR-204 mimics decrease IRF2-inhibited inflammatory cell infiltration in LPS-induced ARDS. We also showed that lncRNA XIST or miR-204 inhibitors aggravated alveolitis and the degree of fibrosis in LPS-induced ARDS. miR-204 mimics silence IRF2-attenuated alveolitis and the degree of fibrosis in LPS-induced ARDS. Mechanistically, lncRNA XIST can significantly upregulate IRF2 expression by miR-204 in LPS-induced ARDS. Based on the interaction between lncRNA XIST and miR-204, and the correlation between miR-204 and IRF2, we hypothesize that lncRNA XIST functions as a ceRNA of miR-204, and aggravates LPS-induced ARDS by upregulation of IRF2.
Conclusions
In summary, lncRNA XIST negatively regulated miR-204, and miR-204 negatively regulated IRF2. LncRNA XIST increased PaO2/FiO2 ratio, and promoted inflammatory reactions and alveolitis in LPS-induced ARDS by the miR-204/IRF2 axis. Therefore, we demonstrated that lncRNA XIST acts as ceRNA of miR-204, and may be a potential biomarker and therapeutic target for ARDS by upregulating IRF2 expression. However, additional studies will be required to confirm the biologic function and precise underlying mechanism of the lncRNA XIST/miR-204/IRF2 axis on LPS-induced ARDS.
Disclosure of conflict of interest
None.
References
- 1.Laffey JG, Misak C, Kavanagh BP. Acute respiratory distress syndrome. BMJ. 2017;359:j5055. doi: 10.1136/bmj.j5055. [DOI] [PubMed] [Google Scholar]
- 2.Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377:562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 3.Dalmedico MM, Salas D, Oliveira AM, Baran FDP, Meardi JT, Santos MC. Efficacy of prone position in acute respiratory distress syndrome: overview of systematic reviews. Rev Esc Enferm USP. 2017;51:e03251. doi: 10.1590/S1980-220X2016048803251. [DOI] [PubMed] [Google Scholar]
- 4.Walkey AJ, Summer R, Ho V, Alkana P. Acute respiratory distress syndrome: epidemiology and management approaches. Clin Epidemiol. 2012;4:159–169. doi: 10.2147/CLEP.S28800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteban A, Anzueto A, Frutos F, Alía I, Brochard L, Stewart TE, Benito S, Epstein SK, Apezteguía C, Nightingale P, Arroliga AC, Tobin MJ Mechanical Ventilation International Study Group. Characteristics and outcomes in adult patients receiving mechanical ventilation: a 28-day international study. JAMA. 2002;287:345–355. doi: 10.1001/jama.287.3.345. [DOI] [PubMed] [Google Scholar]
- 6.Villar J, Blanco J, Anon JM, Santos-Bouza A, Blanch L, Ambros A, Gandia F, Carriedo D, Mosteiro F, Basaldua S, Fernandez RL, Kacmarek RM, Network A. The ALIEN study: incidence and outcome of acute respiratory distress syndrome in the era of lung protective ventilation. Intensive Care Med. 2011;37:1932–1941. doi: 10.1007/s00134-011-2380-4. [DOI] [PubMed] [Google Scholar]
- 7.Mann A, Early GL. Acute respiratory distress syndrome. Mo Med. 2012;109:371–375. [PMC free article] [PubMed] [Google Scholar]
- 8.Guo L, Zhao Y, Yang S, Zhang H, Chen F. An integrated analysis of miRNA, lncRNA, and mRNA expression profiles. Biomed Res Int. 2014;2014:345605. doi: 10.1155/2014/345605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anastasiadou E, Faggioni A, Trivedi P, Slack FJ. The nefarious nexus of noncoding RNAs in cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19072072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592:2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang Y, Wei X, Xue L, Wen F, Gu J, Zheng H. Long non-coding RNA in glioma: target miRNA and signaling pathways. Clin Lab. 2018;64:887–894. doi: 10.7754/Clin.Lab.2018.180107. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Liu J. [Research progress of competing endogenous RNA] . Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2017;34:967–971. doi: 10.7507/1001-5515.201606080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between long noncoding RNAs and MicroRNAs in health and disease. Int J Mol Sci. 2016;17:356. doi: 10.3390/ijms17030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 15.King VM, Borchert GM. MicroRNA expression: protein participants in MicroRNA regulation. Methods Mol Biol. 2017;1617:27–37. doi: 10.1007/978-1-4939-7046-9_2. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Li H, Jin L, Li G, Hu S, Ning C, Guo J, Shuai S, Li X, Li M. Long noncoding RNA GAS5 suppresses 3T3-L1 cells adipogenesis through miR-21a-5p/PTEN signal pathway. DNA Cell Biol. 2018;37:767–777. doi: 10.1089/dna.2018.4264. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Lv G, Wang B, Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503:2555–2562. doi: 10.1016/j.bbrc.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Liang S, Gong X, Zhang G, Huang G, Lu Y, Li Y. The lncRNA XIST interacts with miR-140/miR-124/iASPP axis to promote pancreatic carcinoma growth. Oncotarget. 2017;8:113701–113718. doi: 10.18632/oncotarget.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y, He R, An J, Deng P, Huang L, Yang W. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol Rep. 2017;38:941–948. doi: 10.3892/or.2017.5751. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, Qin G, Yang L, Xiang D, Li S. LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J Cell Physiol. 2019;234:8659–8667. doi: 10.1002/jcp.26327. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi: 10.1016/j.biopha.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Ma C, Liu C, Duan Z, Zhang L. Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem Biophys Res Commun. 2018;503:586–592. doi: 10.1016/j.bbrc.2018.06.042. [DOI] [PubMed] [Google Scholar]
- 23.Vieira RP, Muller T, Grimm M, von Gernler V, Vetter B, Durk T, Cicko S, Ayata CK, Sorichter S, Robaye B, Zeiser R, Ferrari D, Kirschbaum A, Zissel G, Virchow JC, Boeynaems JM, Idzko M. Purinergic receptor type 6 contributes to airway inflammation and remodeling in experimental allergic airway inflammation. Am J Respir Crit Care Med. 2011;184:215–223. doi: 10.1164/rccm.201011-1762OC. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilyugin M, Irminger-Finger I. Long non-coding RNA and microRNAs might act in regulating the expression of BARDS1 mRNAs. Int J Biochem Cell Biol. 2014;54:356–367. doi: 10.1016/j.biocel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Li Q, Wu X. Construction and analysis for differentially expressed long non-coding RNAs and MicroRNAs mediated competing endogenous RNA network in colon cancer. PLoS One. 2018;13:e0192494. doi: 10.1371/journal.pone.0192494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Wang G, Yin R, Qiu M, Xu L. [Comprehensive identification of microRNAs regulated by long non-coding RNA MALAT1] . Zhongguo Fei Ai Za Zhi. 2016;19:247–251. doi: 10.3779/j.issn.1009-3419.2016.05.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, Liao Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci Rep. 2017;37 doi: 10.1042/BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong Q, Zhang S, Liang C, Zhang Y, Kong Q, Chen S, Qin J, Jin Y. LncRNA XIST functions as a molecular sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular carcinoma cell. J Cell Biochem. 2018;119:4458–4468. doi: 10.1002/jcb.26540. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Ma L, Li C, Zhang Z, Yang G, Zhang W. Tumor-targeting TRAIL expression mediated by miRNA response elements suppressed growth of uveal melanoma cells. Mol Oncol. 2013;7:1043–1055. doi: 10.1016/j.molonc.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad R, Katiyar SK. Down-regulation of miRNA-106b inhibits growth of melanoma cells by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget. 2014;5:10636–10649. doi: 10.18632/oncotarget.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragusa M, Barbagallo C, Statello L, Caltabiano R, Russo A, Puzzo L, Avitabile T, Longo A, Toro MD, Barbagallo D, Valadi H, Di Pietro C, Purrello M, Reibaldi M. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biol Ther. 2015;16:1387–1396. doi: 10.1080/15384047.2015.1046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sand M, Skrygan M, Georgas D, Sand D, Gambichler T, Altmeyer P, Bechara FG. The miRNA machinery in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases and benign melanocytic nevi. Cell Tissue Res. 2012;350:119–126. doi: 10.1007/s00441-012-1446-0. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Ma K, Zhang S, Zhang H, Liu J, Wang X, Li S. Pulmonary microRNA expression profiling in an immature piglet model of cardiopulmonary bypass-induced acute lung injury. Artif Organs. 2015;39:327–335. doi: 10.1111/aor.12387. [DOI] [PubMed] [Google Scholar]
- 37.Xin J, Zheng LM, Sun DK, Li XF, Xu P, Tian LQ. miR-204 functions as a tumor suppressor gene, at least partly by suppressing CYP27A1 in glioblastoma. Oncol Lett. 2018;16:1439–1448. doi: 10.3892/ol.2018.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N, Zhang RF, Zhang AN, Dong GX, Suo N, Wu ZP, Liu YM, Wang LT. MiR-204 promotes fracture healing via enhancing cell viability of osteoblasts. Eur Rev Med Pharmacol Sci. 2018;22:29–35. doi: 10.26355/eurrev_201807_15356. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Luo H, Fang Z, Fan Y, Liu X, Zhang Y, Rui S, Chen Y, Hong L, Gao J, Zhang M. MiR-204 acts as a potential therapeutic target in acute myeloid leukemia by increasing BIRC6-mediated apoptosis. BMB Rep. 2018;51:444–449. doi: 10.5483/BMBRep.2018.51.9.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Fajol A, Tu X, Ren B, Shi S. miR-204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7:70058–70065. doi: 10.18632/oncotarget.11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Lv HY, Zhou DM, Zhang EN. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15026415. [DOI] [PubMed] [Google Scholar]
- 42.Chen SJ, Wu P, Sun LJ, Zhou B, Niu W, Liu S, Lin FJ, Jiang GR. miR-204 regulates epithelial-mesenchymal transition by targeting SP1 in the tubular epithelial cells after acute kidney injury induced by ischemia-reperfusion. Oncol Rep. 2017;37:1148–1158. doi: 10.3892/or.2016.5294. [DOI] [PubMed] [Google Scholar]
- 43.Gao PS, Leung DY, Rafaels NM, Boguniewicz M, Hand T, Gao L, Hata TR, Schneider LC, Hanifin JM, Beaty TH, Beck LA, Weinberg A, Barnes KC. Genetic variants in interferon regulatory factor 2 (IRF2) are associated with atopic dermatitis and eczema herpeticum. J Invest Dermatol. 2012;132:650–657. doi: 10.1038/jid.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawasaki A, Furukawa H, Nishida N, Warabi E, Kondo Y, Ito S, Matsumoto I, Kusaoi M, Amano H, Suda A, Nagaoka S, Setoguchi K, Nagai T, Hirohata S, Shimada K, Sugii S, Okamoto A, Chiba N, Suematsu E, Ohno S, Katayama M, Okamoto A, Kono H, Tokunaga K, Takasaki Y, Hashimoto H, Sumida T, Tohma S, Tsuchiya N. Association of functional polymorphisms in interferon regulatory factor 2 (IRF2) with susceptibility to systemic lupus erythematosus: a case-control association study. PLoS One. 2014;9:e109764. doi: 10.1371/journal.pone.0109764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce MM, Burghardt JR, Burghardt RC, Hooper RN, Jaeger LA, Spencer TE, Bazer FW, Johnson GA. Pig conceptuses increase uterine interferon-regulatory factor 1 (IRF1), but restrict expression to stroma through estrogen-induced IRF2 in luminal epithelium. Biol Reprod. 2007;77:292–302. doi: 10.1095/biolreprod.107.060939. [DOI] [PubMed] [Google Scholar]
- 46.Maria A, Agarwal S, Sharma A. Acute respiratory distress syndrome in a neonate due to possible transfusion-related acute lung injury. Asian J Transfus Sci. 2017;11:203–205. doi: 10.4103/ajts.AJTS_120_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerget B, Araz O, Ucar EY, Akgun M, Saglam L. Acute respiratory distress syndrome; A rare complication caused by usage of ruxolitinib. Respir Med Case Rep. 2017;22:243–245. doi: 10.1016/j.rmcr.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ben Salem C. Acute respiratory distress syndrome. N Engl J Med. 2017;377:1904. doi: 10.1056/NEJMc1711824. [DOI] [PubMed] [Google Scholar]
- 49.Bilan N, Dastranji A, Ghalehgolab Behbahani A. Comparison of the spo2/fio2 ratio and the PaO2/FiO2 ratio in patients with acute lung injury or acute respiratory distress syndrome. J Cardiovasc Thorac Res. 2015;7:28–31. doi: 10.15171/jcvtr.2014.06. [DOI] [PMC free article] [PubMed] [Google Scholar]


