Abstract
Introduction: Lymph node metastasis is the primary cause of death in oral squamous-cell carcinoma (OSCC) patients, so understanding the underlying molecular mechanism is critical for treating metastatic OSCC. OVOL1, a transcription factor, functions as a “break” to repress metastasis in breast cancer and prostate cancer. Aims: To explore the roles of OVOL1 in the progression of OSCC, especially during metastasis. Results: The OVOL1 level was increased significantly in non-metastatic OSCC tissues and negatively correlated with ZEB1 level. OVOL1 repressed ZEB1 expression by directly binding to the promoter region of ZEB1. OVOL1 functioned as a tumor suppressor, and suppressed SCC-152 cells proliferation, migration, and invasion and promoted apoptosis. ZEB1 almost fully rescued the overexpressed OVOL1 function in SCC-152 cells. Conclusion: OVOL1 overexpression contributes to the progression of OSCC through inhibiting ZEB1, which may provide a marker for prognosis in OSCC.
Keywords: Oral squamous cell carcinoma, OVOL1, ZEB1, metastasis
Introduction
Oral squamous cell carcinoma (OSCC), a malignant neoplasm of stratified squamous epithelium in the oral cavity, is the most common type of oral cancer, accounting for more than 90% of all oral cancers and 38% of head and neck tumors [1]. Although surgery and chemotherapy have improved over the past decades, oral cancer is still closely related to poor prognosis and low survival rate [2]. Patients with tumor at an advanced stage usually suffer from tumor cell invasion to surrounding tissues, and have an extremely high risk of second malignancy during the patient’s lifetime [3]. Therefore, it is important to understanding the mechanisms driving OSCC progression and metastasis.
Epithelial to mesenchymal transition (EMT) allows cancer cells to dedifferentiate and acquire enhanced migratory and invasive properties which play an important role during tumor metastasis [4]. Hence, deciphering the molecular mechanisms that maintain the hybrid epithelial/mesenchymal phenotype may provide a new choice for anti-metastasis strategies.
OVO-like proteins (OVOL) are a group of zinc finger protein family transcription factors that regulate gene expression in various differentiation processes [5]. Increasing evidence indicates that OVOL genes, especially OVOL1 and OVOL2, are involved in EMT in human cancer including breast cancer and prostate cancer [6]. It is known that OVOL proteins may function as an EMT “break” through repressing ZEB1 and inducing miR-200 family members. However, the role of OVOL proteins during carcinogenesis of OSCC is still unknown.
In this study, we analyzed the expression of OVOL1, OVOL2, and ZEB1 in metastatic and non-metastatic OSCC tissues. The regulatory relationships between OVOL1 and ZEB1 were examined in OSCC cells.
Materials and methods
Patients
Fifty patients with OSCC (25 non-metastasis and 25 lymph node metastasis) were recruited for OVLO1, and OVLO2 levels detection. The clinical characteristics are shown (Table 1). Primary tumor tissue samples were collected from patients undergoing surgical resection of primary tumor at Affiliated Hospital of Stomatology, Nanjing Medical University. Written informed consents were obtained from all patients and the research protocol was approved by review boards and by the Ethics Committee in Research of Affiliated Hospital of Stomatology, Nanjing Medical University. Tumor samples were snap-frozen in liquid nitrogen immediately after surgery. Analysis of hematoxylin and eosin-stained sections by the study pathologists confirmed at least 70% of tumor cells in all OSCC samples.
Table 1.
Clinical characteristics of patients with OSCC in this study
| Patient | Site | Gender | Age (yr) | Pathologic stage | HSV infection |
|---|---|---|---|---|---|
| P0032 | OC-T | M | 63 | T1N0M0 | HSV1 |
| P0052 | OC-T | M | 49 | T2N0M0 | Negative |
| P0043 | OC-FOM | M | 55 | T4N0M0 | Negative |
| P0106 | OC-T | F | 53 | T1N0M0 | Negative |
| P0112 | OC-FOM | M | 69 | T2N0M0 | HSV1 |
| P0156 | OC-FOM | M | 63 | T1N0M0 | Negative |
| P0193 | OC-T | M | 57 | T2N0M0 | Negative |
| P0266 | OC-T | M | 44 | T2N0M0 | Negative |
| P0372 | OC-T | M | 57 | T1N0M0 | Negative |
| P0392 | OC-T | M | 61 | T3N0M0 | HSV1 |
| P0397 | OC-FOM | M | 66 | T1N0M0 | Negative |
| P0432 | OC-FOM | M | 55 | T3N0M0 | HSV2 |
| P0439 | OC-FOM | M | 58 | T2N0M0 | Negative |
| P0444 | OC-T | M | 60 | T2N0M0 | Negative |
| P0531 | OC-T | M | 54 | T4N0M0 | Negative |
| P0536 | OC-T | M | 62 | T2N0M0 | Negative |
| P0621 | OC-FOM | M | 58 | T1N0M0 | HSV1 |
| P0775 | OC-T | F | 71 | T2N0M0 | HSV1 |
| P0812 | OC-T | F | 68 | T2N0M0 | Negative |
| P0843 | OC-T | M | 63 | T3N0M0 | Negative |
| P0915 | OC-FOM | M | 66 | T1N0M0 | Negative |
| P0953 | OC-T | F | 54 | T3N0M0 | Negative |
| P1016 | OC-FOM | M | 59 | T3N0M0 | Negative |
| P1083 | OC-T | M | 63 | T2N0M0 | HSV1 |
| P1106 | OC-T | M | 63 | T1N0M0 | Negative |
| P0008 | OC-FOM | M | 55 | T4N1M1 | Negative |
| P0011 | OC-FOM | M | 58 | T3N1M0 | Negative |
| P0018 | OC-T | M | 62 | T2N1M1 | Negative |
| P0028 | OC-T | M | 55 | T3N2M0 | Negative |
| P0037 | OC-T | F | 66 | T2N2M1 | HSV1 |
| P0055 | OC-T | M | 69 | T3N1M0 | Negative |
| P0059 | OC-FOM | M | 56 | T4N3M1 | Negative |
| P0064 | OC-FOM | M | 58 | T3N1M1 | HSV1 |
| P0066 | OC-FOM | M | 61 | T2N1M0 | Negative |
| P0087 | OC-T | F | 57 | T3N2M1 | Negative |
| P0091 | OC-T | F | 60 | T3N3M1 | Negative |
| P0094 | OC-T | M | 62 | T2N1M0 | HSV2 |
| P0119 | OC-T | M | 59 | T2N2M0 | Negative |
| P0143 | OC-FOM | F | 70 | T4N3M1 | Negative |
| P0195 | OC-T | M | 68 | T3N2M1 | Negative |
| P0229 | OC-T | M | 59 | T3N1M0 | Negative |
| P0398 | OC-FOM | M | 62 | T2N1M0 | Negative |
| P0463 | OC-T | M | 55 | T2N2M1 | Negative |
| P0555 | OC-FOM | M | 47 | T1N3M1 | HSV2 |
| P0588 | OC-T | M | 52 | T2N1M0 | Negative |
| P0667 | OC-T | M | 49 | T1N2M0 | Negative |
| P0687 | OC-T | M | 64 | T2N2M0 | HSV1 |
| P0844 | OC-FOM | M | 63 | T4N2M1 | HSV1 |
| P0933 | OC-T | M | 65 | T3N1M0 | Negative |
| P1089 | OC-T | M | 59 | T3N2M1 | HSV1 |
OC-T: Oral-Cavity - Tongue, OC-FOM: Oral Cavity - Floor of the Mouth. Pathological Stage describes the metastatic status of the samples, with N0 meaning no lymph node metastasis at the time of diagnosis and N>0 indicating the presence of lymph node metastasis.
DNA extraction and PCR amplification
50 micron sections were cut from each tissue specimen block and DNA was extracted using RNAprep pure Tissue Kit (TIANGEN BioTec, Beijing, China). PCR was performed by processing the extracted DNA with primers that are specific for HSV-1 and HSV-2 genes. The primers used for HSV1 are Forward: GTTAGGGAGTTGTTCGGTCATAAGCT, Reverse: TCGGCCATCTTGAGCATCC; the HSV2 primers are: Forward: GTCGGTGTGGTGTTCGGTCATAAGCT, Reverse: GGCTGAATCTGGTAAACACGCTTC.
Cell culture
Oral squamous cell carcinoma (OSCC) SCC-152 cells were obtained from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA). Human embryonic kidney 293T cells were cultured in DMEM medium containing 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 IU/ml penicillin and 10 mg/mL streptomycin. All cells were cultured at 37°C under an atmosphere containing 5% CO2.
Western blotting
To examine the level of target genes, protein was extracted from tissues or cells and then boiled in SDS/β-mercaptoethanol sample buffer. 20 μg protein samples were loaded into each lane of 10% polyacrylamide gels. After electrophoresis, the proteins in the gels were blotted onto PVDF membranes by electrophoretic transfer. Rabbit anti-OVOL1 polyclonal antibody (Abcam, Cambridge, MA, USA), rabbit anti-OVOL2 monoclonal antibody (Abcam, Cambridge, MA, USA) or mouse anti-GAPDH monoclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were used for BRCA1, BRCA2 and β-actin level detection separately. The specific protein-antibody complex was detected by incubating with horseradish peroxidase conjugated goat anti-rabbit (Abcam, Cambridge, MA, USA) or rabbit anti-mouse IgG (Abcam, Cambridge, MA, USA). The chemiluminescence reaction was carried out using the ECL kit (Pierce, Appleton, WI, USA).
Dual luciferase assay
A segment of 332 bp DNA of the ZEB1 promoter region was cloned into upstream of firefly luciferase coding region in pGL3-basic vector (Promega, Madison, WI, USA) to generate luciferase reporter vector.
HEK293T cells were seeded in 48-well plates and allowed to attach overnight. OVOL1 expression vector or siRNA was transfected with 100 ng of reporter vector by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Empty plasmid or sequence-scrambled short RNA was used as control. Cells were harvested, lysed, and assayed using Dual-Luciferase Assay kit (Promega, Madison, WI, USA) two days after transfection. Each treatment was performed in triplicate in three independent experiments. The results were expressed as relative luciferase activity (Firefly LUC/Renilla LUC).
Cell proliferation assay
SCC-152 cells were seeded in 96-well plates at a density of 5×103 cells per well in RPMI-1640 medium, and allowed to attach overnight. OVOL1 expression vector or siRNA was transfected into cells, with empty plasmid or sequence scrambled short RNA as control. Twenty microliters MTT (5 mg/ml) (Sigma, St. Louis, MO, USA) were added into each well two days after transfection, and incubated for further 2 hours. The absorbance was recorded at A570 nm with a 96-well plate reader after DMSO addition.
Cell apoptosis assay
Cell apoptosis was measured using Annexin V-fluorescein isothiocyanate/prepidium iodide (PI) double staining system (BD Bioscience) according to the manufacturer’s procedure.
Cell migration and invasion assay
3×104 SCC-152 cells were seeded into 12-well plates and allowed to attach overnight. The cells were transfected with siRNA, OVOL1, or ZEB1 overexpression vector for 48 hours. Cells were harvested and seeded in the top chamber of a typical 24-well transwell plate, and incubated for 12 hours. Filters were then submerged in formalin for 15 min and cells stained by hematoxylin-eosin and calculated. For cell invasion assays, the procedure was similar to the cell migration assay, except transwell membranes were pre-coated with 24 μg/μl Matrigel (BD Bioscience, Franklin Lakes, NJ, USA).
Chromatin Immunoprecipitation (ChIP)
5×106 cells were used for each assay. Cells were first cross-linked by 1% formaldehyde at room temperature for 5 minutes and then resuspended in lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1)] and incubated for 15 minutes on ice. After being sonicated and centrifuged at 13,000 rpm at 4°C for 10 minutes, the supernatant of the lysate was diluted 10 times in the dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl (pH 8.1)] and was incubated for 1 hour on a rotating platform at 4°C with Protein-A Sepharose. The precleared chromatin was incubated with 1 mg OVOL1 atibody or IgG for 8 hours at 4°C and the immune complexes were collected with Protein-A Sepharose. After three washes with the LiCl Buffer [0.25 M LiCl, 1% Triton X-100, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl (pH 8.1)] and twice with the TE buffer [1 mM EDTA, 10 mM Tris-HCl (pH 8)], DNA was extracted by phenol-chloroform and precipitated by ethanol. Quantitative real-time PCR was performed with SYBR-Green PCR mix.
Statistical analysis
Results were analyzed using SPSS Statistical Package version 15. Student’s t-test was used to analyze independent two groups and χ2-analysis was used to analyze the correlation. P<0.05 was considered significant.
Results
To investigate the roles of OVOL proteins during the progression of OSCC, we collected clinical samples from 50 OSCC patients (25 non-metastasis and 25 lymph node metastasis). Protein was extracted and the levels of OVOL1, OVOL2 and ZEB1 were examined by western blot in 18 OSCC patients (9 non-metastasis and 9 lymph node metastasis) and then the level of OVOL1 and ZEB1 was detected in a further group of 32 OSCC patients (16 non-metastasis and 16 lymph node metastasis). The relative protein levels were quantified and results were analyzed using Student’s t-test. As shown in Figures 1A, 1B, and S1, the OVOL1 level was significantly reduced in the metastatic OSCC samples with a statistical power higher than 0.9. Because of a reported negative regulatory relationship between OVOL1 and ZEB1 in other cancer cells, we wanted to know whether the ZEB1 expression was regulated by OVOL1 in OSCC patients. The ZEB1 protein level was detected and we observed increased ZEB1 level in metastatic OSCC tissues (Figure 1A) and a significant negative correlation existed between OVOL1 and ZEB1 protein level (Figure 1C).
Figure 1.
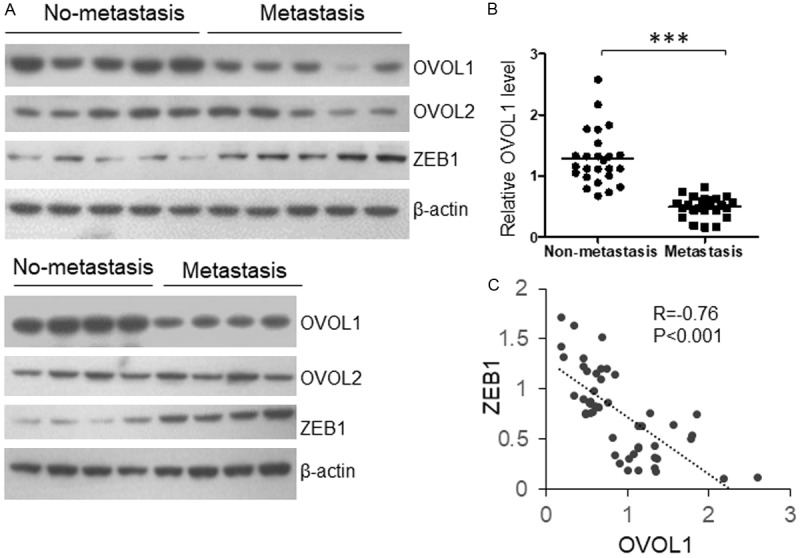
OVOL1 level is reduced in metastatic OSCC samples and negatively correlated with ZEB1 protein level. OSCC tissue samples were collected from 25 metastatic and 25 non-metastatic OSCC patients. The protein levels of OVOL1 and ZEB1 were determined by immunoblotting, and results were quantified by using Quantity One.
It is reported that OVOL1 binds a site in the promoter region of ZEB1, +320 to 340 [4]. We did the Chip assay by using OVOL1 antibody and SCC-152 cells to investigate this relationship in OSCC. As shown in Figure 2A, OVOL1 was successfully recruited by OVOL1 antibody. PCR product b (+382 to +530) was 23.13 fold higher than the IgG control, but PCR product a (-765 to 658) was not recruited by OVOL1 antibody, indicating that the binding site is located near the product b region, probably +320 to 340.
Figure 2.
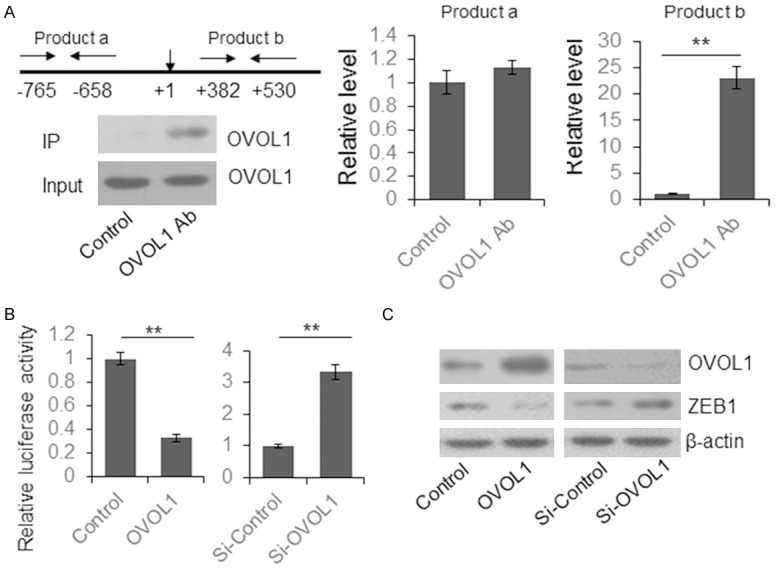
OVOL1 repressed ZEB1 expression by direct binding to the promoter region. A. The binding of OVOL1 to ZEB1 promoter region was detected by ChIP assay. The protein level of OVOL1 in the input and IP samples were determined by immunoblotting. Two segments near the ZEB1 promoter region were quantified by qRT-PCR. B. The effect of OVOL1 binding to the ZEB1 promoter region was determined by dual luciferase assay. C. The OVOL1 was overexpressed or knocked down in SCC-152 cells and the ZEB1 level was determined by immunoblotting.
To further identify the regulatory modules of OVOL1 and ZEB1, a 332 bp region of ZEB1 promoter region containing the OVOL1 binding site was cloned into pGL3-basic vector to construct a reporter plasmid. HEK293T cells were transiently transfected with ZEB1 reporter vector with OVOL1 expression or control vector for 48 hours, and then lysed for dual luciferase assay. We observed a significantly reduced luciferase activity in the OVOL1 overexpression group. When the endogenous OVOL1 was knocked down by siRNA, the luciferase activity was increased (Figure 2B). To further verify the repressive effect of OVOL1 on ZEB1 at the protein level, OVOL1 was overexpressed or knocked down in SCC-152 cells. The OVOL1 and ZEB1 levels were determined by immunoblotting. We observed reduced ZEB1 level in the OVOL1-overexpressed cells and increased ZEB1 level in the OVOL1-knocked down cells (Figure 2C).
To explore the biologic function of OVOL1 in OSCC, OVOL1 was overexpressed or knocked down in SCC-152 cells, and the cell viability was determined by MTT assay. We observed decreased cell viability in the OVOL1 overexpressed cells; and in the OVOL1 knocked down cells, the cell viability was significantly increased (Figure 3A). Cell apoptosis was determined by Annexin V antibody-based flow cytometry. The number of apoptotic cells was increased in the OVOL1 overexpression group and reduced in OVOL1 knocked down group (Figure 3B). Subsequently, the cell migration and invasion were determined by transwell assay. As shown in Figure 3C and 3D, the OVOL1 overexpressed SCC-152 cells have increased migration and invasive cell number, and OVOL1 knocked down cells have decreased migration and invasive cell number. These results indicated that OVOL1 functions as a tumor suppressor in OSCC.
Figure 3.
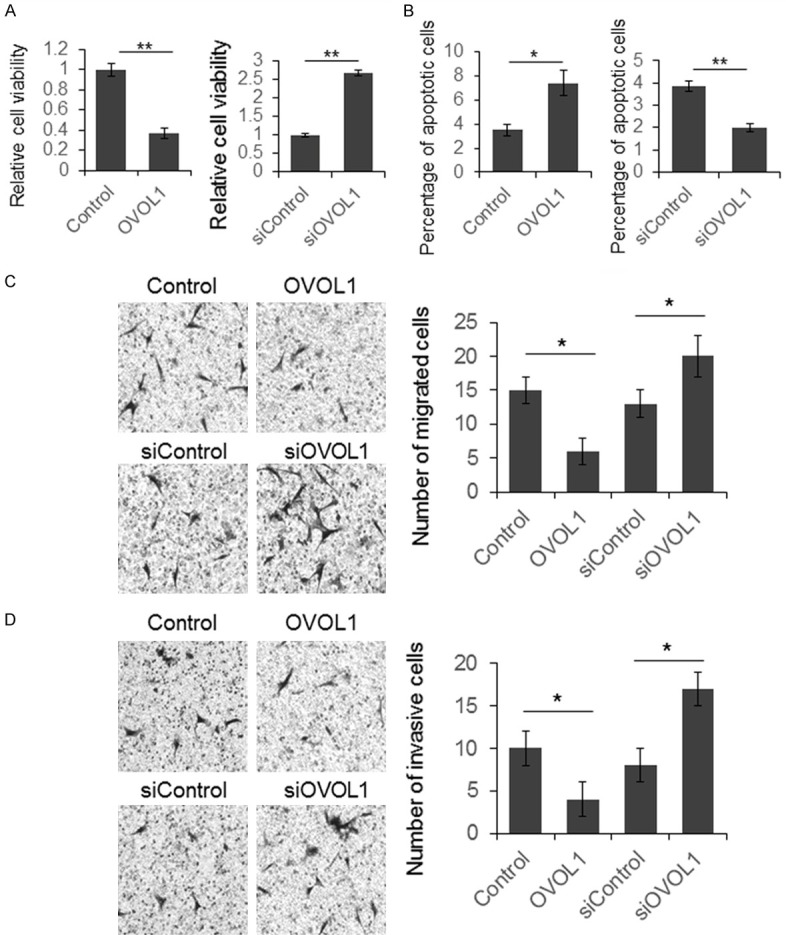
OVOL1 repressed OSCC cell proliferation, migration, invasion, and induced cell apoptosis. SCC-152 cells were transfected with OVOL1 expression vector or siRNA for 48 hours and then the cells were subjected to proliferation (A), apoptosis (B), migration (C) and invasion (D) assays. Results were analyzed by Student t test and P<0.05 was considered significant. *P<0.05, **P<0.01.
To further identify whether OVOL1 represses OSCC growth by regulating ZEB1, we constructed a ZEB1 expression vector. SCC-152 cells were transfected with OVOL1 and ZEB1 expression vector or just OVOL1 expression vector for 48 hours, with cells transfected with empty vector as control. The cell viability, apoptosis, migration, and invasion capacity were detected. As shown in Figure 4A, the repressed cell viability caused by OVOL1 was almost fully rescued by ZEB1. Meanwhile, the up-regulated cell apoptosis by OVOL1 was partially reduced by ZEB1 (Figure 4B). The ZEB1 overexpression also rescued the repressive function of OVOL1 on cell migration and invasion (Figure 4C and 4D).
Figure 4.
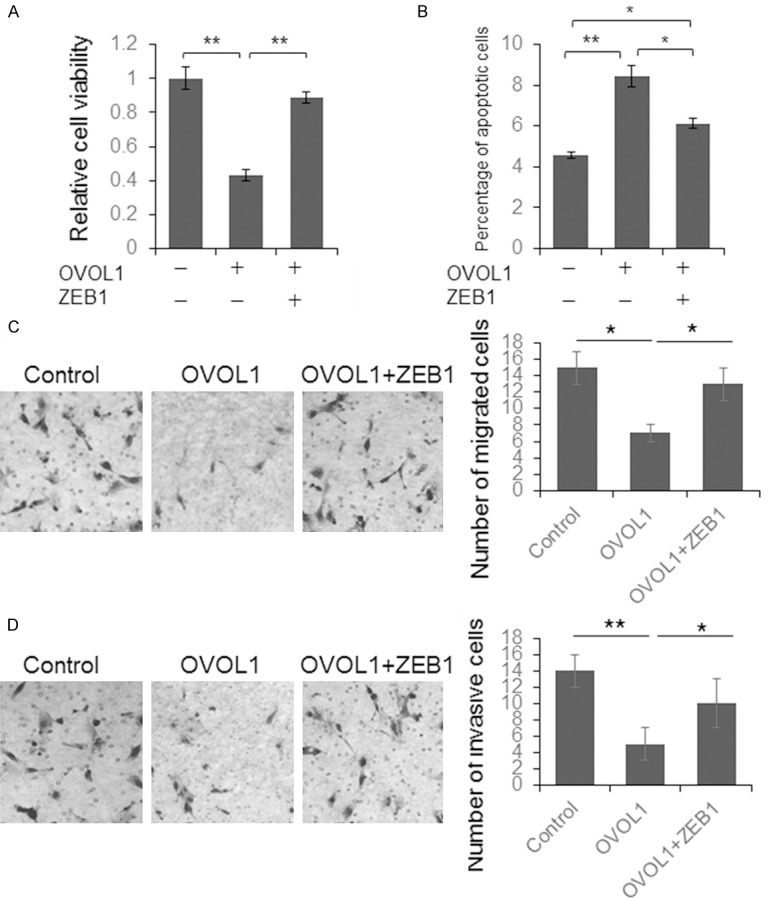
ZEB1 partially rescued the tumor suppressor function. SCC-152 cells were transfected with OVOL1 expression vector, or OVOL1 plus ZEB1 vectors for 48 hours and then the cells were subjected to proliferation (A), apoptosis (B), migration (C) and invasion (D) assay. Results were analyzed by Student t test and P<0.05 was considered significant. *P<0.05, **P<0.01.
Discussion
Lymph node metastasis is the primary cause of death in OSCC patients, so understanding the underlying molecular mechanism is critical. OVOL1 is a transcription factor, that functions as a “break” to repress the cell metastasis in breast and prostate cancers [4]. To explore the role of OVOL1 in the progression of OSCC, especially during the metastasis progress, we first detected the protein level of OVOL1 and OVOL2 in the metastases and non-metastatic OSCC tissues. We found OVOL1 level was increased significantly in the non-metastatic OSCC tissues and negatively correlated with ZEB1 level. Through ChIP and dual luciferase assay, we confirmed that OVOL1 repressed ZEB1 expression by directly binding to the promoter region of ZEB1. Subsequent functional study confirmed that OVOL1 functioned as a tumor suppressor, suppressed SCC-152 cell proliferation, migration, and invasion and promoted cell apoptosis. Interestingly, ZEB1 almost fully rescued the overexpressed OVOL1 function in SCC-152 cells.
ZEB1 is a transcription factor which can inhibit E-cadherin expression. Previous studies indicated that ZEB1 overexpression was significantly correlated with aggressiveness and poor clinical prognosis in many kinds of human malignancies including OSCC [7-9]. Our present study confirmed the negative correlation between OVOL1 and ZEB1 in OSCC clinical samples and cells for the first time. Furthermore, we confirmed that OVOL1 repressed OSCC cell proliferation, migration, and invasion mainly through inhibiting ZEB1 expression.
In summary, our study found that higher OVOL1 expression contributes to the progression of OSCC through inhibiting ZEB1, which may provide markers for prognostic prediction in OSCC.
Acknowledgements
This work is supported by Nature Science Funds of Jiangsu province with the number of BK20151516.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Pereira MC, Oliveira DT, Landman G, Kowalski LP. Histologic subtypes of oral squamous cell carcinoma: prognostic relevance. J Can Dent Assoc. 2007;73:339–344. [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H, Maher CA, Daignault-Newton S, Healy PN, Pienta KJ. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One. 2013;8:e76773. doi: 10.1371/journal.pone.0076773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair M, Teng A, Bilanchone V, Agrawal A, Li B, Dai X. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J Cell Biol. 2006;173:253–264. doi: 10.1083/jcb.200508196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe K, Villarreal-Ponce A, Sun P, Salmans ML, Fallahi M, Andersen B, Dai X. Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell. 2014;29:59–74. doi: 10.1016/j.devcel.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mock K, Preca BT, Brummer T, Brabletz S, Stemmler MP, Brabletz T. The EMT-activator ZEB1 induces bone metastasis associated genes including BMP-inhibitors. Oncotarget. 2015;6:14399–14412. doi: 10.18632/oncotarget.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran J, Lin DL, Wu RF, Chen QH, Huang HP, Qiu NX, Quan S. ZEB1 promotes epithelial-mesenchymal transition in cervical cancer metastasis. Fertil Steril. 2015;103:1606–1614. doi: 10.1016/j.fertnstert.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Yao X, Sun S, Zhou X, Zhang Q, Guo W, Zhang L. Clinicopathological significance of ZEB-1 and E-cadherin proteins in patients with oral cavity squamous cell carcinoma. Onco Targets Ther. 2017;10:781–790. doi: 10.2147/OTT.S111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


