Abstract
Sirt6 is a vital member of the Sirtuin family that plays a key role in cellular apoptosis, aging, DNA damage repair, telomere homeostasis and integrity, energy metabolism, glucose homeostasis, and gene regulation. In recent studies, Sirt6 is down-regulated in several cancers and predicted to be a tumor suppressor, but as a tumor oncogene in other cancers. In this study, we explored the specific role of Sirt6 in human renal cell carcinoma (RCC). We found that Sirt6 was up-expressed in renal tumor tissues and cells. Sirt6 silence in RCC led to G1/S phase arrest, a rise in apoptosis and a decrease in cell viability, as well as an enhancement of chemotherapeutic sensitivity. In conclusion, these findings suggest that Sirt6 acted as an oncogene in human RCC and it could be a potential target for RCC treatment.
Keywords: Sirt6, renal cancer, cell cycle, apoptosis, chemoresistance
Introduction
Renal cell carcinoma (RCC) is one of the most frequent kidney cancer in adults’ malignant tumor [1]. It accounts for approximately 3% of adult malignancies [1]. RCC is the 6th major cause of cancer-related deaths in western countries [2]. To date, although much effort has been paid to develop new treatment of RCC in the past decades, the pathogenesis of RCC is unclear and potential biomarkers for prognosis and novel therapeutic targets are needed to be identified.
Sirtuins are a family of highly conserved gene family which was associated with diverse biological processes and diseases [3,4]. Seven members (SIRT1-7) have been identified in mammalian [5]. Sirtuin 6 (Sirt6), a stress responsive protein deacetylase and mono-ADP ribosyltransferase enzyme, functions in vary biological processes and cell signaling pathways, including DNA damage repair, telomere maintenance, glycolysis, aging, and cancer. Sirt6 deacetylates the histone H3 on acetylated K9, K18, K56 [6-8]. Evidence suggests that the functions of Sirt6 may be tissue and context dependent in carcinogenesis [9], and it plays a crucial and controversial role in the regulation of tumorigenesis through its implication in different biological pathways where it can act as a tumor suppressor or oncogene. There is evidence that Sirt6 deficiency suppresses tumor growth [9-12]. Silence of Sirt6 induced sub-G1 phase arrest, increased apoptosis due to prominent DNA damage, and deregulated BCL2 in human prostate cancer cells [13]. Lee N. et al. observed that Sirt6 depletion inhibited tumor growth by p16/Rb- and p53/p21-independent cellular senescence, G2/M phase arrest, and down-regulated histone variants associated with nucleosome assembly in hepatocellular carcinoma (HCC), which was attributed to DNA damage [14]. In addition, the expression of Sirt6 is also related to the degree of malignancy of the tumors [11,15]. Patients who had Sirt6 expressed high cytoplasm and low nucleus, had more malignant cancer with poor overall survival, and recurrence-free survival, and in vitro analysis revealed that Sirt6 knockdown enhanced paclitaxel sensitivity in lung adenocarcinoma cell [16]. However, few studies have investigated the role of Sirt6 in RCC.
In this study, we have applied both RCC tissues and Renal cancer cell lines to investigate the regulation of Sirt6 in RCC. High levels of Sirt6 expression were observed in RCC tissues and cells, Sirt6 was further overexpressed or silenced in RCC cells to reveal the effects of Sirt6 on RCC, including proliferation, cell cycle, apoptosis, DNA damage, BCL2 gene expression and chemotherapeutic drug resistance. Our findings uncover that Sirt6 is essential for renal cancer progression and might provide a new target for renal cancer therapy.
Materials and methods
Cell culture
Human renal cancer cell lines (786-O) were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were cultured in RPMI 1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY) and 1% penicillin/streptomycin (Hyclone, Logan, UT, USA) at 37°C in a humidified incubator under 5% CO2.
Antibodies and reagents
The primary antibodies were as follows: Sirt6 (D8D12, 1:1000, CST); BCL2 (2872T, 1:1000, CST); p21 (2947, 1:1000, CST); Cas9 (14697, 1:1000, CST) β-Tubulin (KM9003T, 1:2000, Sungene Biotech). The secondary antibodies were obtained from Sungene Biotech (Tianjin, China). Doxorubicin were obtained from Sigma-Aldrich (St. Louis, MO, USA); EDU assay kit was purchased from Guangzhou RiboBio Co. (Guangzhou, China). Annexin V-PE/7-AAD Apoptosis kit was purchased from Key Gene (Nanjing, China).
Western blot
Whole cell proteins were lysed in RIPA buffer containing Protease Inhibitor Cocktail. The supernatant protein concentration was measured by BCA assay (Millipore, CA, USA). Thirty micrograms of total protein were loaded and separated by 12% SDS-PAGE and then transferred to PVDF Membrane (0.22 μm) (Millipore, CA, USA). The membrane was blocked with TBS/T containing 5% non-fat milk at room temperature for 1 h, incubated with primary antibodies with gentle shake at 4°C overnight, and then incubated with appropriate HRP-conjugated secondary antibody at room temperature for 1 h. Target signals were visualized using ECL Western Blotting Substrate (Millipore, CA, USA) and bio-rad ChemiDoc™ image system (Bio-Rad Laboratories, Inc., CA, USA).
Tissue pathological analysis
Hematoxylin and eosin (H&E) staining and Sirt6 IHC were performed by the Immunohistochemistry core facility. Briefly, specimens of kidney tissue were fixed in 10% formalin and then embedded with paraffin. H&E staining and IHC were used for histologic analysis.
Infection and transfection
Sirt6 was overexpressed via infection of ad-Sirt6 (adenovirus-Sirt6) at a cell confluence of 70-80%, and ad-GFP-infected cells as the control. Over 95% of the cells were viable in 24 h and followed by another 3 days’ incubation. Infection efficiency was measured by western blotting. For knockdown of Sirt6, cells at 70% confluency were transfected with 100 nmol/L siRNA by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocols. Target sequence for Sirt6: 5’-GAAUGUGCCAAGUGUAAGATT-3’; negative control: 5’-CGACAUACUGUACAGGCCUTT-3’.
Cell cycle analysis by flow cytometry
After 48 h of Sirt6 siRNA-transfection, 786-O cells were harvested, and fixed in 70% ethanol at 4°C overnight. Then the cells were stained in PBS containing 40 μg/mL propidium iodide (PI), 100 μg/mL RNase A and 0.1% Triton X-100 for 30 min at room temperature. DNA content was measured by Cytoflex (Beckman Coulter, Miami, FL, USA), and analyzed with CytExpert.
Apoptosis detected by flow cytometry
Apoptosis was calculated by the Annexin V/7-AAD double staining Apoptosis Detection Kit (Key Gene, Nanjing, China). Sirt6 siRNA-transfected 786-O cells were harvested, resuspended in binding buffer, stained with Annexin V for 15 min at room temperature, then stained with 7-AAD for 5 min, and subjected to flow cytometer (Beckman Coulter, Miami, FL, USA).
Colony formation assay
786-O cells (500-1000 cells/well) were seeded into 6-well plates and cultured overnight. The cells were transfected with siSirt6- or siCtl-siRNA and cultured for 10 days. The cells were fixed in methanol for 15 min and stained with 0.1% crystal violet for 30 min at room temperature with gentle shake. Images were captured after washing with PBS followed by air-drying.
Cell proliferation assay
786-O cells were seeded at a density of 5 × 104 cells in a 12-well plate and transfected with siRNA. After 48-h incubation, cells were stained with EDU assay kit, and detected by flow cytometry.
Statistical analysis
The data were analyzed using SPSS Statistics v24 (IBM, New York, USA). Comparisons were performed by using unpaired Student’s t-test or one-way ANOVA. P < 0.05 was considered statistically significant difference.
Results
Sirt6 is up-regulated in renal cancer tissues and 786-O cells
To identify Sirt6 expression pattern in human renal tumor tissues and cells. We analyzed Sirt6 expression patterns in two pairs of renal tumor tissues from patients, and the normal tissues para-tumor were used as the control. We observed that Sirt6 protein levels were higher in tumor tissues than the control tissues (Figure 1A). These tissue samples were further stained with a special Sirt6 antibody, Sirt6 expression was revealed to be stronger in renal tumor tissues, compared with the normal renal tissues (Figure 1B). In addition, Sirt6 protein levels were significantly higher in renal cancer cell line 786-O cells than in the control, normal renal cells HK cells (Figure 1C and 1D). These data suggested that Sirt6 was abnormally high expressed in renal cancer tissues and cells.
Figure 1.

Sirt6 mRNA and protein expression were up-regulated in renal cancer tissues and cells. A. Sirt6 protein expression was up-regulated in renal tumor tissues from patients (T) compared to the paired non-tumor tissues (N). B. Immunohistochemistry analysis showed strong positive signal of Sirt6 in tumor tissues (scale bar: 50 μm). C. The expression of Sirt6 was increased in 786-O renal cancer cells, compared with normal control renal cell line HK-2 cells by Western blotting assay. β-tubulin was used as an internal control. D. Sirt6 mRNA expression was up-regulated in 786-O renal cancer cells, compared to control HK-2 cells. All data were presented as mean ± SEM. and analyzed by student’s t-test, *P < 0.05. N, none tumor; T, tumor.
Sirt6 overexpression reduces apoptosis in renal cancer cells
To further investigate the biological role of Sirt6 in the renal cancer, we overexpressed Sirt6 in 786-O cells by infecting adenovirus packaged with Sirt6. By using western blot assay, the protein expression of Sirt6 was identified to be 7.59-fold higher in Sirt6 adenovirus infected 786-O cells over Ad-GFP-treated control cells (Figure 2A). Then, we detected the ratios of apoptotic cells stained with Annexin V and 7-AAD by using a flow cytometry assay. The apoptotic cells in Ad-Sirt6 treated cells were significantly less than in Ad-GFP-treated control cells (4.37% vs 7.30%, n = 3, P < 0.05, Figure 2B). These data suggested that overexpression of Sirt6 in renal cancer cells significantly inhibited cell apoptosis.
Figure 2.
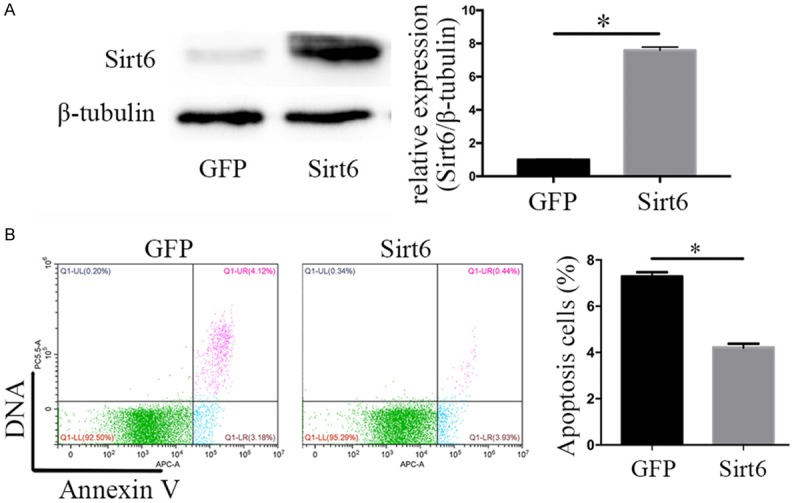
Overexpression of Sirt6 inhibited 786-O renal cancer cell apoptosis. A. Western blot and qPCR tested the efficiency of Adenovirus-Sirt6 in 786-O renal cancer cells. B. Overexpression of Sirt6 reduced ratios of apoptotic cells stained with apoptotic double staining and detected by using a flowcytometry. All data were presented as mean ± SEM. and analyzed by student’s t-test, *P < 0.05.
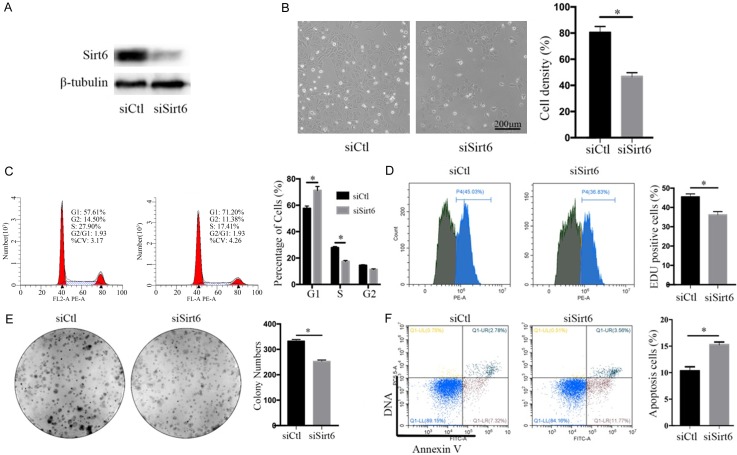
Sirt6 silence promotes apoptosis and cell cycle arrest in renal cancer cells
On the contrary, to examine whether Sirt6 deficiency accelerates renal cancer cell growth, we designed siRNA sequences targeting to Sirt6 (siSirt6). By using a western blot assay, we observed that Sirt6 expression in 786-O cells was significantly reduced by siSirt6 transfection (Figure 3A), which was accompanied with a significant decrease in cell growth by 41.9%, compared with the negative control siRNA (siCtl)-treated cells (siSirt6 vs siCtl: 46.7 ± 1.75% vs 80.4 ± 2.65%, n = 3, P < 0.05, Figure 3B). Flow cytometry assay indicated that Sirt6 silence induced the arrest of G1/S transition in 786-O cells, accompanied with an increase in the percentage of G1 phase (siCtl vs siSirt6: 57.61% vs 71.20%, n = 3, P < 0.05) and a decrease in the percentage of S phase (siCtl vs siSirt6: 27.90% vs 17.41%, n = 3, P < 0.05, Figure 3C). To further investigate the impact of Sirt6 on renal cancer cell growth, we detected DNA Synthesis stage (S phase) in cell cycle by using pulse-labeling EDU incorporation assay and colony formation assay. We observed that EDU-positive cells were significantly reduced in siSirt6-treated 786-O cells by 9.30%, compared with siCtl-treated control cells (36.1 ± 0.99% vs 45.4 ± 0.92%, n = 3, P < 0.05) (Figure 3D). In addition, the colony number of the siSirt6-treated 786-O cells was significantly less by 24.0% than that of siCtl-treated control cells (siCtl vs siSirt6: 331.7 ± 4.06 vs 252.0 ± 3.61, n = 3, P < 0.05, Figure 3E). Furthermore, the ratios of apoptotic cells in siSirt6-treated 786-O cells were significantly more by 5.2% than those in siCtl-treated control cells (siCtl vs siSirt6: 10.1% vs 15.3%, n = 3, P < 0.05, Figure 3F). These data suggested that knockdown of Sirt6 in renal cancer cells significantly inhibited cell cycle progression and promoted cell apoptosis.
Figure 3.
Sirt6 silence reduced 786-O renal cancer cell proliferation. A. Western blot tested the efficiency of small interfering RNA (siSirt6) and non-sense control (siCtl) in 786-O renal cancer cells. B. Images presented the cell density after siCtl or siSirt6 transfection for 48 h (scale bar: 200 μm). C. G1/S phase of cell cycle arrest were analyzed by using a flowcytometry after 786-O renal cancer cells were stained with PI. D. S phase cells were analyzed by using a flowcytometry after 786-O renal cancer cells were labeled by EDU for 2 hours. E. Colony number decreased after siSirt6 transfection. F. Ratios of apoptosis cells increased in 48-h siSirt6-transfected 786-O renal cancer cells analyzed by using a flowcytometry with apoptotic double staining. All data were presented as mean ± SEM. and analyzed by student’s t-test, *P < 0.05.
Sirt6 silence actives signaling pathways involved in G1/S transit and cell apoptosis
To reveal the underlying mechanisms involved in the regulation of Sirt6 on renal cancer cells, we further evaluated the change in key regulators of cell cycle and apoptosis. Sirt6 silence reduced S-phase in renal cancer cells and impaired G1/S transition in cell cycle. Cyclin D1 is a mediator of G1/S transition and accumulated in G1 phase, while p21 is a CDK inhibitor to prevent cells from entering S phase. Our results showed that Sirt6 silence induced a significant increase in both Cyclin D1 and p21 protein levels as expected, compared with the control cells (Figure 4). In addition, Sirt6 silence induced an increase in the ratios of apoptotic cells, which was parallel to the changes in the apoptotic pathway, including a decreased BCL-2 (an apoptotic inhibitor) and an increased cleaved caspase 8 (an apoptotic activator) protein levels (Figure 4). These data suggested that Sirt6 played an important role in regulation of cell cycle and apoptosis.
Figure 4.
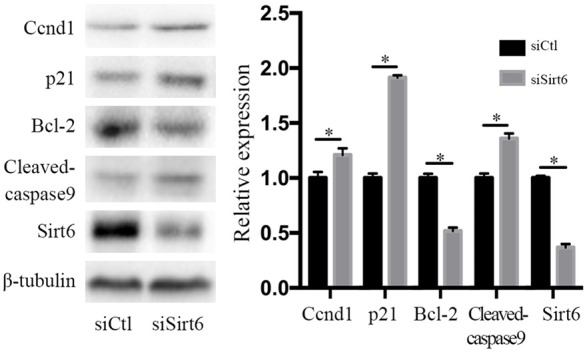
Sirt6 regulated cell cycle and apoptosis signaling pathways. Sirt6 silence induced an increase in protein levels of Ccnd1, p21 and Cleaved-caspase 9, but a decrease in Bcl-2, detected by using western blot assay. All data were presented as mean ± SEM. and analyzed by student’s t-test, *P < 0.05.
Sirt6 silence enhances chemotherapy sensitivity
To test whether Sirt6 deficiency increase chemotherapy sensitivity, Annexin V/7-AAD double stain assay was used to examine cell apoptosis in doxorubicin (Dox) combined with Sirt6 silence treated renal cancer cells. We found that apoptosis was increased by 5.23% in Sirt6-silenced 786-O cells, compared to the control cells (P < 0.05). However, Sirt6 deficiency increased the cell apoptosis in Dox-treated 786-O cells by 58.9%, compared with siCtl-treated cells with DOX challenge (17.58 ± 0.84% and 27.94 ± 0.71% in siCtl + Dox- and siSirt6 + Dox-treated cells, respectively, n = 3, P < 0.05, Figure 5). Taken together, Sirt6 silence enhances doxorubicin sensitivity to renal cancer cells.
Figure 5.
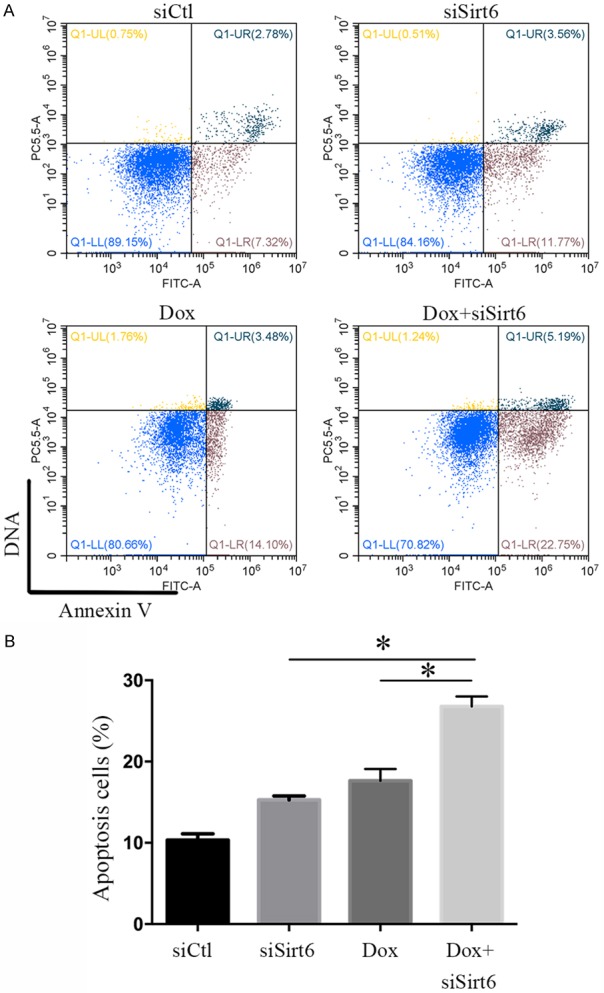
Sirt6 silence enhanced doxorubicin sensitivity to renal cancer cells. A. Ratios of apoptotic cells increased after combination of siSirt6 transfection and Dox treatment for 48 h, analyzed by a flowcytometry assay with apoptotic double staining. B. Apoptotic index. All data were presented as mean ± SEM. and analyzed by one-way ANOVA, *P < 0.05.
Discussion
Sustaining activation of proliferative signaling, escaping from growth suppression, avoiding cell death are three hallmarks of cancer [17]. RCC has the highest mortality in genitourinary cancers and the incidence has increased stably [2]. The causes of RCC are not completely known [18]. Now, only a few etiologic factors have been clinically identified as risk factors for RCC [18]. Age, sex, race, excess body weight, genetic and epigenetic changes are important factors in RCC development [19]. Previously studies show that sirtuins regulate several cellular processes, including DNA damage repair, telomere balance, mitochondrial function, glycolysis, and nutrient metabolism [3,4]. Sirt6 has also been involved in colorectal cancer, non-small-cell lung cancer, cervical carcinoma, and melanoma [20,21]. The role of Sirt6 in vary cancers remains controversial, it has been verified as a tumor suppressor or oncogene [11,14,21-23]. Thus, to reveal the specific role of Sirt6 in malignancies was urgent and meaningful, which might help the accurately prediction, detection, diagnosis, stage and treatment of cancer. Our study facilitated to identify the specific role of Sirt6 in the pathogenesis of renal cancer. We observed that Sirt6 protein expression in renal tumor tissues or RCC cell line cells was much higher than paratumor control tissues or normal renal cells, which indicated that the high expression of SIRT6 may be involved in RCC development and suggested a predictor role of Sirt6 for renal tumor diagnosis. In order to discern the real biological role of Sirt6 in renal tumors, we further silenced Sirt6 in human renal cancer cells, which led to an arrest of cell cycle at the G1/S phase, a decline in cell viability and an elevation in apoptosis. These results confirmed that the high Sirt6 expression promoted RCC development.
The function of Sirt6 in cancer is probably context- and tissue-specific. Sirt6 acts as a tumor suppressor in liver and intestinal cancer through regulation of cancer metabolism [21], repressing the oncogene survivin [24], and controlling oncogenic molecular pathways [25]. Otherwise, Sirt6 is down-regulated in human pancreatic and colon cancers. It regulates aerobic glycolysis process by repressing MYC activity [21]. Enhanced Sirt6 prompts apoptosis in cancer cells but not in normal cells [26]. Sirt6 is also shown to be a tumor suppressor through positive role in DNA damage repair and telomere balance [27-29]. However, Sirt6 acts as an oncogene in skin cancer, increasing cell survival and proliferation in the skin epidermis [9]. Indeed, Sirt6 was shown to be up-regulated in human squamous cell carcinoma [30] and prostate cancer [13] and is repressed by miR-34a in keratinocytes [30]. Cancer is biologically a disease of uncontrolled cell division [17]. Its initiation and progression are usually connected to dysregulation of cell cycle regulators [31]. p21, a cyclin-dependent kinase inhibitor, is regulated through p53-dependent or -independent pathways [32]. It primarily inhibits apoptosis by causing cell cycle arrest and DNA repair. Cyclin D1 is an important regulator of cell cycle progression and performs as a transcriptional co-regulator [32]. It is a mediator of G1/S transition and accumulated in G1 phase. The overexpression of Cyclin D1 has been associated to the development and progression of several types of cancer [33]. In our study, we further probed the regulation mechanism of Sir6 on cell cycle in renal cancer. Sirt6 silence impaired G1/S transition through the upregulation of p21 and the accumulation of Cyclin D1. Silence of Sirt6 repressed the growth of HCC by increasing cellular senescence and DNA damage [14], promoting the expression of p-ATRSer428 and p-chk2Thr68 [34]. The serine/threonine protein kinases ATM and ATR were triggered as response for genotoxic stress and induced phosphorylation of chk2, chk1, and H2AX [34]. On DNA damage response, p21 induction is essential for cell cycle arrest and cell senescence, performing as an inhibitor of cell proliferation. These findings are consistent with our results.
Besides, prolonged cell cycle arrest and unrepaired DNA damage can activate cell apoptotic pathways [35,36], p21 accumulation leads to cell cycle arrest and induces cell apoptosis. Sirt6 via its deacetylase activity suppresses cell apoptosis through the induction of the BCL2-associated X protein signaling pathway [37]. In our study, Sirt6 deficiency led to a reduction in BCL-2 levels, while increased the protein levels of cleaved caspase 8. These data suggested that Sirt6 promoted cell cycle progression and proliferation, and inhibited apoptosis in renal cancer cells.
Chemotherapeutic agent resistance is one of the major obstacles in various cancer treatment, such as RCC, which finally leads to cancer recurrence and cancer associated death. Therefore, the improvements of chemo-sensitivity and determination of its mechanism in RCC may facilitate the treatments in RCC patients in the future. Sirt6-deficiency decreased cell viability and enhanced chemotherapeutic sensitivity of Taxol in prostate cancer [13] and non-small-cell lung cancer [16]. In HCC, Sirt6 deficiency suppressed MDR1 transcription by targeting its promoter when co-treated with chemotherapeutic drugs, including doxorubicin, cisplatin, and sorafenib, through regulating of CCAAT/enhancer binding protein β (CEBPβ) [38]. Several functions of Sirt6 are determined by its activity in chromatin, via deacetylation of histone H3 on acetylated K9, K56 and K18, allowing the proper unwinding of chromatin and the docking of DNA damage interacting factors [8,39]. Consistent with our study, doxorubicin treatment induced more apoptosis in Sirt6-selenced RCC, suggesting that the inhibition of Sirt6 benefits to enhance chemotherapeutic sensitivity. In summary, these findings suggest that Sirt6 acted as an oncogene in human renal cell carcinoma and Sirt6 could be a potential therapeutic target for the treatment for RCCs.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81870221, 81670249, 31271226 and 31071001 to Dr. Wei Jiang).
Disclosure of conflict of interest
None.
References
- 1.Protzel C, Maruschke M, Hakenberg OW. Epidemiology, aetiology, and pathogenesis of renal cell carcinoma. European Urology Supplements. 2012;11:52–59. [Google Scholar]
- 2.Jeh SU, Park JJ, Lee JS, Kim DC, Do J, Lee SW, Choi SM, Hyun JS, Seo DH, Lee C, Kam SC, Chung KH, Hwa JS. Differential expression of the sirtuin family in renal cell carcinoma: aspects of carcinogenesis and prognostic significance. Urol Oncol. 2017;35:675, e9–e15. doi: 10.1016/j.urolonc.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 5.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014;33:1609–1620. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ming M, Han W, Zhao B, Sundaresan NR, Deng CX, Gupta MP, He YY. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014;74:5925–5933. doi: 10.1158/0008-5472.CAN-14-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. J Biol Chem. 2015;290:9604–9613. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khongkow M, Olmos Y, Gong C, Gomes AR, Monteiro LJ, Yague E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong S, Koo CY, Harada-Shoji N, Tsang JW, Coombes RC, Schwer B, Khoo US, Lam EW. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34:1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 12.Feng XX, Luo J, Liu M, Yan W, Zhou ZZ, Xia YJ, Tu W, Li PY, Feng ZH, Tian DA. Sirtuin 6 promotes transforming growth factor-beta1/H2O2/HOCl-mediated enhancement of hepatocellular carcinoma cell tumorigenicity by suppressing cellular senescence. Cancer Sci. 2015;106:559–566. doi: 10.1111/cas.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Xie QR, Wang B, Shao J, Zhang T, Liu T, Huang G, Xia W. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell. 2013;4:702–710. doi: 10.1007/s13238-013-3054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee N, Ryu HG, Kwon JH, Kim DK, Kim SR, Wang HJ, Kim KT, Choi KY. SIRT6 depletion suppresses tumor growth by promoting cellular senescence induced by DNA damage in HCC. PLoS One. 2016;11:e0165835. doi: 10.1371/journal.pone.0165835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cagnetta A, Caffa I, Acharya C, Soncini D, Acharya P, Adamia S, Pierri I, Bergamaschi M, Garuti A, Fraternali G, Mastracci L, Provenzani A, Zucal C, Damonte G, Salis A, Montecucco F, Patrone F, Ballestrero A, Bruzzone S, Gobbi M, Nencioni A, Cea M. APO866 increases antitumor activity of cyclosporin-a by inducing mitochondrial and endoplasmic reticulum stress in leukemia cells. Clin Cancer Res. 2015;21:3934–3945. doi: 10.1158/1078-0432.CCR-14-3023. [DOI] [PubMed] [Google Scholar]
- 16.Azuma Y, Yokobori T, Mogi A, Altan B, Yajima T, Kosaka T, Onozato R, Yamaki E, Asao T, Nishiyama M, Kuwano H. SIRT6 expression is associated with poor prognosis and chemosensitivity in patients with non-small cell lung cancer. J Surg Oncol. 2015;112:231–237. doi: 10.1002/jso.23975. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Mehdi A, Riazalhosseini Y. Epigenome aberrations: emerging driving factors of the clear cell renal cell carcinoma. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabanathan D, Park JJ, Marquez M, Francisco L, Byrne N, Gurney H. Cure in advanced renal cell cancer: is it an achievable goal? Oncologist. 2017;22:1470–1477. doi: 10.1634/theoncologist.2017-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kugel S, Feldman JL, Klein MA, Silberman DM, Sebastian C, Mermel C, Dobersch S, Clark AR, Getz G, Denu JM, Mostoslavsky R. Identification of and molecular basis for sirt6 loss-of-function point mutations in cancer. Cell Rep. 2015;13:479–488. doi: 10.1016/j.celrep.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, Zoppoli G, Cea M, Feldmann G, Mostoslavsky R, Ballestrero A, Patrone F, Bruzzone S, Nencioni A. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287:40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Li C, Zhang X. The promoter methylation status and mRNA expression levels of CTCF and SIRT6 in sporadic breast cancer. DNA Cell Biol. 2014;33:581–590. doi: 10.1089/dna.2013.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, Zatloukal K, Hui L, Wagner EF. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 25.Marquardt JU, Fischer K, Baus K, Kashyap A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, Thorgeirsson SS, Galle PR, Strand S. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. 2013;58:1054–1064. doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: an update. J Biol Chem. 2012;287:42444–42452. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennen RI, Chua KF. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci. 2011;36:39–46. doi: 10.1016/j.tibs.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, Albinger-Hegyi A, Hoetzenecker W, Kolfschoten I, Wagner EF, Werner S, Dotto GP. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shostak A. Circadian clock, cell division, and cancer: from molecules to organism. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, Hinds PW. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng L, Yang Q, Li C, Dai L, Yang Y, Wang Q, Ding Y, Zhang J, Liu L, Zhang S, Fan P, Hu X, Xiang R, Yu D, Wei Y, Deng H. DDA1, a novel oncogene, promotes lung cancer progression through regulation of cell cycle. J Cell Mol Med. 2017;21:1532–1544. doi: 10.1111/jcmm.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol. 2009;11:580–591. doi: 10.1038/ncb1864. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH, Zhou L, Tang H, Chen X, Chen K, Li WY, Huang AL, Chen J. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated x protein-dependent apoptotic pathway. Clin Cancer Res. 2016;22:3372–3382. doi: 10.1158/1078-0432.CCR-15-1638. [DOI] [PubMed] [Google Scholar]
- 38.Xia YQ, Hua RJ, Juan C, Zhong ZH, Tao CS, Fang R, Lin H, Rui G, Yong C. SIRT6 depletion sensitizes human hepatoma cells to chemotherapeutics by downregulating MDR1 expression. Front Pharmacol. 2018;9:194. doi: 10.3389/fphar.2018.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D’Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]