Abstract
The development of cancer occurs with various genomic and epigenetic modifications that act as indicators for early diagnosis and treatment. Recent data have shown that the abnormal expression of the claudin (CLDN) tight junction (TJ) proteins is involved in the tumorigenesis of numerous human cancers. Real-time quantitative PCR and western blotting were used to explore the differences in the expression of the CLDN TJ proteins in breast carcinoma tissues and non-neoplastic tissues. The results showed that CLDN5, CLDN9, CLDN12 and CLDN13 were not expressed in breast carcinoma tissues or non-neoplastic tissues. CLDN1, CLDN3, CLDN8 and CLDN10 were expressed in breast carcinoma and non-neoplastic tissues, but there was no significant difference between the expression of these CLDN proteins among them. The expression of CLDN2, -6, -11 and -14 varied between the breast carcinoma and non-neoplastic tissues. Moreover, 86 samples of breast carcinoma and non-neoplastic tissues were examined for the expression of CLDN2, -6, -11 and -14 by streptavidin-peroxidase immunohistochemical staining. The data revealed that the CLDN2, CLDN6, and CLDN14 were expressed in the cell membrane and the expression levels of these proteins were downregulated in breast carcinoma. The CLDN11 was expressed in cell cytoplasm and the expression level of CLDN11 was upregulated compared with those in non-neoplastic tissues. Consistent with these findings, the expression of CLDN2, CLDN6 and CLDN14 were downregulated, while the expression of CLDN11 was upregulated in breast carcinoma compared with those in non-neoplastic tissues. Furthermore, the associations between these CLDNs and clinicopathologic indicators were analyzed, and these CLDN expressions were revealed to be associated with distant metastasis and to predict a poor prognosis. In conclusion, our data showed that the expression levels of CLDN2, -6, -11 and -14 differed between breast carcinoma tissues and histologically non-neoplastic tissues, and the expression levels of these CLDNs may be useful as molecular markers for the diagnosis of breast carcinoma as well as for the determination of metastasis and prognosis.
Keywords: Tight junction, claudin-2, claudin-6, claudin-11, claudin-14, breast carcinoma
Introduction
Tight junctions (TJs), in cooperation with adherens junctions and desmosomes, form the apical junctional complex in epithelial and endothelial cellular sheets [1]. TJs are crucial for the tight fastening of cellular sheets, which allows monitoring of the paracellular ion flux and consequently maintains tissue homeostasis [2]. More than 40 diverse proteins have been identified in the TJs of epithelia and endothelia to date [3,4]. The TJs have a cement-like function and can prevent cell detachment in epithelial cells [5]. A vital phase in the initiation of cancer metastases is contact with and dissemination of the vascular endothelium by disconnected tumor cells [6]. TJs are consequently the first obstacle that tumor cells must overcome for metastasis [7]. TJs consist of three major types of fundamental membrane proteins: occludin, claudin (CLDN), and junctional adhesion molecules (JAM) [8,9]. Although the precise characteristics of these proteins are not entirely clear, increased information on the molecular construction of TJs led to the development of the aforementioned biological models that show TJs to be morphologic features in diverse tissues and to respond to fluctuating natural, pathological or experimental surroundings [10,11].
The CLDN protein family, which is expressed in the transmembrane area, plays a critical role in the foundation of TJs and comprises approximately 27 members, most of which can strongly bind with PDZ-containing proteins [12,13]. These interesting findings have altered the view of TJs as a purely paracellular barrier to being considered as a composite structure participating in signaling cascades that control cell proliferation and differentiation [14]. Hence, CLDNs are linked to multimolecular multiplexes and the transduction of cell signaling pathways [15-17]. Due to the ability of CLDNs to interact with signaling proteins, CLDNs have been shown to be associated with the regulation of cell proliferation, differentiation, and other cellular functions [18,19]. Although the expression profile of CLDNs is tissue-specific, most tissues express manifold CLDNs, which can recruit both homotypic and heterotypic CLDNs to constitute TJ components [20]. The precise mixture of CLDNs inside an assumed tissue can determine the selectivity and strength of the TJs. CLDNs are polymerized together inside acell and can cooperate with the CLDNs of histologically normal cells to form an adhesive structure. The expression of CLDNs has been revealed as altered in numerous tumor types [21]. Tumor cells commonly display an uncharacteristic CLDN expression profile, along with reduced differentiation and cell polarity [22,23]. CLDN1 has been shown to be downregulated in colon cancer [24], These studies revealing decreased TJ protein expression in cancer is consistent with the commonly recognized idea that tumorigenesis is associated with the interruption of TJs, which may play a vital role in the damaged interconnection and deficient differentiation detected in tumor cells.
In contrast, most studies conducted on this topic thus far have identified the upregulation of CLDNs in tumor cells. One of the initial reports on this finding described a serial analysis of gene expression (SAGE) study showing that both CLDN3 and CLDN4 were amon the most highly increased genes in ovarian cancer [25,26]. As revealed previously, numerous CLDNs, such as CLDN3 and CLDN4, are characteristically increased in various cancers, revealing that these proteins may promote the tumorigenesis of human cancers. Thus, the roles of CLDNs may be extremely tissue-specific and may depend on the precise molecular circuitry of the cell. Changes in CLDN expression may be associated with the tumorigenesis and progression of human tumors. However, the precise expression profiles of the CLDNs in breast carcinoma have not yet been defined. Thus, the objectives of this study were to explore the expression of CLDNs in histologically non-neoplastic tissues and breast carcinoma tissues, and to detect the associations between alterations of CLDNs and patient clinicopathologic characteristics in breast carcinoma.
Materials and methods
Patients
Biopsies were gathered from 86 patients with an average age of 54 years, who received treatment at Jilin Cancer Hospital or The First Hospital of Jilin University between June 2007 and May 2012 with a pathologically confirmed diagnosis of breast carcinoma. The patients were carefully chosen based on the following criteria: no history of radiotherapy and chemotherapy and no prior cancer. The grade and classification of the breast carcinoma patients were defined according to the American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) staging system [27]. Histologically non-neoplastic breast tissues were also collected from patients with breast hyperplasia who received treatment at Jilin Cancer Hospital between July 2006 and April 2013, and these tissues were verified to be pathologically non-neoplastic.
Real-time reverse transcription polymerase chain reaction (real-time PCR)
Real-time PCR was utilized to detect the expression of CLDNs in breast carcinoma tissues and non-neoplastic tissues. Total RNA was extracted using a Perfect Pure RNA Cultured Cell Kit (Thermo Fisher Scientific, Waltham, MA) as described by the manufacturer’s protocol. Real-time PCR was carried out as previously described [28]. The RT cDNA reaction products were subjected to quantitative real-time PCR using a CTFX 96 Real-time system (Bio-Rad, Hercules, CA, USA) and SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocol. The primer pairs targeting CLDNs and glyceraldehyde phosphate dehydrogenase (GAPDH) were as follows: CLDN1 forward (5’-GCCACAGCAAGGTATGGTAAC-3’) and reverse (5’-AGTAGGGCACCTCCCAGAAG-3’); CLDN2 forward (5’-TTCATCGGCAACAGCATCG-3’) and reverse (5’-GGTTATAGAAGTCCCGGATGA-3’); CLDN3 forward (5’-AGTGCAAGGTGTACGACTC-3’) and reverse (5’-AGTCCCGGATAATGGTGTTG-3’); CLDN4 forward (5’-TTGTCACCTCGCAGACCATC-3’) and reverse (5’-GCAGCGAGTCGTACACCTTG-3’); CLDN5 forward (5’-AACATCGTGACGGCGCAGACCA-3’) and reverse (5’-TCAGAGCCAGCACCGAGTCGTACA-3’); CLDN6 forward (5’-GGCAACAGCATCGTCGTGG-3’) and reverse (5’-GAAGTCCTGGATGATAGAGTGGGC-3’); CLDN7 forward (5’-TTTTCATCGTGGCAGGTCTT-3’) and reverse (5’-GGCCAAACTCATACTTAATGTTGG-3’); CLDN8 forward (5’-TCTGCAGTAGGACATAGAAACCCCTAA-3’) and reverse (5’-CGTTTAGGGGTTTCTATGTCCTACTGC3’); CLDN9 forward (5’-CTAGCACTAGTTTCGAAATGGCTTCGACCGGCTTAG-3’) and reverse (5’-TCTCGAGCTAGTCGACTCACACGTAGTCCCTCTTGTC-3’); CLDN10 forward (5’-GGAGGCTCCGATAAAGCCAA-3’) and reverse (5’-GTGGCCCCGTTGTATGTGTA-3’); CLDN11 forward (5’-TGACCTGCAGCTACACCATC-3’) and reverse (5’-GGG GTT TGC AGT GGT AGA GA-3’); CLDN12 forward (5’-CCGTGATGTCCTTCTTGGCTTTC-3’) and reverse (5’-CTCTGATGATGGCATTGGCAACC-3’); CLDN13 forward (5’-TAGTGTTGGCCTTCTGATGC-3’) and reverse (5’-AGCCAAGCAATGGGTTAAAG-3’); CLDN14 forward (5’-TGGCTGGGCTGGGTGGTCTC-3’) and reverse (5’-AGCGGCCATGAGCTGGTGTG-3’) and GAPDH forward (5’-AACGTGTCAGTCGTGGACCTG-3’) and reverse (5’-AGTGGGTGTCGCTGTFGAAGT-3’).
Western blotting
Western blotting was utilized to detect the expression of CLDNs in all the breast carcinoma and non-neoplastic tissues. The cells were washed with ice-cold phosphate-buffered saline (PBS) 3 times, and cell lysates were prepared with a lysis buffer containing 10 mM Tris-HCl (pH 7.4), 1% SDS, and 1 mM Na3VO4. A BCA Protein Assay Kit (Pierce Chemical Co., Rockford, Illinois, USA) was utilized to detect the protein concentration. Total protein (thirty micrograms) was separated in a 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane (Millipore, Temecula, California, USA). Next, the membrane was blocked and incubated with the following primary antibodies: rabbit anti-human CLDN2 (ab53032, Abcam Company, USA), rabbit anti-human CLDN6 (ab107059, Abcam Company, USA), rabbit anti-human CLDN11 (ab53041, Abcam Company, USA), or rabbit anti-human CLDN14 (ab19035, Abcam Company, USA) and mouse anti-human β-actin (ab8226, Abcam Company, USA) at a 1:1000 dilution at room temperature. Subsequently, the membranes were washed 3 times with PBS and incubated with horseradish peroxidase-conjugated secondary antibodies (1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Immunoreactive bands were detected using ECL Western blotting reagents (GE, Fairfield, Connecticut, USA) and were analyzed with Image Lab 6.0.1 software (Bio-Rad Laboratories).
Immunohistochemistry
Immunohistochemistry was utilized to explore the expression patterns of CLDNs in breast carcinoma tissues and non-neoplastic tissues. The experimental method was the same as that described previously [29], and the slides were incubated at 4°C overnight with a rabbit anti-human CLDN1 antibody (sc-28670, Santa Cruz Biotechnology, USA), rabbit anti-human CLDN3 antibody (sc-66834, Santa Cruz Biotechnology, USA), rabbit anti-human CLDN7 antibody (ab27487, Abcam Company, USA), or rabbit anti-human CLDN8 antibody (ab183738, Abcam Company, USA), which were diluted 1:400, 1:300, 1:400, and 1:300, respectively. The evaluation of protein expression levels was based on the percentage of positively stained tumor cells together with the staining intensity, as previously described [30].
Follow-up
The pathologically confirmed diagnosis of breast carcinoma was followed-up from the beginning of diagnosis to 60 months to assess occurrence and metastasis and determine survival. The living status of the patients was determined either through a telephone interview or on an outpatient basis before July 2017.
Statistical methods
All experiments were repeated 3 times, and all of the data are based on the mean ± SD of at least 3 experimental results. The results were analyzed by a paired Student’s t-test, and P < 0.05 was considered significant. The Chi-square test/Chi-square goodness-of-fit test was applied for correlation analysis with clinical case indicators. In addition, the correlations between CLDNs and clinical survival were analyzed by Kaplan-Meier method and compared with log-rank tests.
Results
Expression levels of CLDN family members in breast carcinoma tissues and non-neoplastic tissues
Real-time quantitative PCR and western blotting were used to detect the expression of CLDN family members in breast carcinoma and non-neoplastic tissues. As shown in Figure 1, neither the mRNAs nor the proteins of CLDN5, CLDN9, CLDN12 and CLDN13 were expressed in the samples of breast carcinoma and non-neoplastic tissues; CLDN1, CLDN3 CLDN8 and CLDN10 were expressed in breast carcinoma and non-neoplastic tissues, but there was no significant difference between the expression of these proteins in breast carcinoma and non-neoplastic tissues. How-ever, the mRNA and protein expression of CLDN2, CLDN6 and CLDN14 was downregulated in breast carcinoma, while the mRNA and protein expression of CLDN11 was upregulated compared with non-neoplastic tissues.
Figure 1.

Expression of CLDNs in human breast carcinoma and histologically non-neoplastic tissues. A. Real-time quantitative PCR analysis of CLDN mRNA expression in breast carcinoma andnon-neoplastic tissues. B. Western blotting was used to investigate significant differences in the expression of CLDNs in breast carcinoma and non-neoplastic tissues. C. Corresponding statistical analysis of protein expression. **P < 0.01 vs. non-neoplastic tissues.
The expression of CLDN2, CLDN6, and CLDN14 was downregulated in breast carcinoma
CLDN1 expression was further evaluated in 86 breast carcinoma tissue specimens and in 86 specimens of histologically non-neoplastic tissues. As shown in Figure 2A and 2B, CLDN2 were expressed in the cell membrane. Positive protein expression was found in 30.2% (26/86) of breast carcinoma and in 60.4% (52/86) of histologically non-neoplastic tissues (Table 1). The expression of CLDN2 was significantly lower in breast carcinoma than in histologically non-neoplastic tissues (Chi-square test/Chi-square goodness-of-fit test, χ2 = 8.673, P < 0.01). As shown in Table 1, the expression of CLDN2 was not correlated with age (P = 0.116), the expression of Ki 67 (P = 0.158), histologic grade (P = 0.552) but associated with distant metastasis (P = 0.003).
Figure 2.

Immunohistochemical demonstration of CLDN expression in human breast carcinoma and histologically non-neoplastic tissues. A. CLDN2 expression in non-neoplastic tissue (400×). B. CLDN6 expression in non-neoplastic tissue (400×). C. CLDN11 expression in non-neoplastic tissue (400×). D. CLDN14 expression in non-neoplastic tissue (400×). E. CLDN2 expression in breast carcinoma (400×). F. CLDN6 expression in breast carcinoma (400×). G. CLDN11 expression in breast carcinoma. H. CLDN14 expression in breast carcinoma (400×).
Table 1.
Expression of CLDN2 and CLDN6 clinicopathologic characteristics in breast carcinoma patients
| Item | n | CLDN2 (+) | CLDN2 (-) | χ2 | P | n | CLDN6 (+) | CLDN6 (-) | χ2 | P |
|---|---|---|---|---|---|---|---|---|---|---|
| Breast carcinoma tissue | 86 | 26 | 60 | 8.673 | < 0.01 | 86 | 30 | 56 | 8.493 | < 0.01 |
| Histologically non-neoplastic tissue | 86 | 52 | 34 | 86 | 58 | 28 | ||||
| Age (year) | ||||||||||
| ≤ 60 | 54 | 15 | 39 | 0.116 | 0.764* | 54 | 18 | 46 | 2.422 | 0.162* |
| > 60 | 32 | 11 | 21 | 32 | 12 | 20 | ||||
| Histologic grade | ||||||||||
| Well-differentiated | 48 | 14 | 34 | 0.324 | 0.552* | 48 | 16 | 32 | 1.276 | 0.231* |
| Moderately and poorly differentiated | 38 | 12 | 26 | 38 | 14 | 24 | ||||
| Distant metastasis | ||||||||||
| + | 47 | 10 | 31 | 6.827 | < 0.01 | 47 | 7 | 40 | 9.276 | < 0.01 |
| - | 39 | 16 | 23 | 39 | 23 | 16 | ||||
| Ki67 | ||||||||||
| + | 51 | 17 | 34 | 2.234 | 0.158* | 51 | 18 | 33 | 0.186 | 0.726* |
| - | 35 | 9 | 26 | 35 | 12 | 23 |
No statistical significance.
As shown in Table 1, membrane expression of the CLDN6 protein was found in 34.9% (30/86) of breast carcinoma and in 67.4% (56/86) of histologically non-neoplastic tissues. The expression of CLDN6 in breast carcinoma was significantly lower than in histologically non-neoplastic tissues (Chi-square test/Chi-square goodness-of-fit test, χ2 = 8.493, P < 0.01) (Figure 2E, 2F). As shown in Table 1, the expression of CLDN6 was not correlated with age (P = 0.162), histologic grade (P = 0.231), or the expression of Ki 67 (P = 0.726) but was correlated with distant metastasis (P = 0.001).
As shown in Figure 2G and 2H, positive membrane expression of CLDN14 protein was found in 27.9% (24/86) of breast carcinoma and in 54.7% (47/86) of histologically non-neoplastic tissues (Table 1). The expression of CLDN14 was significantly lower in breast carcinoma tissues than in histologically non-neoplastic tissues (Chi-square test/Chi-square goodness-of-fit test, χ2 = 8.163, P < 0.01). As shown in Table 1, the expression of CLDN14 was not correlated with age (P = 0.926), the expression of Ki 67 (P = 0.354), histologic grade (P = 0.175) but associated with distant metastasis (P = 0.002).
The expression of CLDN11 was increased in breast carcinoma
The cytoplasm staining of CLDN11 was strong in breast carcinoma and weak in histologically non-neoplastic tissues. Positive cytoplasm expression of CLDN11 was expressed in 53.5% (54/86) of breast carcinoma tissues. Cells were positive for CLDN11 in 25.6% (22/86) of histologically non-neoplastic tissues (Figure 2E, 2F). We concluded that CLDN11 expression was significantly higher in breast carcinoma than in histologically non-neoplastic breast carcinoma tissues (Chi-square test/Chi-square goodness-of-fit test, χ2 = 7.265, P < 0.01). As shown in Table 2, the expression of CLDN11 was not correlated with age (P = 0.324), but was correlated with histologic grade (P = 0.024), the expression of Ki67 (P = 0.027) and distant metastasis (P = 0.001).
Table 2.
Expression of CLDN11 and CLDN14 and clinicopathologic characteristics in breast carcinoma patients
| n | CLDN11 (+) | CLDN11 (-) | χ2 | P | n | CLDN14 (+) | CLDN14 (-) | χ2 | P | |
|---|---|---|---|---|---|---|---|---|---|---|
| Breast carcinoma tissue | 86 | 46 | 40 | 7.265 | < 0.01 | 86 | 24 | 62 | 8.163 | < 0.01 |
| Non-neoplastic tissue | 86 | 22 | 64 | 86 | 47 | 39 | ||||
| Age (year) | ||||||||||
| ≤ 60 | 54 | 27 | 27 | 0.912 | 0.324* | 54 | 13 | 41 | 0.086 | 0.926* |
| > 60 | 32 | 19 | 13 | 32 | 11 | 21 | ||||
| Histologic grade | ||||||||||
| Well-differentiated | 48 | 21 | 27 | 5.243 | < 0.05 | 48 | 10 | 28 | 2.126 | 0.175 |
| Moderately and poorly differentiated | 38 | 25 | 13 | 38 | 14 | 24 | ||||
| Distant metastasis | ||||||||||
| + | 47 | 32 | 15 | 8.742 | < 0.01 | 47 | 7 | 40 | 7.936 | < 0.01 |
| - | 39 | 14 | 25 | 39 | 17 | 22 | ||||
| Ki67 | ||||||||||
| + | 51 | 29 | 22 | 6.214 | < 0.05 | 51 | 14 | 37 | 0.724 | 0.354* |
| - | 35 | 17 | 1 8 | 35 | 10 | 25 |
No statistical significance.
CLDN2 and CLDN6 are concurrently expressed in breast carcinoma tissue
As revealed in Table 3, a correlation between CLDN2 and CLDN6 expression was detected in breast carcinoma tissues (Chi-square test/Chi-square goodness-of-fit test, φ = 0.879).
Table 3.
Correlation between the expression of CLDN2 and CLDN6 in breast carcinoma
| Item | CLDN6 (+) | CLDN6 (-) | φ* | P |
|---|---|---|---|---|
| CLDN2 (+) | 20 | 6 | 0.879 | < 0.01 |
| CLDN2 (-) | 10 | 50 |
Statistical significance was observed.
Associations with survival and clinical outcomes
As shown in Figure 3, patients with tumors that were positive for the CLDN2, CLDN6, and CLDN14 proteins (median survival, 59.12 months; median survival, 58.73 months; median survival, 58.69 months, respectively) exhibited a significantly longer survival time (P = 0.002, P = 0.018 and P = 0.012, respectively) than those whose tumors were negative for the CLDN2, CLDN6 and CLDN14 proteins (median survival, 55.23 months; median survival, 54.36 months; median survival, 55.12 months, respectively). Patients with tumors that were positive for the CLDN11 proteins (median survival, 54.83 months) had a significantly shorter survival time (P = 0.012) than those whose tumors were negative for the CLDN11 proteins (median survival, 58.72 months).
Figure 3.
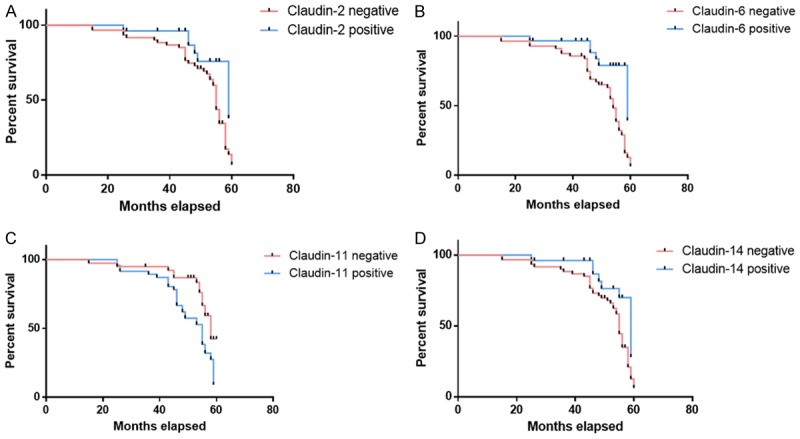
Association between the expression of CLDNs and patient survival. Kaplan-Meier analysis was utilized for survival analysis in breast carcinoma patients. (A) CLDN2 curvature, (B) CLDN6 curvature, (C) CLDN11 curvature and (D) CLDN14 curvature.
Discussion
Damage to cell-to-cell adhesion is commonly recognized as an initial event in metastasis, permitting the release of separate tumor cells from the primary tumor [6]. Cell-to-cell adhesion in epithelial cells is maintained primarily through adherens junctions and TJs [1,3]. Many studies have focused on the adherens junction protein E-cadherin [31]. Deficiencies in E-cadherin function have been shown to lead to cell diffusion and confer invasive abilities in various cell types [32,33]. Because of these functions, E-cadherin is an accepted tumor suppressor in many tissues and has been demonstrated to be a valuable prognostic marker for several human cancers [34], indicating a vital role of cell-to-cell adhesion proteins in tumor tumorigenesis.
Interruption of TJs, which play crucial roles in cell penetrability and polarity, is thought to lead to epithelial tumorigenesis as well. The exact characteristics of alterations in the structure and function of TJs have been detected in multiple types of tumor. The CLDNs are the main components of TJs, and anomalous expression of these proteins can lead to disruption of TJs and therefore damage to cell polarity and interconnection [35]. Deficiency or abnormal expression of CLDNs has been suggested to be associated with harmful pathophysiologic consequences. CLDN1-deficient mice show no survival within one day of birth as a consequence of the loss of epidermal barrier function [36]. Abnormality of TJ integrity triggered by anomalous CLDN expression might play a major role in permitting the dispersion of nutrients and other factors essential for the maintenance and progress of the tumor cells [37]. As described above, disruption of CLDNs in tumors has been suggested to be a mechanism underlying deficient cell adhesion and a vital event in the evolution of tumor cells toward metastasis [38-40]. Consistent with this theory, a recent study demonstrated that the expression of CLDN4 in pancreatic carcinoma cells decreases the metastatic phenotype of these cells. Additionally, CLDN1 re-expression in tumor cells can contribute to enhanced apoptosis in three-dimensional cultures [41]. Although the typical ratio of CLDNs plays a vital role in maintaining the structure and function of TJs in epithelial cells [19], the mechanisms by which CLDN expression and damage to TJs enhance tumor formation and the consequences of these alterations for tumor progression have not been explored comprehensively.
The principal reason for cancer-related death is metastasis of tumor cells from primary tumor locations to distant organs. The crucial event that is believed to allow tumor cells to metastasize is the epithelial to mesenchymal transition (EMT) [42,43]. However, downregulation of certain claudins in tumors is accompanied by the interruption of TJs during tumorigenesis and EMT [44,45]. Furthermore, enforced initiation of EMT in epithelial cells leads to a notable loss of function of TJs and abnormal expression of CLDNs [46]. Moreover, CLDNs have been revealed to interact with the TJ protein ZO-1 through their carboxyl terminus [15]. Fascinatingly, ZO-1 binds to numerous proteins that participate in cell signaling and transcriptional regulation [47,48]. These reports suggested that CLDNs may play an auxiliary role in cell signaling and transcriptional regulation. In view of the specificity of CLDN expression profiles in tumors, it has been proposed that CLDNs may be valuable indicators of diverse tumor types. For instance, a set of four indicators including CLDN3 was reported to be sufficient to precisely categorize all 158 ovarian cancers assessed [49]. Moreover, CLDNs may serve as a prognostic marker. CLDN1 expression is correlated with a poor prognosis in stage II colon cancer. Furthermore, CLDN10 expression was shown to be an autonomous prognostic indictor of hepatocellular carcinoma recurrence [50]. However, little information is available on the functional correlation between breast carcinoma carcinogenesis and CLDNs. In the present study, the expression of CLDNs was examined in breast carcinoma samples and histologically non-neoplastic tissues from 86 patients. We demonstrated that the expression of CLDN2, CLDN6 and CLDN14 was decreased, while the expression of CLDN11 was increased in breast carcinoma compared with histologically non-neoplastic tissues. Although functional roles of CLDN2, -6, -11 and -14 in breast carcinoma have not yet been determined, there is evidence to support their role in cell-to-cell adhesion, indicating that abnormalities in these proteins may contribute to metastasis.
In conclusion, the current work shows that the expression of CLDN2, -6, -11 and -14 differs between human breast carcinoma and histologically non-neoplastic tissues. These CLDN expressions were found to be associated with distant metastasis and to predict a poor prognosis. In addition, CLDN2 and CLDN6 were shown to be simultaneously expressed in breast carcinoma. However, the detailed mechanisms underlying these observations remain to be determined.
Acknowledgements
We would like to thank American Journal Experts (AJE) for help with this manuscript.
Disclosure of conflict of interest
None.
References
- 1.Brandner JM, Haftek M, Niessen CM. Adherens junctions, desmosomes and tight junctions in epidermal barrier function. Open Dermatology Journal. 2010:4. [Google Scholar]
- 2.Green KJ, Getsios S, Troyanovsky S, Godsel LM. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a000125. doi: 10.1101/cshperspect.a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niessen CM. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 4.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–28. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 5.Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira SS, Morgado-Díaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64:17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 2009;1788:872–91. doi: 10.1016/j.bbamem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Van Itallie CM, Anderson JM. The molecular physiology of tight junction pores. Physiology (Bethesda) 2004;19:331–338. doi: 10.1152/physiol.00027.2004. [DOI] [PubMed] [Google Scholar]
- 9.Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–42. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Günzel D, Alan S. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–69. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, de Prost Y, Munnich A, Hadchouel M. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 2004;127:1386–1390. doi: 10.1053/j.gastro.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 13.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96:511–6. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778:729–56. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–28. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the β-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–76. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 18.Angelow S, Ahlstrom R, Alan S. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–76. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–45. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Günzel D, Fromm M. Claudins and other tight junction proteins. Compr Physiol. 2012;2:1819–52. doi: 10.1002/cphy.c110045. [DOI] [PubMed] [Google Scholar]
- 21.Escudero-Esparza A, Jiang WG, Martin TA. The claudin family and its role in cancer and metastasis. Front Biosci (Landmark Ed) 2011;16:1069–83. doi: 10.2741/3736. [DOI] [PubMed] [Google Scholar]
- 22.Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10:235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouban A, Ahmed AA. Claudins in human cancer, a review. Histol Histopathol. 2010;25:83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 24.Morohashi S, Kusumi T, Sato F, Odagiri H, Chiba H, Yoshihara S, Hakamada K, Sasaki M, Kijima H. Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int J Mol Med. 2007;20:139–43. [PubMed] [Google Scholar]
- 25.Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–85. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 26.Rangel LB, Agarwal R, D’Souza T, Pizer ES, Alò PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 27.Edge SB, Compton CC. The American Joint committee on cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) nethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, Yang YD, Fu L, Xu W, Liu D, Liang Q, Zhang X, Xu L, Guan XY, Wu B, Sung JJ, Yu J. CLDN3 inhibits cancer aggressiveness via wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5:7663–7676. doi: 10.18632/oncotarget.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao M, Li W, Wang H, Wang G. The distinct expression patterns of claudin-10, -14, -17 and E-cadherin between adjacent non-neoplastic tissues and gastric cancer tissues. Diagn Pathol. 2013;8:205. doi: 10.1186/1746-1596-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berx G, Becker KF, Höfler H, Van Roy F. Mutations of the human E-cadherin (CDH1) gene. Hum Mutat. 1998;12:226–37. doi: 10.1002/(SICI)1098-1004(1998)12:4<226::AID-HUMU2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 32.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 33.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–293. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osanai M, Takasawa A, Murata M, Sawada N. Claudins in cancer: bench to bedside. Pflugers Arch. 2017;469:55–67. doi: 10.1007/s00424-016-1877-7. [DOI] [PubMed] [Google Scholar]
- 36.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier a lesson from claudin-1-deficient mice. J Cell Biol. 2002;156:1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61:431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med. 2006;8:1–11. doi: 10.1017/S1462399406000056. [DOI] [PubMed] [Google Scholar]
- 41.Chang TL, Ito K, Ko TK, Liu Q, Salto-Tellez M, Yeoh KG, Fukamachi H, Ito Y. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology. 2010;138:255–265. e253. doi: 10.1053/j.gastro.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 42.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 43.Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhat A, Pope J, Smith J, Ahmad R, Chen X, Washington M, Beauchamp R, Singh A, Dhawan P. Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene. 2015;34:4570–80. doi: 10.1038/onc.2014.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317:1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 47.Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem. 2002;277:455–461. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- 49.Bose CK, Mukhopadhyay A. Claudin and ovarian cancer. J Turk Ger Gynecol Assoc. 2010;11:48–54. [PMC free article] [PubMed] [Google Scholar]
- 50.Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, Fan ST, So S. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11:551–556. [PubMed] [Google Scholar]


