Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most fatal malignancies due to its high morbidity and mortality. DNA methylation exerts a vital part in the development of PDAC. However, a mechanistic role of mutual interactions between DNA methylation and mRNA as epigenetic regulators on transcriptomic alterations and its correlation with clinical outcomes such as survival have remained largely uncovered in cancer. Therefore, elucidation of aberrant epigenetic alteration in the development of PDAC is an urgent problem to be solved. In this work, we conduct an integrative epigenetic analysis of PDAC to identify aberrant DNA methylation-driven cancer genes during the occurrence of cancer.
Methods
DNA methylation matrix and mRNA profile were obtained from the TCGA database. The integration of methylation and gene expression datasets was analyzed using an R package MethylMix. The genes with hypomethylation/hypermethylation were further validated in the Kaplan–Meier analysis. The correlation analysis of gene expression and aberrant DNA methylation was also conducted. We performed a pathway analysis on aberrant DNG methylation genes identified by MethylMix criteria using ConsensusPathDB.
Results
188 patients with both methylation data and mRNA data were considered eligible. A mixture model was constructed, and differential methylation genes in normal and tumor groups using the Wilcoxon rank test was performed. With the inclusion criteria, 95 differential methylation genes were detected. Among these genes, 74 hypermethylation and 21 hypomethylation genes were found. The pathway analysis revealed an increase in hypermethylation of genes involved in ATP-sensitive potassium channels, Robo4, and VEGF signaling pathways crosstalk, and generic transcription pathway.
Conclusion
Integrated analysis of the aberrant epigenetic alteration in pancreatic ductal adenocarcinoma indicated that differentially methylated genes could play a vital role in the occurrence of PDAC by bioinformatics analysis. The present work can help clinicians to elaborate on the function of differentially methylated expressed genes and pathways in PDAC. CDO1, GJD2, ID4, NOL4, PAX6, TRIM58, and ZNF382 might act as aberrantly DNA-methylated biomarkers for early screening and therapy of PDAC in the future.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is still one of the primary health problems due to high mortality and incidence worldwide. PDAC remains the primary cause of cancer-related mortality worldwide. It is reported that a 5-year survival rate remains lower, and the average survival time is no more than six months [1]. PDAC is the fourth primary cause of cancer death affecting 56,670 new patients in 2017 in the USA [2, 3]. Although the advances in surgical techniques and chemoradiotherapy protocols had largely improved, the overall survival of PDAC patients remains poor. Meanwhile, due to resistant to radiotherapy and chemotherapy in patients with PDAC, little progress has been made related to its therapy in the past decades [4]. Therefore, to reduce mortality and improve the treatment of PDAC, we need to find new early diagnostic biomarkers and therapeutic targets for early detection and risk classification of PDAC.
DNA methylation has previously been found to be a valuable biomarker for several cancers [5–7]. The epigenetic variations usually suppress protein translation and gene transcription in human carcinogenesis. Several studies have demonstrated that DNA methylation exerted an early event, and new efforts are focused on finding biomarkers for early disease detection, prognostication, and treatment selection, especially in multiple cancers [8–11]. Therefore, elaborating the potential mechanisms during the initiation and development of cancer would greatly improve the diagnosis, treatment, and prognosis evaluation. Abnormal methylation could affect the functions of crucial genes by altering their expression. In this study, we utilized systemic analysis to identify a group of novel gene signatures, which may be regulated by DNA methylation. In addition, the present study can help clinicians to elaborate on the function of DMGs in PDAC. Our study might be the groundwork for further elucidation of the PDAC mechanism and screening of the diagnostic biomarkers for the early stage of PDAC.
2. Materials and Methods
2.1. Data Source and Data Processing
In the current study, the mRNA expression and DNA methylation data of the PDAC cohort were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/, August 28, 2018). The 4 adjacent nontumor pancreatic tissues and 187 PDAC samples were included in the gene expression profiles, where the mRNA microarray employed IlluminaHiSeq RNA-Seq array, while 10 adjacent nontumor control tissues and 178 PDAC tissues were included in the gene methylation dataset, where the methylation microarray used Illumina HumanMethylation 450 BeadChip.
The DEGList and calcNormFacors functions in the edgeR package were employed to normalize the RNA sequence data and DNA methylation data [12]. Both tumor samples and normal samples were used in the same way.
2.2. Integrative Analysis
Through the integration of gene expression and DNA methylation datasets, the MethylMix package in R software was employed to recognize DNA methylation-driven cancer genes [13]. There are three steps to detect DNA methylation-driven cancer genes between the DNA methylation and gene expression datasets. First, the correlation between gene methylation and gene expression level was imputed, and significant correlation genes were found. Second, a beta mixture model was constructed to determine a methylation state across multiple patients. Third, the Wilcoxon rank sum test was employed to compare DNA methylation states between tumor and normal samples. A cutoff of 0.05 was considered statistically significant. The hypomethylation genes were defined as positive differential methylation (DM), while hypermethylation genes were regarded as negative DM.
2.3. Survival Analysis
To further explore the correlation of DNA hypermethylation or hypomethylation genes with overall analysis, the Kaplan–Meier survival analysis and univariate Cox regression analysis were conducted to analyze DNA methylation genes. The log-rank test was employed to compare the survival difference between the PDAC and nontumor samples. A two-sided P value of <0.05 was defined as statistically significant. The R “Survival” package was used to identify independent prognostic variables.
2.4. Pathway Analysis
The pathway analysis was analyzed by the ConsensusPathDB website (http://cpdb.molgen.mpg.de/), which integrated interaction networks in Homo sapiens including protein-protein, gene regulatory, genetic, signaling, metabolic, and drug-target interactions, as well as biochemical pathways [14]. The pathway analysis was performed using the prognostic DNA methylation-driven gene lists produced by MethylMix. The pathway analysis was conducted on the hypermethylation genes and hypomethylation genes, respectively.
3. Results
3.1. Demography
After excluding those patients with a survival of less than one month, 178 patients were included in the study. The clinical and pathological information of the cohort study is exhibited in Table 1. In the whole cohort, 1.12% of patients were less than 35–39 years old, 10.11% were 40–49 years old, 20.79% were 50–59 years old, 29.78% were 60–69 years old, 29.21% were 70–79 years old, and 8.99% were above 80 years old. The median follow-up duration was 46.0 months (range, 2–119 months). There were, respectively, 19 PDCA patients with pathologic TNM stage I, 147 patients with pathologic TNM stage II, 4 patients with pathologic TNM stage III, 5 patients with pathologic TNM stage IV, and 3 patients with an unknown TNM stage in our study. By the end of the last follow-up, 94 (52.81%) patients of the entire population had died.
Table 1.
Clinical characteristics.
| Clinical variables | Clinical values (N = 185) |
|---|---|
| Sex (male/female) | 98/80 |
| Age (mean ± std) | 64.70 ± 11.13 |
| Race (Asian/black/white/NA) | 11/7/155/14 |
| Pathologica stage (I/II/III/IV/V/NA) | 19/21/147/4/5/3 |
| Pathologic_T stage (T1/T2/T3/T4/NA) | 7/21/144/4/2 |
| Pathologic_N stage (N0/N1/NA) | 48/125/5 |
| Pathologic_N stage (M0/M1/NA) | 83/5/90 |
| Grade (G1/G2/G3/G4/Gx/NA) | 29/94/50/2/3 |
3.2. Identifying Methylation-Driven Cancer Genes
A combined approach was utilized to assess the epigenetic alterations that may be involved in the occurrence of the PDAC. The DNA methylation-driven cancer genes were screened using the MethylMix package in R software. The 95 genes were recognized as differential DNA methylation genes when adjusted P value <0.05 and corP value <−0.3 were set as the threshold for differential methylation genes (DMGs). Among these genes, 74 genes (77.89%) were hypermethylation genes, and the remainder of genes were hypomethylation genes (Supplementary ). The heat map is shown in Figure 1.
Figure 1.
Representative heat map of the 74 differential methylation genes. Red represents upregulation; blue represents downregulation.
3.3. Correlation Analysis between DNA Methylation Genes and mRNA
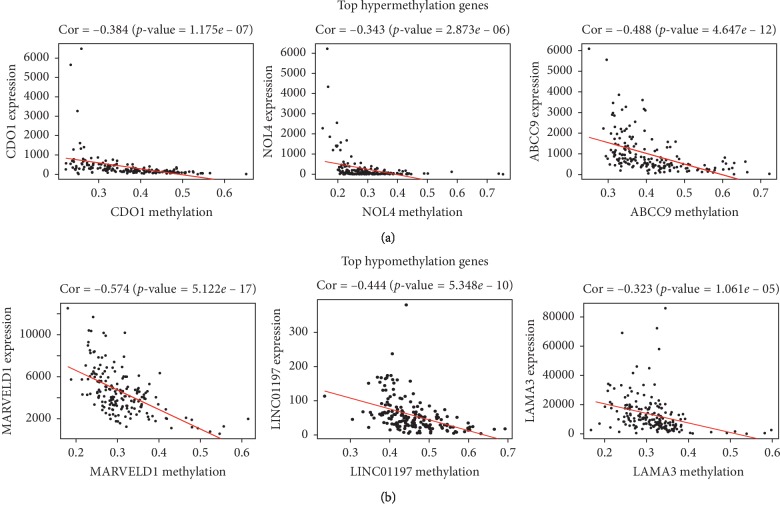
Among 95 differential methylation genes, 74 genes exhibited higher methylation levels in tumor samples compared with normal samples and were referred to as hypermethylation genes, while 21 genes were defined as hypomethylation genes. The top five hypermethylated/hypomethylated genes are shown in Figure 2. All methylation-driven cancer genes showed a negative association between DNA methylation genes and mRNA. The top five hypermethylated/hypomethylated genes are also exhibited in Figure 3.
Figure 2.
Summary of (a) top three hypermethylated and (b) top three hypomethylated genes. The abscissa is the degree of methylation, the ordinate is the number of methylated samples, the histogram represents the methylation distribution of the tumor samples, and the curve demonstrates the simulated trend curve of the methylation distribution in the tumor samples. The black horizontal line above the graph is the methylation level distribution of the normal samples. The red line represents the distribution of methylation in tumor samples.
Figure 3.
Correlation analysis between gene expression and hypermethylated/hypomethylated genes.
3.4. Survival Analysis
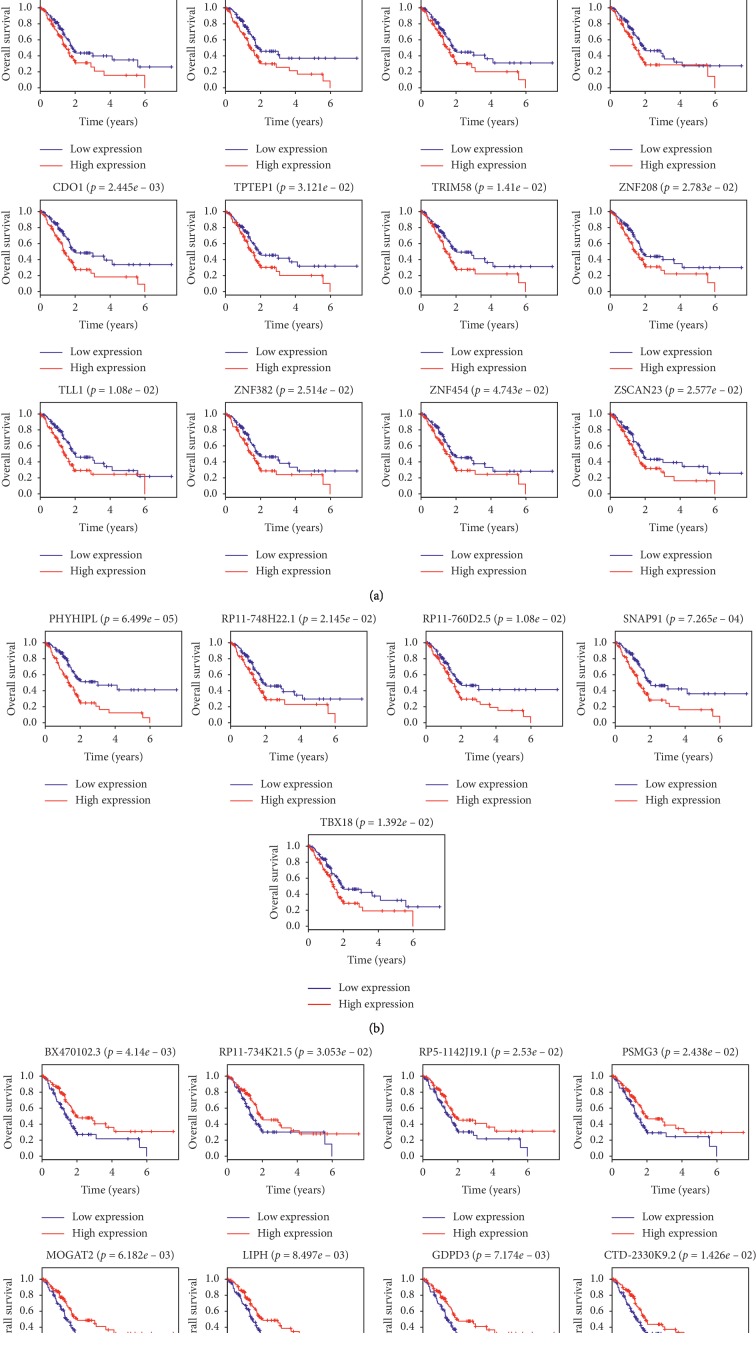
In order to evaluate the effect of differential genes on PDAC patient's prognosis, we conducted the Kaplan–Meier survival analysis and univariate Cox regression analysis. The findings indicated that 25 out of 74 hypermethylation genes and 10 out of 21 hypomethylation were associated with the patient's overall analysis (Table 2). Patients with higher expression in the hypermethylation group exhibited poorer OS than those who have lower expression. However, patients with lower expression in the hypomethylation group demonstrated poorer OS than those who have lower expression. Kaplan–Meier curves for the high-risk and low-risk groups are observed in Figure 4.
Table 2.
Prognostic hypermethylation/hypomethylation genes for PDAC in Kaplan–Meier survival analysis and univariate Cox regression analysis.
| Gene | P value (KM) | HR | Low 95% | High 95% | P value (Cox) |
|---|---|---|---|---|---|
| ID4 | 0.042672 | 3.630678 | 0.520149 | 25.34241 | 0.193383 |
| CBLN4 | 0.023498 | 8.850718 | 1.048889 | 74.68398 | 0.045088 |
| NOL4 | 0.004024 | 4.335809 | 0.684543 | 27.46246 | 0.119342 |
| ZSCAN23 | 0.025771 | 2.565136 | 0.291866 | 22.54433 | 0.395617 |
| ZNF208 | 0.027825 | 4.378881 | 0.610815 | 31.39183 | 0.14171 |
| TPTEP1 | 0.031211 | 9.031061 | 0.565839 | 144.1401 | 0.119457 |
| CTD-2554C21.2 | 0.02653 | 3.226591 | 0.659874 | 15.77709 | 0.148008 |
| HMGCLL1 | 0.032094 | 5.418468 | 0.99398 | 29.53761 | 0.050821 |
| TBX18 | 0.013916 | 1.719498 | 0.399601 | 7.399053 | 0.466623 |
| CDO1 | 0.002445 | 9.153049 | 1.051298 | 79.69033 | 0.044934 |
| GJD2 | 0.03223 | 2.621186 | 0.475785 | 14.44059 | 0.268377 |
| KCNJ8 | 0.047027 | 2.946227 | 0.607266 | 14.29399 | 0.179935 |
| ZNF382 | 0.025139 | 2.887076 | 0.625426 | 13.32725 | 0.174279 |
| RP11-748H22.1 | 0.021448 | 2.767153 | 0.603237 | 12.69342 | 0.190326 |
| AC005498.3 | 0.037114 | 3.33757 | 0.887535 | 12.55091 | 0.074517 |
| KCNA3 | 0.017149 | 1.742724 | 0.2681 | 11.3282 | 0.560838 |
| TLL1 | 0.010797 | 5.624124 | 0.745679 | 42.41878 | 0.093876 |
| ZNF454 | 0.047434 | 2.430637 | 0.414374 | 14.25765 | 0.325139 |
| GRIA2 | 0.007244 | 4.082081 | 0.778325 | 21.40929 | 0.096198 |
| SNAP91 | 0.000726 | 4.182435 | 0.957687 | 18.26563 | 0.057108 |
| PHYHIPL | 6.50E-05 | 4.99462 | 0.648914 | 38.44306 | 0.122433 |
| PAX6 | 0.045649 | 3.477117 | 0.489556 | 24.69657 | 0.212806 |
| TRIM58 | 0.014099 | 2.686691 | 0.452754 | 15.9431 | 0.276686 |
| RP11-760D2.5 | 0.010799 | 3.659025 | 0.898893 | 14.89439 | 0.070119 |
| PABPC5 | 0.029366 | 2.600122 | 0.768849 | 8.79319 | 0.124263 |
| PSMG3 | 0.024383 | 0.070082 | 0.005646 | 0.869985 | 0.038608 |
| BX470102.3 | 0.00414 | 0.056038 | 0.003752 | 0.836902 | 0.036704 |
| CTD-2330K9.2 | 0.014257 | 0.234518 | 0.064259 | 0.855889 | 0.028124 |
| RP11-734K21.5 | 0.030528 | 0.143102 | 0.033485 | 0.611557 | 0.008702 |
| GDPD3 | 0.007174 | 0.056417 | 0.005122 | 0.621427 | 0.018844 |
| C19orf33 | 0.042501 | 0.239202 | 0.03509 | 1.630583 | 0.144101 |
| MOGAT2 | 0.006182 | 0.040403 | 0.00416 | 0.392444 | 0.005669 |
| RP5-1142J19.1 | 0.025301 | 0.111422 | 0.015353 | 0.808636 | 0.030006 |
| C11orf53 | 0.013738 | 0.265261 | 0.031938 | 2.20313 | 0.219203 |
| LIPH | 0.008497 | 0.087046 | 0.010817 | 0.700467 | 0.021758 |
Figure 4.
Kaplan–Meier survival curves for overall survival outcomes according to the risk cutoff point for prognostic hypermethylated/hypomethylated genes. The P value of the log-rank test is less than 0.01.
3.5. Pathway Analysis
To explore the potential functional implication of DNA methylation-driven cancer genes, we performed the pathway analysis by ConsensusPathDB. Several pathways are identified in Figure 5. For hypermethylated genes, pathways were mainly enriched in Robo4 and VEGF signaling pathways crosstalk, ATP-sensitive potassium channels, and generic transcription pathway. For hypomethylated genes, a total of 4 pathways focusing on the biological pathways were enriched, including a6b1 and a6b4 Integrin signaling, metabolism of lipids, and phospholipid metabolism reactome.
Figure 5.
The pathways enriched for hypermethylated and hypomethylated genes in the TCGA PDAC cohort.
4. Discussion
The PDCA is characterized by late diagnosis, poor prognosis, low rates of overall survival, and locoregional recurrences. The primary validated treatment selection remains surgical resection. Local recurrence is a primary cause of failure to treatment [15]. Despite several factors were identified biomarkers for early detection and develop new treatments in PDAC, the overall survival rate and prognosis remain poor [16, 17]. Meanwhile, due to an absence of particular symptoms at an early stage, along with resistance to therapies, high metastatic ability, and lack of diagnostic biomarkers and screening methods, early diagnosis remains the primary treatment option in PDAC. Therefore, it was urgent to explore the potential mechanisms and pathogenesis during the development and progression of PDCA and to uncover new biomarkers and therapeutic targets.
Epigenetic altercation exerts a vital part in carcinogenesis and tumor development progression. Aberrant methylation could affect the functions of crucial genes by altering their expression. Several studies have demonstrated that DNA methylation is referred to as an early phenomenon, and new efforts are focused on recognizing biomarkers of early disease detection, prognostication, and treatment option selection, especially in PDCA [5–7, 18, 19]. DNA hypomethylation has also been documented to be involved in the occurrence of tumors and alters genome rearrangement and chromosomal instability [20, 21]. Therefore, elaborating on the potential mechanisms of development of PDCA would largely elevate the diagnosis and improve the treatment and prognosis evaluation.
In current works, we integrated DNA methylation data and mRNA data and screen DNA methylation-driven cancer genes, and Kaplan–Meier survival analysis was further validated these prognostic results. Compared to normal groups, 95 differential methylation genes (74 hypermethylation genes and 21 hypomethylation gens) were found in the tumor group. We also found that patients with hypermethylation yielded poor-prognosis modifications, demonstrating that many combinations of hypermethylation modifications contribute to poor prognosis. The pathway analysis was also performed, and the results indicated that Robo4 and VEGF signaling pathways crosstalk and ATP-sensitive potassium channels may be related to the development and progression of PDAC. One important result from the pathway analysis was involved in the vascular endothelial growth factor (VEGF) pathway among hypermethylated genes. It is widely accepted that VEGF is a vital driver of the angiogenic modification in physiological and pathological processes in both embryo and adult. VEGF is often found overexpressed in tumors [22]. VEGF exerts a crucial role in vascular homeostasis and the maintenance of vascular integrity. The VEGF signal transduction pathway has identified as an important therapeutic target for patients with many cancers [23, 24]. The two hypermethylated genes (SLIT2 and KDR) were enriched in the pathway. The methylation of SLIT2 was associated with the development and progression of hepatocellular carcinoma [25], dysplasia of pancreatic cystic neoplasms [26], breast cancer [27], and nasopharyngeal carcinoma [28]. The methylation of KDR was also correlated with the development and progression of oral squamous cell carcinoma [29].
Several prognostic hypermethylated genes had been shown to be correlated with a variety of cancers in prior studies (Table 3). A growing body of evidence indicated that CDO1 promoter methylation was correlated with many cancers. Kojima et al. suggested that the hypermethylated gene of CDO1 served as biomarkers and contributed to colorectal cancer [30]. Brait et al. reported that CDO1 serves as a tumor suppressor and is deactivated by promoter methylation in several tumors [31]. Jeschke et al. demonstrated that the silence of CDO1 may account for the survival of breast cancer cells and resistance to anthracyclines [32]. Yang et al. reported the methylation status of the CDO1 promoter to become a diagnostic biomarker for hepatitis B virus-related HCC [33]. CDO1 promoter methylation was also associated with the risk of gastric cancer [34], breast cancer [35], hepatocellular carcinoma [36], and prostate cancer [37]. Sirnes et al. reported that GJC1 promoter methylation played a crucial role in colorectal cancer [38] and follicular lymphoma [39]. ID4 serves as hypermethylation gene and tumor suppressor gene in breast cancer [40, 41] and acute leukemia [42, 43]. ID4 promoter methylation was also correlated with the risk of prostate cancer [44, 45]. Meanwhile, NOL4, PAX6, TRIM58, and ZNF382 promoter methylation was also associated with the occurrence of many cancers [46–55].
Table 3.
Literature search of key hypermethylation genes screened by MethylMix criteria.
| Gene symbol | Gene name | Chromosome | Tumor suppressor gene in cancer | Hypermethylated gene in cancer | Altered pathways | Cancer type |
|---|---|---|---|---|---|---|
| CDO1 | Cysteine dioxygenase type 1 | 5q22.3 | Hypermethylation | Viral mRNA translation; metabolism | Colorectal cancer; hepatocellular carcinoma; gastric cancer; prostate cancer; esophageal squamous cell carcinoma | |
| GJD2 | Gap junction protein delta 2 | 15q14 | Hypermethylation | Gap junction; G-beta gamma signaling |
Colorectal cancer | |
| ID4 | Inhibitor of DNA binding 4, HLH protein | 6p22.3 | Tumor suppressor gene | Hypermethylation | TGF-beta signaling pathway (KEGG); signaling pathways regulating pluripotency of stem cells | Breast cancer; acute leukemia |
| NOL4 | Nucleolar protein 4 | 18q12.1 | Tumor suppressor gene | Head and neck cancer; cervical cancer | ||
| PAX6 | Paired box 6 | 11p13 | Hypermethylation | Gastric cancer; breast cancer | ||
| TRIM58 | Tripartite motif containing 58 | 1q44 | Hypermethylation | Colorectal cancer; lung squamous cell carcinoma; hepatocellular carcinoma | ||
| ZNF382 | Zinc finger protein 382 | 19q13.12 | Tumor suppressor gene | Hypermethylation | Generic transcription pathway | Gastric cancer; pediatric acute myeloid leukemia |
Integrated analysis of the aberrant epigenetic alteration in PDAC indicated that differentially methylated genes may be involved in the occurrence of PDAC. Moreover, the present study can help clinicians to elaborate on the function of differentially methylated expressed genes in PDAC. Our study might be the groundwork for further mechanisms elucidation of PDAC and identification of the diagnostic biomarkers for an early stage of PDAC.
Contributor Information
Yikun Peng, Email: 1076770900@qq.com.
Jukun Song, Email: songjukun@163.com.
Data Availability
The data used to support the findings of this study could be obtained from the TCGA website.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
JKS, QYX, JGZ, YKP, GLH, and RX wrote the main manuscript text. JKS and XHY prepared Figures 1–5. JKS, QYX, and JGZ contributed to data analysis. All the authors reviewed the manuscript.
Supplementary Materials
Supplementary Table 1: identifying methylation-driven cancer genes in PDAC.
References
- 1.Vogelzang N. J., Benowitz S. I., Adams S., et al. Clinical cancer advances 2011: annual report on progress against cancer from the American society of clinical oncology. Journal of Clinical Oncology. 2012;30(1):88–109. doi: 10.1200/jco.2011.40.1919. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Zhang S., Zhao P., Zeng H., Zou X. Report of cancer incidence and mortality in China, 2010. The Annals of Translational Medicine. 2014;2:p. 61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Jones S., Zhang X., Parsons D. W., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishihara H., Yamashita S., Fujii S., Tanabe K., Mukai H., Ushijima T. DNA methylation marker to estimate the breast cancer cell fraction in DNA samples. Medical Oncology. 2018;35(11):p. 147. doi: 10.1007/s12032-018-1207-3. [DOI] [PubMed] [Google Scholar]
- 6.Lin R. K., Hung W. Y., Huang Y. F., et al. Hypermethylation of BEND5 contributes to cell proliferation and is a prognostic marker of colorectal cancer. Oncotarget. 2017;8(69):113431–113443. doi: 10.18632/oncotarget.22266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shindo T., Shimizu T., Nojima M., et al. Evaluation of urinary DNA methylation as a marker for recurrent bladder cancer: a 2-center prospective study. Urology. 2018;113:71–78. doi: 10.1016/j.urology.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Joosten S. C., Deckers I. A., Aarts M. J., et al. Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics. 2017;9(9):1243–1257. doi: 10.2217/epi-2017-0040. [DOI] [PubMed] [Google Scholar]
- 9.Wong C. C., Li W., Chan B., Yu J. Epigenomic biomarkers for prognostication and diagnosis of gastrointestinal cancers. Seminars in Cancer Biology. 2019;55:90–105. doi: 10.1016/j.semcancer.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Jouinot A., Assie G., Libe R., et al. DNA methylation is an independent prognostic marker of survival in adrenocortical cancer. The Journal of Clinical Endocrinology & Metabolism. 2017;102:923–932. doi: 10.1210/jc.2016-3205. [DOI] [PubMed] [Google Scholar]
- 11.Tian Y., Arai E., Gotoh M., Komiyama M., Fujimoto H., Kanai Y. Prognostication of patients with clear cell renal cell carcinomas based on quantification of DNA methylation levels of CpG island methylator phenotype marker genes. BMC Cancer. 2014;14:p. 772. doi: 10.1186/1471-2407-14-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy D. J., Chen Y., Smyth G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Research. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gevaert O. MethylMix: an R package for identifying DNA methylation-driven genes. Bioinformatics. 2015;31(11):1839–1841. doi: 10.1093/bioinformatics/btv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamburov A., Wierling C., Lehrach H., Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Research. 2009;37(suppl_1):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H.-B., Zhou J., Zhao F.-Q. A prognostic nomogram for disease-specific survival in patients with pancreatic ductal adenocarcinoma of the head of the pancreas following pancreaticoduodenectomy. Medical Science Monitor. 2018;24:6313–6321. doi: 10.12659/msm.909649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunne R. F., Hezel A. F. Genetics and biology of pancreatic ductal adenocarcinoma. Hematology/Oncology Clinics of North America. 2015;29(4):595–608. doi: 10.1016/j.hoc.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barhli A., Cros J., Bartholin L., Neuzillet C. Prognostic stratification of resected pancreatic ductal adenocarcinoma: past, present, and future. Digestive and Liver Disease. 2018;50(10):979–990. doi: 10.1016/j.dld.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Dorantes M., Tellez-Ascencio N., Cerbon M. A., Lopez M., Cervantes A. DNA methylation: an epigenetic process of medical importance. Revista de Investigación Clínica. 2004;56(1):56–71. [PubMed] [Google Scholar]
- 19.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods in Molecular Biology. 2009;507:3–20. doi: 10.1007/978-1-59745-522-0_1. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe Y., Maekawa M. Methylation of DNA in cancer. Advances in Clinical Chemistry. 2010;52:145–167. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S., Kelly T. K., Jones P. A. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehghani S., Nosrati R., Yousefi M., et al. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): a review. Biosensors and Bioelectronics. 2018;110:23–37. doi: 10.1016/j.bios.2018.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Hopirtean C., Nagy V. Optimizing the use of anti VEGF targeted therapies in patients with metastatic colorectal cancer: review of literature. Medicine and Pharmacy Reports. 2018;91(1):12–17. doi: 10.15386/cjmed-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vachhani P., George S. VEGF inhibitors in renal cell carcinoma. Clinical Advances in Hematology & Oncology. 2016;14:1016–1028. [PubMed] [Google Scholar]
- 25.Sun G., Zhang C., Feng M., et al. Methylation analysis of p16, SLIT2, SCARA5, and Runx3 genes in hepatocellular carcinoma. Medicine (Baltimore) 2017;96 doi: 10.1097/md.0000000000008279.e8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hata T., Dal Molin M., Hong S.-M., et al. Predicting the grade of dysplasia of pancreatic cystic neoplasms using cyst fluid DNA methylation markers. Clinical Cancer Research. 2017;23(14):3935–3944. doi: 10.1158/1078-0432.ccr-16-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G.-E., Lee K. H., Choi Y. D., et al. Detection of Slit2 promoter hypermethylation in tissue and serum samples from breast cancer patients. Virchows Archiv. 2011;459(4):383–390. doi: 10.1007/s00428-011-1143-5. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Zhou C., Wang G., et al. Promoter hypermethylation of SLIT2 is a risk factor and potential diagnostic biomarker for nasopharyngeal carcinoma. Gene. 2018;644:74–79. doi: 10.1016/j.gene.2017.10.059. [DOI] [PubMed] [Google Scholar]
- 29.Tsai B. S., Villani-Price D., Keith R. H., et al. SC-41930: an inhibitor of leukotriene B4-stimulated human neutrophil functions. Prostaglandins. 1989;38(6):655–674. doi: 10.1016/0090-6980(89)90048-8. [DOI] [PubMed] [Google Scholar]
- 30.Kojima K., Nakamura T., Ohbu M., et al. Cysteine dioxygenase type 1 (CDO1) gene promoter methylation during the adenoma-carcinoma sequence in colorectal cancer. PLoS One. 2018;13(5) doi: 10.1371/journal.pone.0194785.e0194785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brait M., Ling S., Nagpal J. K., et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0044951.e44951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeschke J., O’Hagan H. M., Zhang W., et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clinical Cancer Research. 2013;19(12):3201–3211. doi: 10.1158/1078-0432.ccr-12-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., Fan Y.-C., Gao S., et al. Methylated cysteine dioxygenase-1 gene promoter in the serum is a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. The Tohoku Journal of Experimental Medicine. 2014;232(3):187–194. doi: 10.1620/tjem.232.187. [DOI] [PubMed] [Google Scholar]
- 34.Ushiku H., Yamashita K., Ema A., et al. DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer. Gastric Cancer. 2017;20(5):784–792. doi: 10.1007/s10120-017-0697-6. [DOI] [PubMed] [Google Scholar]
- 35.Minatani N., Waraya M., Yamashita K., et al. Prognostic significance of promoter DNA hypermethylation of cysteine dioxygenase 1 (CDO1) gene in primary breast cancer. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0144862.e0144862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J.-I., Cho E.-H., Kim S. B., et al. Promoter methylation of cysteine dioxygenase type 1: gene silencing and tumorigenesis in hepatocellular carcinoma. Annals of Hepato-Biliary-Pancreatic Surgery. 2017;21(4):181–187. doi: 10.14701/ahbps.2017.21.4.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meller S., Zipfel L., Gevensleben H., et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11(12):871–880. doi: 10.1080/15592294.2016.1241931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirnes S., Honne H., Ahmed D., et al. DNA methylation analyses of the connexin gene family reveal silencing of GJC1(Connexin45) by promoter hypermethylation in colorectal cancer. Epigenetics. 2011;6(5):602–609. doi: 10.4161/epi.6.5.15237. [DOI] [PubMed] [Google Scholar]
- 39.Bennett L. B., Schnabel J. L., Kelchen J. M., et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes, Chromosomes and Cancer. 2009;48(9):828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umetani N., Mori T., Koyanagi K., et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24(29):4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 41.Nasif D., Campoy E., Laurito S., et al. Epigenetic regulation of ID4 in breast cancer: tumor suppressor or oncogene? Clinical Epigenetics. 2018;10(1):p. 111. doi: 10.1186/s13148-018-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Zhong W. W., Kang H. Y., et al. Clinical significance of ID4 methylation detection by quantitative methylation-specific PCR in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22:675–680. doi: 10.7534/j.issn.1009-2137.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y., Kang H. Y., Wang L. L., Lu X. C., Zhu H. L., Yu L. Establishment of methylation-specific quantitative PCR system for ID4 gene in acute leukemia cells and its specificity and sensitivity. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22:269–274. doi: 10.7534/j.issn.1009-2137.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Carey J. P., Asirvatham A. J., Galm O., Ghogomu T. A., Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:p. 173. doi: 10.1186/1471-2407-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinarskaja A., Goering W., Ingenwerth M., Schulz W. A. ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World Journal of Urology. 2012;30(3):319–325. doi: 10.1007/s00345-011-0750-8. [DOI] [PubMed] [Google Scholar]
- 46.Demokan S., Chuang A. Y., Pattani K. M., Sidransky D., Koch W., Califano J. A. Validation of nucleolar protein 4 as a novel methylated tumor suppressor gene in head and neck cancer. Oncology Reports. 2014;31(2):1014–1020. doi: 10.3892/or.2013.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S. S., Smiraglia D. J., Wu Y.-Z., et al. Identification of novel methylation markers in cervical cancer using restriction landmark genomic scanning. Cancer Research. 2008;68(7):2489–2497. doi: 10.1158/0008-5472.can-07-3194. [DOI] [PubMed] [Google Scholar]
- 48.Mzik M., Chmelarova M., John S., et al. Aberrant methylation of tumour suppressor genes WT1, GATA5 and PAX5 in hepatocellular carcinoma. Clinical Chemistry and Laboratory Medicine (CCLM) 2016;54(12):1971–1980. doi: 10.1515/cclm-2015-1198. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q., Shao Y., Shi J., et al. Concomitant PIK3CA amplification and RASSF1A or PAX6 hypermethylation predict worse survival in gastric cancer. Clinical Biochemistry. 2014;47(1-2):111–116. doi: 10.1016/j.clinbiochem.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Moelans C. B., Verschuur-Maes A. H., van Diest P. J. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. The Journal of Pathology. 2011;225(2):222–231. doi: 10.1002/path.2930. [DOI] [PubMed] [Google Scholar]
- 51.Liu M., Zhang X., Cai J., et al. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncology Reports. 2018;40:1251–1260. doi: 10.3892/or.2018.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu X., Huang Y., Zhou Y., Zheng F. Aberrant methylation of TRIM58 in hepatocellular carcinoma and its potential clinical implication. Oncology Reports. 2016;36(2):811–818. doi: 10.3892/or.2016.4871. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W., Cui Q., Qu W., Ding X., Jiang D., Liu H. TRIM58/cg26157385 methylation is associated with eight prognostic genes in lung squamous cell carcinoma. Oncology Reports. 2018;40:206–216. doi: 10.3892/or.2018.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei L., He X., Li S., et al. KRAB zinc-finger protein 382 regulates epithelial-mesenchymal transition and functions as a tumor suppressor, but is silenced by CpG methylation in gastric cancer. International Journal of Oncology. 2018;53:961–972. doi: 10.3892/ijo.2018.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao Y.-F., Hu S.-Y., Lu J., et al. Zinc finger protein 382 is downregulated by promoter hypermethylation in pediatric acute myeloid leukemia patients. International Journal of Molecular Medicine. 2014;34(6):1505–1515. doi: 10.3892/ijmm.2014.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: identifying methylation-driven cancer genes in PDAC.
Data Availability Statement
The data used to support the findings of this study could be obtained from the TCGA website.