Abstract
Objective: Mucolipidosis II and III alpha/beta (ML II & ML III alpha/beta) are rare autosomal recessive lysosomal storage disorders. ML II is clinically evident from birth with a progressive course and fatal outcome in childhood. The typical phenotypes of ML II include limited statural growth, craniofacial abnormality, skeletal malformation, intelligence developmental deficiency and visceral organ abnormality. ML III is milder than ML II. Mutations in GNPTAB cause the ML II/III. Methods: Two families with ML II/III (initially undiagnosed) were recruited. We applied whole-exome sequencing (WES) and filtered mutations by genes causing lysosomal storage diseases with skeletal involvement. Mutational analysis and co-segregation confirmation were then performed. Results: We presented two families with ML II or ML III alpha/beta. By WES, the compound heterozygosity of GNPTAB (c.2404C>T, p.Q802* and c.2590dup, p.E864Gfs*4) is identified in a family with ML II, and c.1364C>T, p.A455V and c.2715+1G>A are detected in a family with ML III alpha/beta. Conclusion: We detected the causative mutations in two ML II/III families by WES and confirmed their diagnosis of the diseases. The present identification of mutations expands the spectrum of known GNPTAB mutations and it may contribute to novel approaches to genetic diagnosis and counseling for patients with ML II/III.
Keywords: GNPTAB, ML II, ML III, GlcNAc-1-PT, whole-exome sequencing, lysosomal storage disorders
Introduction
Mucolipidosis II alpha/beta ML II; OMIM 252500) and Mucolipidosis III alpha/beta (ML III alpha/beta; OMIM 252600) are very rare autosomal recessive lysosomal storage disorders, originally called Inclusion Cell Disease (I-cell disease) [1,2]. Both of them are severe multi-organ diseases and result from a deficiency of N-acetylglucosamine-1-phosphotransferase (GlcNAc-1-PT or GNPT) [3]. Symptoms of ML II are observed at birth and cause progressive deterioration, and may result in death in childhood (most often during the first decade of life) [4]. The typical phenotypes of ML II include limited statural growth, craniofacial abnormality (including facial coarseness, prominent forehead, shallow orbits, depressed nasal bridge and hyperrophic gingiva), skeletal malformation (including scoliosis, hip dislocation, long bone deformities, hand contracture and clubfeet), intelligence deficiency and visceral organ abnormality [5-7]. ML III is milder than ML II, with an onset in early childhood, slowly progressive course, and fatal outcome from early adulthood [8,9].
GlcNAc-1-PT is a Golgi-resident enzyme complex and catalyzes the initial step in the production of the mannose 6-phosphate (M6P) recognition marker, which is essential for lysosomal hydrolase transport into lysosomes [10,11]. GlcNAc-1-PT contains two α, two β and two γ polypeptides [12]. GNPTAB gene encodes the α- and β-subunits, and GNPTG encodes the γ-subunit [13,14]. Human GNPTAB is located on chromosome 12q23.3 (13), it spans approximately 80 kb of cDNA consisting of 21 exons, and encodes a 1256 amino acid (AAs) precursor protein (16). This precursor protein needs to be processed into mature α- and β-polypeptides by proteolytic cleavage at the Lys928-Asp929 bond [10,15]. Mutations in the GNPTAB lead to ML II and ML III alpha/beta, and GNPTG mutations cause ML III gamma (OMIM 252605) [16].
In this study, we present two families from Central-South region of China, one with ML II and the other with ML III alpha/beta. In the first family, a compound heterozygosity of GNPTAB (c.2404C>T, p.Q802* and c.2590dup, p.E864Gfs*4) is identified, and we found c.1364C>T, p.A455V and c.2715+1G>A in the second family.
Materials and methods
Patients and subjects
The Review Board of Xiangya Hospital of the Central South University has approved this research. All subjects have consented to this study. Blood was collected from the proband and related family members. Segregation analysis was applied to all family members according to the WES results.
Whole-exome sequencing
Genomic DNA was extracted with a DNeasy blood and tissue kit (Qiagen, Valencia, Calif., USA). The Novogene Bioinformatics Institute (Beijing, China) provided the exome capture, high throughput sequencing and common filtering. All the exomes were captured by means of Agilent SureSelect Human All Exon V5 kits and were sequenced with an Illumina HiSeq2000 platform. Filtering the common variants (frequency >0.05) from the 1000 Genomes Project (https://www.genome.gov/27528684/1000-genomes-project/), YH (http://yh.genomics.org.cn/), dbSNP (https://www.ncbi.nlm.nih.gov/SNP/) and ESP (http://evs.gs.washington.edu/EVS/) databases, unique single-nucleotide polymorphisms (SNPs) were detected in subjects. Potential causative variants were screened by the list of genes about lysosomal storage diseases with skeletal involvement (Table S1) and then predicted by bioinformatics programs including MutationTaster (http://www.mutationtaster.org/), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://provean.jcvi.org/index.php). The analyses of gene function, inheritance pattern, and clinical phenotype were conducted by Online Mendelian Inheritance in Man (OMIM) (https://www.omim.org).
Sanger sequencing
Primer pairs were designed by DNASTAR and the sequences of primers will be provided upon request. The target fragments were amplified with polymerase chain reaction (PCR) and determined by the ABI 3100 Genetic Analyzer (ABI, Foster City, CA).
Result
Clinical features
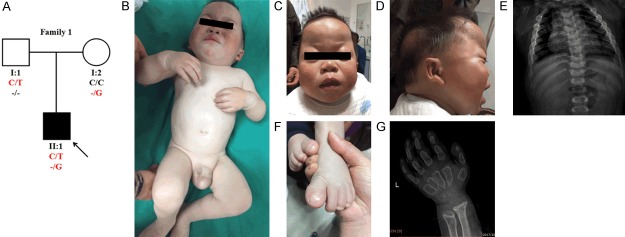
We first identified a family with ML II (Family 1) (Figure 1A). The proband (II:1) is a 3 year and 8 months old boy. He was born at term and diagnosed with inguinal hernia. One year after his birth, he had coarse facial features and no ability to speak and walk. Now, the proband has a short stature (75.3 cm, 10.4 kg), facial coarseness with depressed nasal bridge and shallow orbits, metopic prominence, thickened alveolar ridges, cup-ear and sparse hair in a part of the head, scoliosis, hand contracture, broad thumbs and big toes with abnormal nails, bowed lower limbs, intelligence developmental deficiency (his verbal expression was still limited to a few words) and inguinal hernia (Figure 1B-G). The family went to our department because the proband failed in unaided walking. We primarily diagnosed him with lysosomal storage disorder, most likely Mucopolysaccharidosis type 1H (MPS-1H, also named Hurler syndrome; OMIM 607015), and confirmed he had ML II after genetic diagnosis. His parents (I:1 and I:2) were not affected.
Figure 1.
(A) Pedigree of the Family 1 with segregation analysis. Family members are identified by their generation (indicated by Roman numerals) and a number. Squares represent male family members and circles, female members. The black symbols represent a member affected and the white symbols represent unaffected members. The arrow indicates the proband. Genotype is identified by letters and slash, red represents mutations. (B-G) Phenotypes of the proband of Family 1 (B) is a full body shot of the proband of Family 1, who has coarse facial features (C), cup-ear (D), mild scoliosis (E), broad thumbs and big toes with abnormal nails (F, G) and inguinal hernia (B).
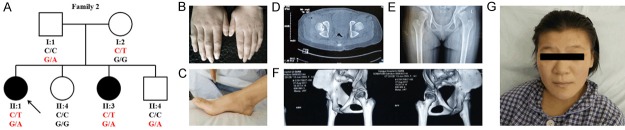
In the second family (Family 2) (Figure 2A), the proband (II:1) is a 14-year-old girl (150 cm, 42 kg) diagnosed with ML III alpha/beta, without intelligence developmental deficiency. No abnormalities were present at her birth and she walked independently at 2 and half years old. She was observed to have bilateral contracture of 2-5 fingers and talipes cavus before age 3 years (Figure 2B, 2C). The proband presented to our department for the pain of hip joints and was diagnosed with congenital dislocation of the hip (CDH) presenting unusual symptoms (shallow acetabulum and caput femoris, rugged articulatio coxae with low density focus) by the X-ray, CT and 3-D image (Figure 2D-F). Also, the proband had minor facial changes (Figure 2G). We initially diagnosed her with MPS or other lysosomal storage disorder. We thought her sister (II:3), a 4-year-old girl with bilateral contracture of digital joints and having the ability to walk independently at 3 years, might suffer from the same disease. Other members (I:1, I:2, II:2 and II:4) were not affected.
Figure 2.
(A) Pedigree of the Family 2 with segregation analysis. The meanings of all symbols are same as Figure 1A. (B-G) Phenotypes of the proband of Family 2. The proband of Family 2 with ML III has bilateral contracture of 2-5 fingers (B), talipes cavus (C), congenital dislocation of the hip (D-F), and minor facial changes (G).
Genetic analysis
By WES, we detected 1156 unique SNPs in the proband of Family 1 and 1064 SNPs in the proband of Family 2. Variants were filtered by genes for lysosomal storage diseases with skeletal involvement. 2 GNPTAB variants were identified in each of two families, without other variants of genes of lysosomal storage diseases (Table 1). The bioinformatic prediction about pathogenicity and the analysis of inheritance pattern, OMIM clinical phenotypes and American College of Medical Genetics classification all suggested these mutations causing these two families’ diseases.
Table 1.
Variants identified by WES in combination with lysosomal storage diseases (skeletal involvement)-related gene-filtering in the present families
| Family | Gene | Variant | MutationTaster | Polyphen-2 | SIFT | OMIM clinical phenotype | American College of Medical Genetics classification |
|---|---|---|---|---|---|---|---|
| 1 | GNPTAB | NM_024312: c. 2404C>T: p.Q802* | D (1.000) | - | - | AR; Mucolipidosis II alpha/beta/AR; Mucolipidosis III alpha/beta | PVS1 |
| 1 | GNPTAB | NM_024312: c.2590dupG: p.E864fs* | D (1.000) | - | - | PVS1 | |
| 2 | GNPTAB | NM_024312: c.2715+1G>A | D (1.000) | - | - | PVS1 | |
| 2 | GNPTAB | NM_024312: c. 1364C>T: p.A455V | D (1.000) | D (0.973) | D (0.000) | PM5 |
D, disease causing; AR, autosomal recessive; PVS, pathogenic very strong; PM, pathogenic moderate.
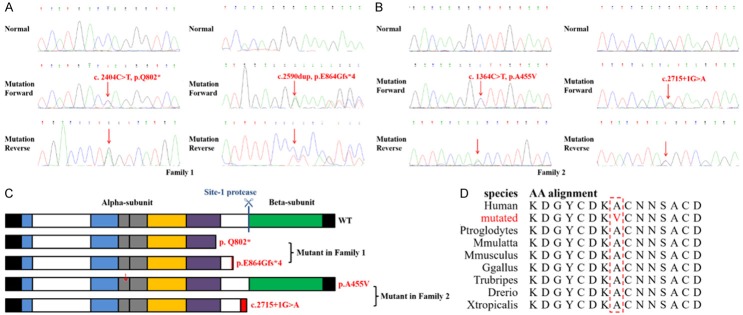
In Family 1, Sanger sequencing showed that a nonsense mutation (c.2404C>T, p.Q802*) of GNPTAB in the proband was inherited from his father and a frameshift mutation (c.2590dup, p.E864Gfs*4) from his mother (Figure 3A). Similarly, in Family 2, the mutations (c.1364C>T, p.A455V and c.2715+1G>A) of the proband were inherited from parents respectively, and co-segregated with the affected family members (Figure 3B). Neither of the mutations was identified in the 200 control cohorts that our group studied previously. The final diagnosis was that proband of Family 1 (II:1) has ML II and patients in Family 2 (II:1 and II:3) have ML III.
Figure 3.
(A, B) Sequencing results of the GNPTAB mutations. Sequence chromatograms indicate compound heterozygosity (c.2404C>T, p.Q802* and c.2590dup, p.E864Gfs*4) in the proband of Family 1 (A) and indicate compound heterozygosity (c.1364C>T, p.A455V and IVS13+1G>A) in the proband of Family 2 (B). (C) A schematic presentation of the domain of the wild-type (indicated by “WT”) GNPTAB precursor and mutant GNPTAB precursor proteins in the present study (indicated by red words). The rectangular box represents the precursor protein with the N-terminus on the left and C-terminus on the right. The black box represents the transmembrane domain, blue represents the Stealth domain, gray represents the Notch-repeat-like domain, yellow represents the γ-binding domain, purple represents the DMAP domain and green represents a conserved region. The position of the site-1 protease cleavage site is indicated by a blue vertical bar. Incorrect amino acids sequence in frameshift mutants are indicated by the red box, and the red arrows represent a mutated AA site. (D) This site of the α subunit with A455V is highly evolutionarily conserved across species. Red grapheme represents mutated amino acid, red box emphasizes these sites cross species to compare.
Discussion
In this study, we identified 4 mutations of GNPTAB in two families with ML II/III. GNPTAB precursor includes two transmembrane domains, two Stealth domains, Notch-repeat-like domains, a γ-binding domain, a DMAP domain and a conserved region (Figure 3C) [1,17]. In the patient of Family 1, the nonsense mutation (c.2404C>T, p.Q802*) is expected to produce truncated protein and the frameshift mutation (c.2590dup, p.E864Gfs*4) transcribe into an incorrect transcript with a premature stop codon, which results in the GlcNAc-1-PT with deficiency or lack of biological activity and causes ML II (Figure 3C). Both have been described by Wang et al. (2018) and Liu et al. (2015) respectively in Chinese families with ML II/III [18,19]. The splice-site mutation (c.2715+1G>A) has described already in patients from Japan, Korea and China [19-22]. In fact, it is the second most common mutation and appears to be quite frequent in Chinese patients. The mutation produces a transcript without the 13rd exon (1103 bases) and formations a new stop codon. The missense mutation (c.1364C>T, p.A455V) happens at Notch-repeat-like domains and the site is predicted to be evolutionarily conserved (Figure 3C, 3D).
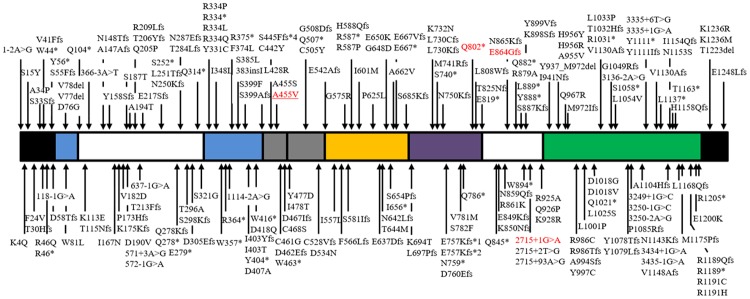
At least 207 variants/mutations of GNPTAB has been reported (Figure 4). Summarizing these variants/mutations and referencing the previous predictions, it suggests that 1) most GNPTAB genotypes in the patients of ML II are homozygous or compound heterozygous either for frameshift mutations or for nonsense mutations; 2) the patients of ML III alpha/beta carry homozygous or compound heterozygous mutations at least including one missense or splice-site mutation [8]. Nonsense mutations produce a truncated protein, and frameshift mutations result in a premature stop codon or an entirely different following sequence. These “null” or “amorph” alleles would lead to endoplasmic reticulum (ER) retention of the mutant proteins [1,17]. Therefore these mutants are always inactive and cause the severe ML II phenotypes. ML III patients mostly carry mutations that induce only partial loss of activity. Given II:3 in Family 2 harboring a missense mutation and a splice-site mutation of GNPTAB and beginning to present slight symptoms, we diagnosed her with ML III alpha/beta in the early stage and advised her to receive treatment, for example physical therapy, to slow down the progression. This showed the superiority of genetic diagnosis in the early diagnosis of inherited progressive disease. Prediction is helpful to genetic diagnosis and also needs more cases to demonstrate it.
Figure 4.
GNPTAB mutations identified in the alpha/beta-subunit precursor proteins of the GlcNAc-1-PT. The rectangular box represents the precursor protein with the N-terminus on the left and C-terminus on the right, and the meanings are same as Figure 3C. Mutations are grouped based on the mutation site. The red words represent the present mutations. Underline represents the mutation that is the first reported.
The symptoms of ML II and ML III are similar with MPS-diseases, including onset in the early period of life, facial coarseness, skeletal dysplasia, dysgnosia and visceral organ abnormality [23,24]. But their disease-causing genes are different; for example, MPS-IH is caused by homozygous or compound heterozygous mutations of IDUA (encoding alphe-L-iduronidase) [25]. Genetic diagnosis is one of the best ways to distinguish between ML II/III and MPS-diseases, which can be widely used to distinguish various lysosomal storage disorders. Urine screens support the diagnosis. In patients with MPS-diseases, urinary excretion of glycosaminoglycans (GAG) increases. But GAG excretion of ML II/III is normal, while oligosaccharide may be excessive [4]. As to our patient of Family 1, urinary screening by toluidine blue test for GAG was negative. ML II and ML III are pathologically similar, but ML III is milder than ML II [2]. In the present study, the symptoms of the patient of Family 1 were more severe than patients of Family 2.
In conclusion, the present study identified two families from Central-South region of China, one with ML II and the other with ML III alpha/beta. In the first family, a compound heterozygosity of GNPTAB (c.2404C>T, p.Q802* and c.2590dup, p.E864Gfs*4) was identified, and we found c.1364C>T, p.A455V and c.2715+1G>A in the second family. The mutation c.1364C>T, p.A455V is first reported here. The present identification of mutations expands the spectrum of known GNPTAB mutations and it may contribute to novel approaches to genetic diagnosis and counseling for patients with ML II/III.
Acknowledgements
This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (2017ZX10103005-006), the National Natural Science Foundation of China (81370394), the Scientific Research Plan Projects of Health and Family Planning Commission of Hunan Province (C2017002) and the Fundamental Research Funds for Central Universities of Central South University (2019zzts048, XCX20190747). We thank the patients and their families for participating in this study.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ludwig NF, Velho RV, Sperb-Ludwig F, Acosta AX, Ribeiro EM, Kim CA, Gandelman Horovitz DD, Boy R, Rodovalho-Doriqui MJ, Lourenco CM, Santos ES, Braulke T, Pohl S, Schwartz IVD. GNPTAB missense mutations cause loss of GlcNAc-1-phosphotransferase activity in mucolipidosis type II through distinct mechanisms. Int J Biochem Cell Biol. 2017;92:90–94. doi: 10.1016/j.biocel.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho MF, Encarnacao M, Laranjeira F, Lacerda L, Prata MJ, Alves S. Solving a case of allelic dropout in the GNPTAB gene: implications in the molecular diagnosis of mucolipidosis type III alpha/beta. J Pediatr Endocrinol Metab. 2016;29:1225–1228. doi: 10.1515/jpem-2016-0173. [DOI] [PubMed] [Google Scholar]
- 3.Cury GK, Matte U, Artigalas O, Alegra T, Velho RV, Sperb F, Burin MG, Ribeiro EM, Lourenco CM, Kim CA, Valadares ER, Galera MF, Acosta AX, Schwartz IV. Mucolipidosis II and III alpha/beta in Brazil: analysis of the GNPTAB gene. Gene. 2013;524:59–64. doi: 10.1016/j.gene.2013.03.105. [DOI] [PubMed] [Google Scholar]
- 4.Cathey SS, Leroy JG, Wood T, Eaves K, Simensen RJ, Kudo M, Stevenson RE, Friez MJ. Phenotype and genotype in mucolipidoses II and III alpha/beta: a study of 61 probands. J Med Genet. 2010;47:38–48. doi: 10.1136/jmg.2009.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang M, Cho SY, Park HD, Choi R, Kim YE, Kim J, Lee SY, Ki CS, Kim JW, Sohn YB, Song J, Jin DK. Clinical, biochemical and molecular characterization of Korean patients with mucolipidosis II/III and successful prenatal diagnosis. Orphanet J Rare Dis. 2017;12:11. doi: 10.1186/s13023-016-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibazaki T, Hirabayashi K, Saito S, Shigemura T, Nakazawa Y, Sakashita K, Takagi M, Shiohara M, Adachi K, Nanba E, Sakai N, Koike K. Clinical and laboratory outcomes after umbilical cord blood transplantation in a patient with mucolipidosis II alpha/beta. Am J Med Genet A. 2016;170A:1278–1282. doi: 10.1002/ajmg.a.37563. [DOI] [PubMed] [Google Scholar]
- 7.Hashemi-Gorji F, Ghafouri-Fard S, Salehpour S, Yassaee VR, Miryounesi M. A novel splice site mutation in the GNPTAB gene in an Iranian patient with mucolipidosis II alpha/beta. J Pediatr Endocrinol Metab. 2016;29:991–993. doi: 10.1515/jpem-2016-0032. [DOI] [PubMed] [Google Scholar]
- 8.Cathey SS, Leroy JG, Wood T, Eaves K, Simensen RJ, Kudo M, Stevenson RE, Friez MJ. Phenotype and genotype in mucolipidoses II and III alpha/beta: a study of 61 probands. J Med Genet. 2010;47:38–48. doi: 10.1136/jmg.2009.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara M, Inokuchi T, Taniwaki T, Otomo T, Sakai N, Matsuishi T, Yoshino M. An adult patient with mucolipidosis III alpha/beta presenting with parkinsonism. Brain Dev. 2013;35:462–465. doi: 10.1016/j.braindev.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Marschner K, Kollmann K, Schweizer M, Braulke T, Pohl S. A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science. 2011;333:87–90. doi: 10.1126/science.1205677. [DOI] [PubMed] [Google Scholar]
- 11.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Velho RV, De Pace R, Klunder S, Di Lorenzo G, Schweizer M, Braulke T, Pohl S. Site-1 protease and lysosomal homeostasis. Biochim Biophys Acta Mol Cell Res. 2017;1864:2162–2168. doi: 10.1016/j.bbamcr.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Gowda VK, Raghavan VV, Bhat M, Benakappa A. Mucolipidosis type II secondary to GNPTAB gene deletion from India. J Pediatr Neurosci. 2017;12:115–116. doi: 10.4103/1817-1745.205656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arowolo AT, Adeola HA, Khumalo NP. Next generation sequencing identifies mutations in GNPTG gene as a cause of familial form of scleroderma-like disease. Pediatr Rheumatol Online J. 2017;15:88. doi: 10.1186/s12969-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiede S, Storch S, Lubke T, Henrissat B, Bargal R, Raas-Rothschild A, Braulke T. Mucolipidosis II is caused by mutations in GNPTA encoding the alpha/beta GlcNAc-1-phosphotransferase. Nat Med. 2005;11:1109–1112. doi: 10.1038/nm1305. [DOI] [PubMed] [Google Scholar]
- 16.Qian Y, van Meel E, Flanagan-Steet H, Yox A, Steet R, Kornfeld S. Analysis of mucolipidosis II/III GNPTAB missense mutations identifies domains of UDP-GlcNAc:lysosomal enzyme GlcNAc-1-phosphotransferase involved in catalytic function and lysosomal enzyme recognition. J Biol Chem. 2015;290:3045–3056. doi: 10.1074/jbc.M114.612507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velho RV, De Pace R, Klunder S, Sperb-Ludwig F, Lourenco CM, Schwartz IV, Braulke T, Pohl S. Analyses of disease-related GNPTAB mutations define a novel GlcNAc-1-phosphotransferase interaction domain and an alternative site-1 protease cleavage site. Hum Mol Genet. 2015;24:3497–3505. doi: 10.1093/hmg/ddv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Wei X, Kong X, Guo X, Sun Y, Man J, Du L, Zhu H, Qu Z, Tian P, Mao B, Yang Y. Targeted next-generation sequencing for clinical diagnosis of 561 mendelian diseases. PLoS One. 2015;10:e0133636. doi: 10.1371/journal.pone.0133636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Ye J, Qiu WJ, Han LS, Gao XL, Liang LL, Gu XF, Zhang HW. Identification of predominant GNPTAB gene mutations in Eastern Chinese patients with mucolipidosis II/III and a prenatal diagnosis of mucolipidosis II. Acta Pharmacol Sin. 2019;4:279–287. doi: 10.1038/s41401-018-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik KH, Song SM, Ki CS, Yu HW, Kim JS, Min KH, Chang SH, Yoo EJ, Lee IJ, Kwan EK, Han SJ, Jin DK. Identification of mutations in the GNPTA (MGC4170) gene coding for GlcNAc-phosphotransferase alpha/beta subunits in Korean patients with mucolipidosis type II or type IIIA. Hum Mutat. 2005;26:308–314. doi: 10.1002/humu.20205. [DOI] [PubMed] [Google Scholar]
- 21.Otomo T, Muramatsu T, Yorifuji T, Okuyama T, Nakabayashi H, Fukao T, Ohura T, Yoshino M, Tanaka A, Okamoto N, Inui K, Ozono K, Sakai N. Mucolipidosis II and III alpha/beta: mutation analysis of 40 Japanese patients showed genotype-phenotype correlation. J Hum Genet. 2009;54:145–151. doi: 10.1038/jhg.2009.3. [DOI] [PubMed] [Google Scholar]
- 22.Zhan T, Cui X, Xing X, Ren A, Gan G, Liu Y, Zhang J, Tang Z, Liu M. Mucolipidosis in a Chinese family with compound heterozygous mutations at the GNPTAB gene. Clin Chim Acta. 2011;412:1469–1471. doi: 10.1016/j.cca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Leroy JG, Sillence D, Wood T, Barnes J, Lebel RR, Friez MJ, Stevenson RE, Steet R, Cathey SS. A novel intermediate mucolipidosis II/IIIalphabeta caused by GNPTAB mutation in the cytosolic N-terminal domain. Eur J Hum Genet. 2014;22:594–601. doi: 10.1038/ejhg.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehtonen A, Rust S, Jones S, Brown R, Hare D. Social functioning and behaviour in mucopolysaccharidosis IH [Hurlers Syndrome] . JIMD Rep. 2018;39:75–81. doi: 10.1007/8904_2017_47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngiwsara L, Ketudat-Cairns JR, Sawangareetrakul P, Charoenwattanasatien R, Champattanachai V, Kuptanon C, Pangkanon S, Tim-Aroon T, Wattanasirichaigoon D, Svasti J. p. X654R IDUA variant among Thai individuals with intermediate mucopolysaccharidosis type I and its residual activity as demonstrated in COS-7 cells. Ann Hum Genet. 2018;82:150–157. doi: 10.1111/ahg.12236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.