Abstract
Purpose: To investigate the role of the autophagy-related genes Beclin1 and LC3 in the prognosis of pancreatic cancer. Methods: A total of 86 pancreatic cancer tissues and 84 paired, adjacent normal pancreatic tissues were collected from 86 patients who underwent pancreatic resection surgery in our hospital from January 2009 to August 2011. Demographic data including age, gender, family cancer history, and clinic-pathological characteristics, including tumor diameter, differential, TNM staging and lymphatic metastasis were collected. The expressions of Beclin1 and LC3 were determined using both immunohistochemistry (IHC) and RT-qPCR. Results: The expression levels of both Beclin1 and LC3 mRNA and proteins were significantly up-regulated in the tumor tissues compared with the normal tissues. Higher expressions of Beclin1 and LC3 were found in the tumor tissues of patients with TNM stages III~IV, patients with lymphatic metastasis, and patients who died. Meanwhile Beclin1 and LC3 correlated with TNM stage, differential condition, and the patients’ lymphatic metastasis rates. A survival analysis showed that patients with low expressions of Beclin1 and LC3 had longer survival times, and both the Beclin1 and LC3 genes were independent risk factors for 5-year mortality in pancreatic cancer patients. Conclusion: The Beclin1 and LC3 genes correlate with the tumor stage, metastasis conditions, and pancreatic cancer patients’ mortality.
Keywords: Autophagy, pancreatic cancer, Beclin1, LC3, prognosis
Introduction
Pancreatic cancer, one of the leading causes of cancer related death, has a high incidence, with >40,000 cases diagnosed each year and a very low 5-year survival rate estimated to be <5% [1,2]. Due to the lack of effective treatment options, late diagnosis, aggressive growth, metastasis and resistance to most therapeutic methods such as cytotoxic chemotherapies and radiotherapy, patients with pancreatic cancer always have a very poor prognosis [3,4]. Thus new treatment methods and a deeper understanding of cancer development mechanisms are always needed.
Recently, the relationship between autophagy and cancers has attracted much attention [5]. Autophagy is a cellular pathway which is associated with the degradation of cytoplasmic macromolecules and organelles and is also related to the progression of cell death [6,7]. However, autophagy in cancer development demonstrates very complicated functions, and it is thought to be involved in both tumorigenesis and tumor suppression, depending on the type of cancer [8-10]. Studies show that in several cancers, autophagy can suppress tumor development, such as in prostate cancer [11] and liver cancer [12]. Meanwhile, in some cancers autophagy is thought to promote tumor growth and is associated with poor prognosis, such as in colorectal cancer [13,14] and pancreatic cancer [15]. Studies demonstrate that autophagy is activated in pancreatic cancer cells [16] and the promotion of autophagy can decrease gemcitabine-induced apoptosis in pancreatic cancer cells [17].
LC3 and Beclin-1 are central autophagy-related genes involved in the autophagy flux [18]. Though some studies have demonstrated that the LC3 protein is associated with the prognosis of pancreatic cancer, few studies have focused on the relationship between the Beclin-1 and LC3 genes and the prognosis of pancreatic cancer. In the present study, we aimed to investigate the role of the autophagy related genes Beclin1 and LC3 in the prognosis of pancreatic cancer. This study may give more clinical evidence for autophagy in pancreatic cancer and may provide some new therapeutic targets for the treatment of pancreatic cancer patients.
Methods and materials
Patient samples and data collection
In the present study, 86 pancreatic cancer tissues and 84 paired adjacent normal pancreatic tissues were collected from 86 patients who underwent pancreatic resection surgery in our hospital from January 2009 to August 2011. All patients were diagnosed with primary pancreatic ductal adenocarcinoma which was confirmed by pathology according to the WHO classification. No patients received chemotherapy or radiotherapy before the surgery. All tissues were fresh frozen until used.
Demographic data including age, gender, and family cancer history, and clinicopathological characteristics including tumor diameter, differential, TNM staging, and lymphatic metastasis were obtained by reviewing the medical records and contacting the physicians in charge. Informed consents were obtained from all patients. The present study was approved by ethics committee of Xuzhou Central Hospital.
Immunohistochemistry (IHC)
For immunohistochemistry, 20 cancer tissues and matching normal tissues were randomly selected. The tissues were fixed with 10% formalin, embedded in paraffin and sectioned. HE staining was then conducted and the samples were immersed with 3% H2O2 and incubated with anti-LC3II or anti-Beclin1 primary antibodies (both purchased from Abcam, Cambridge, MA, USA) at 4°C overnight. The tissues were then incubated with a corresponding second antibody (Abcam) at 37°C for 30 min and stained with diaminobenzidine (DAB). The IHC scores were calculated using the following method: the sum of the staining intensity 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining), and the percentage scores of the stained area, 0 (none); 1 (<1/100); 2 (1/100 to 1/10); 3 (1/10 to 1/3); 4 (1/3; to 2/3); and 5 (>2/3). The final IHC score was the product of the above.
Quantitative real time PCR (RT-qPCR)
The expressions of Beclin1 and LC3 were determined using real-time PCR. Briefly, total RNA was extracted from the tissues using Trizol reagent (Invitrogen, Carlsbad, CA). The Prime-ScriptTM one step RT-PCR kit (TAKARA, Dalian, China) was used to convert RNA into cDNA. RT-qPCR reactions were performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, USA). The primer sequences used in this study were as follows: Beclin1, forward 5’-GGCTGAGAGACTGGATCAGG-3’ and reverse 5’-CTGCGTCTGGGCATAACG-3-3’; LC3-II, forward 5’-GAGAAGCAGCTTCCTGTTCTGG-3’ and reverse 5’-GTGTCCGTTCACCAACAGGAAG-3’; GAPDH, forward 5’-GGACTGACCTGCCGTCTAG-3’ and reverse 5’-TAGCCCAGGATGCCCTTGAG-3’. The Beclin1 and LC3 mRNA levels were normalized to GAPDH. Relative mRNA levels were calculated using the 2-ΔΔCq method.
Statistical analysis
Comparisons between two groups were performed using Student’s t-test or the Mann-Whitney U-test when appropriate. A chi square test was used to compare the categorical variables. The correlation between Beclin1 and LC3 was determined using Spearman’s correlation analysis. For the 5-year survival analysis, the overall survival time was calculated from the date of diagnosis to the date of death or the date of last contact. A Kaplan-Meier curve was created using a log-rank test. For the logistic analysis, both expressions of Beclin1 and LC3 were chosen in a logistic regression model using the stepwise method. A P-value <0.05 was considered to be statistically significant. All calculations were made using SPSS 20.0.
Results
Basic clinical information for all participants
As shown in Table 1, the present study included a total of 86 patients, with a mean age of 60.79±6.27, the female:male ratio was 53:33, and the mean tumor diameter size was 2.92±1.10 cm. Among all the patients, 28 cases were divided into TNM stages I~II, and 58 cases were in TNM stages II~IV. The lymphatic metastasis rate was 67.4%, and 59 cases died during the follow-up for a mortality rate of 68.6%.
Table 1.
Basic clinical information for all patients
| Variables | All patients, n=86 |
|---|---|
| Age, year | 60.79±6.27 |
| Gender, male:female | 53:33 |
| Family cancer history, n (%) | 18 (20.9) |
| Tumor diameter, cm | 2.92±1.10 |
| <3 cm, n (%) | 45 (52.3) |
| ≥3 cm, n (%) | 41 (47.7) |
| Differential | |
| Well to moderate, n (%) | 32 (37.2) |
| Poor, n (%) | 54 (62.8) |
| TNM staging | |
| I and II, n (%) | 28 (32.6) |
| II and IV, n (%) | 58 (67.4) |
| Lymphatic metastasis, n (%) | 58 (67.4) |
| Mortality, n (%) | 59 (68.6) |
The expressions of Beclin1 and LC3 were up-regulated in the pancreatic tumor tissues
The expressions of the Beclin1 and LC3 genes were determined in both tumor tissues and normal tissues using RT-qPCR. As shown in Figures 1 and 2, the expressions of both the Beclin1 and LC3 mRNAs were significantly up-regulated in the tumor tissues compared with the normal tissues (P<0.05), which was consistent with the IHC results. Meanwhile, Spearman’s correlation analysis showed that expressions of Beclin1 and LC3 were significantly correlated in the tumor tissues (P<0.05).
Figure 1.
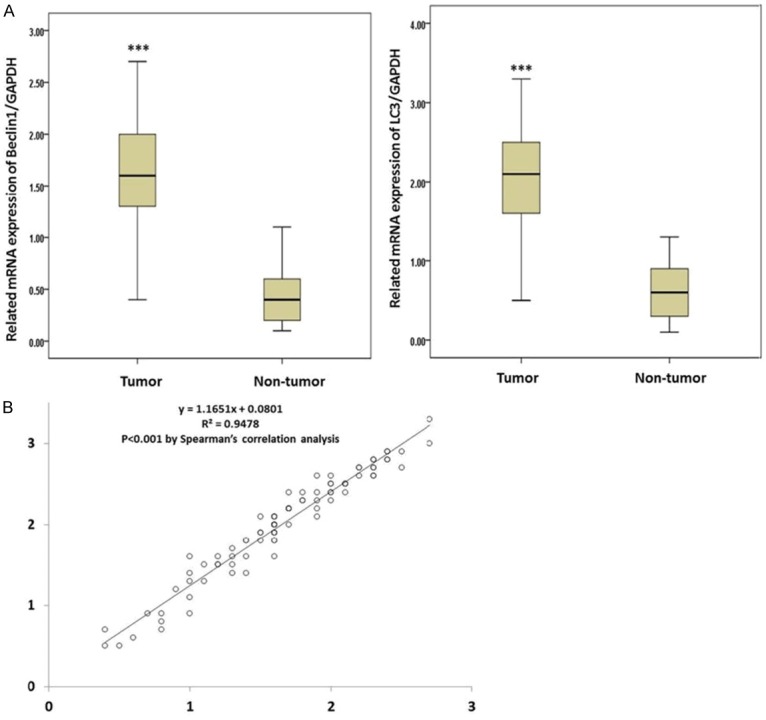
A. Related expressions of Beclin1 and LC3 in both tumor tissues and normal tissues by RT-qPCR; B. The correlation of the mRNA levels of Beclin1 and LC3 in tumor tissues. *(P<0.05).
Figure 2.
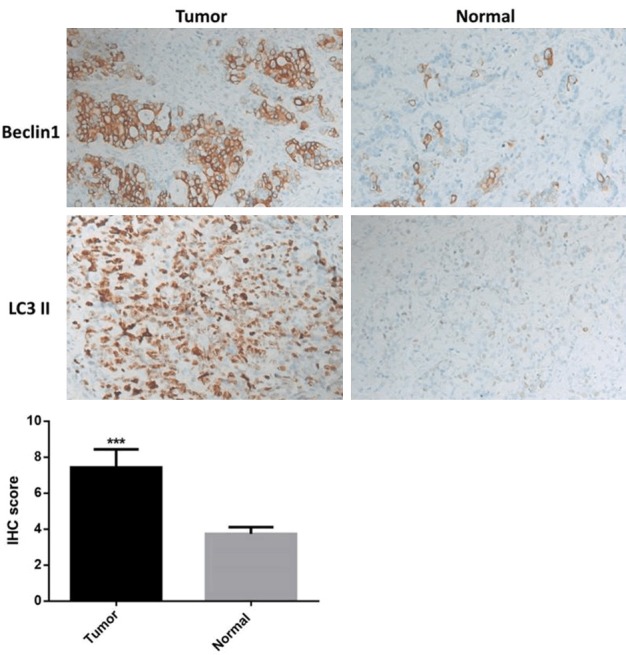
The IHC analysis also showed that Beclin1 and LC3II were elevated in pancreatic cancer tissues.
The expressions of Beclin1 and LC3 were associated with the outcomes of the pancreatic cancer patients
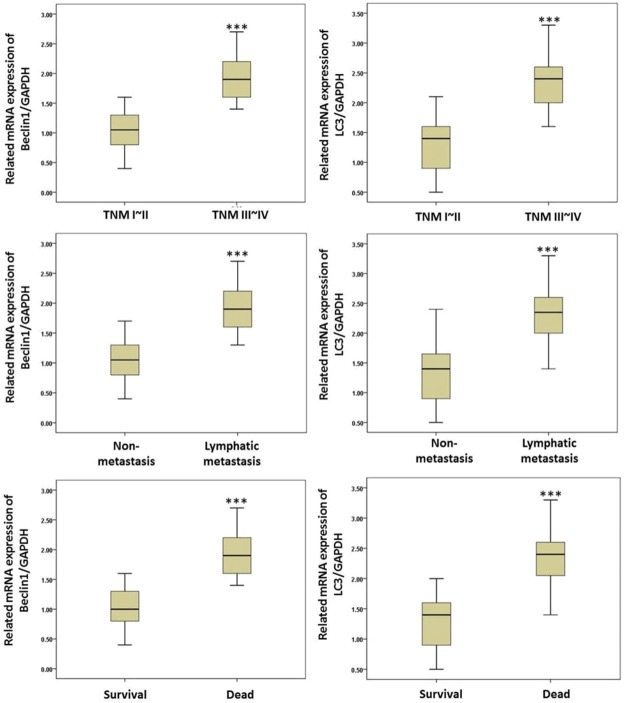
To further study the relationship of the Beclin1 and LC3 genes in pancreatic cancer, the expressions of Beclin1 and LC3 in different kinds of patients were investigated. The results showed that in patients with TNM stages III~IV, the expressions of Beclin1 and LC3 were significantly higher than in the patients with TNM stages I~II (P<0.05) (Figure 3). Meanwhile the expressions of Beclin1 and LC3 in the lymphatic metastasis and deceased patients was also higher than they were in the non-metastasis or surviving patients, respectively (P<0.05), indicating that Beclin1 and LC3 were associated with tumor stage, metastasis, and the mortality of the patients.
Figure 3.
The expressions of Beclin1 and LC3 in different patients. *(P<0.05). Beclin1 and LC3 were significantly higher in patients with TNM stages III~IV and lymphatic metastasis and deceased patients.
Then patients were divided into 4 groups (high/low Beclin1/LC3) according to the median values of the expressions of Beclin1 and LC3, and the clinical outcomes were analyzed. As shown in Table 2, in both the highly expressed Beclin1 and LC3 patients, the TNM stage, differential condition, and lymphatic metastasis rates showed a significant difference compared with the low expression groups (P<0.05). These results suggest that patients with low expressions of Beclin1 and LC3 might have a better prognosis.
Table 2.
Relationship of expression of Beclin1 and LC3 with clinical outcomes of pancreatic cancer patients, n (%)
| Outcomes | Low Beclin1, n=45 | High Beclin1, n=41 | P value | Low LC3, n=47 | High LC3, n=39 | P value |
|---|---|---|---|---|---|---|
| Age | 0.322 | 0.571 | ||||
| ≥60 | 25 (55.6) | 20 (48.8) | 26 (55.3) | 20 (51.3) | ||
| <60 | 20 (44.4) | 21 (51.2) | 21 (44.7) | 19 (48.7) | ||
| Gender | 0.884 | 0.467 | ||||
| Male | 28 (62.2) | 25 (61.0) | 30 (63.8) | 23 (59.0) | ||
| Female | 17 (37.8) | 16 (39.0) | 17 (36.2) | 16 (41.0) | ||
| Family cancer history | 10 (22.2) | 8 (19.5) | 0.702 | 11 (23.4) | 7 (17.9) | 0.381 |
| Tumor diameter | 0.671 | 0.671 | ||||
| ≥3 cm | 22 (48.9) | 19 (46.3) | 23 (48.9) | 18 (46.2) | ||
| <3 cm | 23 (51.1) | 22 (53.7) | 24 (51.1) | 21 (53.8) | ||
| TNM stage | <0.001 | <0.001 | ||||
| I and II | 28 (62.2) | 0 (0) | 28 (59.6) | 0 (0) | ||
| III and IV | 17 (37.8) | 41 (100) | 19 (40.4) | 39 (100) | ||
| Differential | <0.001 | <0.001 | ||||
| Well to moderate | 25 (55.6) | 7 (17.1) | 24 (51.1) | 8 (20.5) | ||
| Poor | 20 (44.4) | 34 (82.9) | 23 (48.9) | 31 (79.5) | ||
| Lymphatic metastasis | <0.001 | <0.001 | ||||
| Yes | 18 (40) | 40 (97.6) | 20 (42.6) | 38 (97.4) | ||
| No | 27 (60) | 1 (0.4) | 27 (57.4) | 1 (0.6) |
The expressions of Beclin1 and LC3 are associated with the mortality of pancreatic cancer patients
Finally, we analyzed the relationship of Beclin1 and LC3 with the mortality of pancreatic cancer patients. As shown in Figure 4, the K-M curve showed that patients with lowly expressed Beclin1 and LC3 had longer survival times, but patients with highly expressed Beclin1 and LC3 had shorter survival times (P<0.05). Additionally, logistic regression showed that expressions of the Beclin1 and LC3 genes were both independent risk factors of 5-year mortality for pancreatic cancer patients (Table 3).
Figure 4.
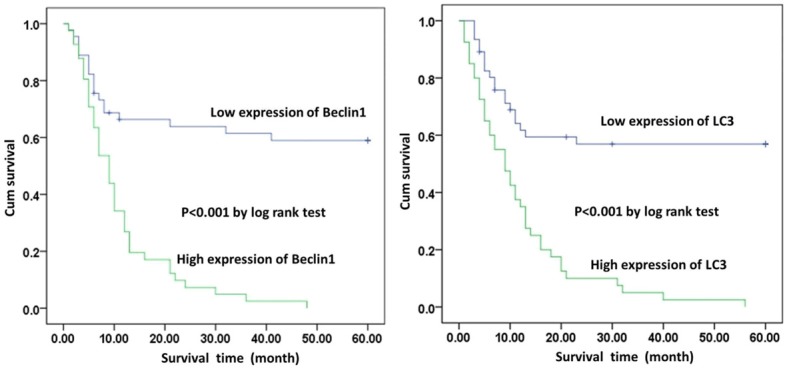
A K-M curve for 5-year mortality for pancreatic cancer patients. Patients with highly expressed Beclin1 and LC3 had shorter survival times (P<0.05).
Table 3.
The correlation between the Beclin1 and LC3 genes and the 5-year mortality for pancreatic cancer patients using logistic regression analysis
| Wald | Odds ratio | 95% CI | P value | |
|---|---|---|---|---|
| Beclin1 | 12.358 | 13.655 | 8.519 (4.207~17.250) | <0.001 |
| LC3 | 13.643 | 9.628 | 1.518 (0.917~2.513) | <0.001 |
Discussion
Due to late diagnoses, most pancreatic cancer patients are found to have an aggressive state of the disease when diagnosed [19]. Moreover, since pancreatic cancer is easily resistant to most chemotherapy, surgical resection is the only curative treatment for the disease [20]. However, less than 20% of tumors are resectable at the time of diagnosis [21]. Thus patients with pancreatic cancer often have very poor prognoses, with a 5-year survival rate <5% [1]. Autophagy is found to play an important role in the tumor development of many cancers and its functions differ among different cancer types. Studies show that in many cancers autophagy is considered to promote cancer development, and the inhibition of autophagy could inhibit tumor growth and enhance tumor cell apoptosis. Hwang et al. showed that the inhibition of autophagy could enhance the apoptosis of malignant mesothelioma and non-small cell lung cancer cells [22]. Mikhaylova et al. found that miR-204 could suppress the tumor growth of renal clear-cell carcinoma through the inhibition of autophagy [23].
In addition, autophagy is also thought to promote tumorigenesis in pancreatic cancer. Zhang et al. demonstrated that the inhibition of Beclin1-mediated autophagy could enhance the radiosensitivity of pancreatic cancer cells [24]. Fujii et al. showed that autophagy is activated in pancreatic cancer cells [16]. Yang et al. found that pancreatic cancer cells require autophagy for tumor growth, and the inhibition of autophagy leads to significant growth suppression of pancreatic cancer cells in vitro [25]. Despite the studies above, few studies have focused on the autophagy related genes Beclin1 and LC3 in the prognosis of pancreatic cancer. In the present study, we demonstrated for the first time that the Beclin1 and LC3 genes were correlated with the prognoses of pancreatic cancer patients.
First we demonstrated that both Beclin1 and LC3 were up-regulated in pancreatic tumor tissues. The expressions of Beclin1 and LC3 are up-regulated in many cancers. Fujii et al. found that the LC3 protein is over-expressed in pancreatic cancer patients [16]. Schmitz et al. showed that Beclin1 and LC3 are both overexpressed in colorectal cancer [26]. All these results are consistent with our findings. Then we demonstrated that the Beclin1 and LC3 genes correlate with the tumor stage, metastasis conditions, and the mortality of the pancreatic cancer patients. Wang et al. studied Beclin-1 and LC3 in human hypopharyngeal squamous cell carcinoma and found that the expressions of Beclin-1 and LC3 are correlated with the poor prognoses of patients with hypopharyngeal squamous cell carcinoma [27]. Guo et al. demonstrated that Beclin-1 and LC3 could predict cetuximab efficacy in advanced colorectal cancer [28]. Moreover, in a recent study, Song et al. demonstrated that the levels of Beclin1 and Bcl-2 expressions were evaluated in pancreatic neoplasms and that high Beclin1 and low Bcl-2 expressions were significantly correlated with poor differentiation and distant metastasis, which was also consistent with our findings [29]. Though autophagy shows different roles in cancer development, promoting or suppressing it, in this study we confirm again that autophagy in pancreatic cancer is a tumor promoter, which of course needs more studies to give more evidence. The present study also has some limitations. This is a single-center study with only a small sample size. We tried to use the TCGA database to confirm our results, but because both LC3 and Beclin1 are autophagy-related genes, such data were not found.
In conclusion, the present study investigated the role of the autophagy-related genes Beclin1 and LC3 in the prognosis of pancreatic cancer patients. The results showed that the Beclin1 and LC3 genes were correlated with tumor stage, metastasis conditions, and the mortality of the pancreatic cancer patients. This study may give more clinical evidence for autophagy in pancreatic cancer and may provide some new therapeutic targets for the treatment of pancreatic cancer patients.
Acknowledgements
The present study was approved by ethics committee of Xuzhou Central Hospital.
Informed consents were obtained from all patients.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Kaistha BP, Honstein T, Müller V, Bielak S, Sauer M, Kreider R, Fassan M, Scarpa A, Schmees C, Volkmer H, Gress TM, Buchholz M. Key role of dual specificity kinase TTK in proliferation and survival of pancreatic cancer cells. Br J Cancer. 2014;111:1780–7. doi: 10.1038/bjc.2014.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 5.Bhutia SK, Mukhopadhyay S, Sinha N, Das DN, Panda PK, Patra SK, Maiti TK, Mandal M, Dent P, Wang XY, Das SK, Sarkar D, Fisher PB. Autophagy: cancer’s friend or foe? Adv Cancer Res. 2013;118:61–95. doi: 10.1016/B978-0-12-407173-5.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–20. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorin S, Hamai A, Mehrpour M, Codogno P. Autophagy regulation and its role in cancer. Semin Cancer Biol. 2013;23:361–79. doi: 10.1016/j.semcancer.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Roy S, Debnath J. Autophagy and tumorigenesis. Semin Immunopathol. 2010;32:383–96. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kung CP, Budina A, Balaburski G, Bergenstock MK, Murphy M. Autophagy in tumor suppression and cancer therapy. Crit Rev Eukaryot Gene Expr. 2011;21:71–100. doi: 10.1615/critreveukargeneexpr.v21.i1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar D, Shankar S, Srivastava RK. Rottlerin induces autophagy and apoptosis in prostate cancer stem cells via PI3K/Akt/mTOR signaling pathway. Cancer Lett. 2014;343:179–89. doi: 10.1016/j.canlet.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–71. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 13.Yang M, Zhao H, Guo L, Zhang Q, Zhao L, Bai S, Zhang M, Xu S, Wang F, Wang X, Zhao B. Autophagy-based survival prognosis in human colorectal carcinoma. Oncotarget. 2015;6:7084–103. doi: 10.18632/oncotarget.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2013;32:1570–9. doi: 10.1038/onc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong J, Munoz AR, Chan D, Ghosh R, Kumar AP. STAT3 down regulates LC3 to inhibit autophagy and pancreatic cancer cell growth. Oncotarget. 2014;5:2529–41. doi: 10.18632/oncotarget.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813–9. doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yan J, Wang L, Xiao F, Yang Y, Guo X, Wang H. Beclin1 inhibition promotes autophagy and decreases gemcitabine-induced apoptosis in Miapaca2 pancreatic cancer cells. Cancer Cell Int. 2013;13:26. doi: 10.1186/1475-2867-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R, Peng J, Yuan P, Xu F, Wei W. Divergent roles of BECN1 in LC3 lipidation and autophagosomal function. Autophagy. 2015;11:740–7. doi: 10.1080/15548627.2015.1034404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada S, Fujii T, Sugimoto H, Nomoto S, Takeda S, Kodera Y, Nakao A. Aggressive surgery for borderline resectable pancreatic cancer: evaluation of National Comprehensive Cancer Network guidelines. Pancreas. 2013;42:1004–10. doi: 10.1097/MPA.0b013e31827b2d7c. [DOI] [PubMed] [Google Scholar]
- 20.Morris S, Gurusamy KS, Sheringham J, Davidson BR. Cost-effectiveness of diagnostic laparoscopy for assessing resectability in pancreatic and periampullary cancer. BMC Gastroenterol. 2015;15:44. doi: 10.1186/s12876-015-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donadelli M, Dando I, Zaniboni T, Costanzo C, Dalla Pozza E, Scupoli MT, Scarpa A, Zappavigna S, Marra M, Abbruzzese A, Bifulco M, Caraglia M, Palmieri M. Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism. Cell Death Dis. 2011;2:e152. doi: 10.1038/cddis.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang KE, Kim YS, Jung JW, Kwon SJ, Park DS, Cha BK, Oh SH, Yoon KH, Jeong ET, Kim HR. Inhibition of autophagy potentiates pemetrexed and simvastatin-induced apoptotic cell death in malignant mesothelioma and non-small cell lung cancer cells. Oncotarget. 2015;6:29482–96. doi: 10.18632/oncotarget.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, Czyzyk-Krzeska MF. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–46. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Shi H, Lin S, Ba M, Cui S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep. 2015;34:1557–64. doi: 10.3892/or.2015.4078. [DOI] [PubMed] [Google Scholar]
- 25.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz KJ, Ademi C, Bertram S, Schmid KW, Baba HA. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J Surg Oncol. 2016;14:189. doi: 10.1186/s12957-016-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng L, Jin T. Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PLoS One. 2013;8:e69038. doi: 10.1371/journal.pone.0069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH, Chen XX, Wang F, Xia LP. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol. 2011;17:4779–86. doi: 10.3748/wjg.v17.i43.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song S, Wang B, Gu S, Li X, Sun S. Expression of Beclin1 and Bcl-2 in pancreatic neoplasms and its effect on pancreatic ductal adenocarcinoma prognosis. Oncol Lett. 2017;14:7849–7861. doi: 10.3892/ol.2017.7218. [DOI] [PMC free article] [PubMed] [Google Scholar]