Abstract
Objective
Ulcerative colitis (UC) is a chronic idiopathic inflammatory bowel disease whose treatment strategies remain unsatisfactory. This study aims to investigate the mechanisms of Quyushengxin formula acting on UC based on network pharmacology.
Methods
Ingredients of the main herbs in Quyushengxin formula were retrieved from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database. Absorption, distribution, metabolism, and excretion properties of all ingredients were evaluated for screening out candidate bioactive compounds in Quyushengxin formula. Weighted ensemble similarity algorithm was applied for predicting direct targets of bioactive ingredients. Functional enrichment analyses were performed for the targets. In addition, compound-target network, target-disease network, and target-pathway network were established via Cytoscape 3.6.0 software.
Results
A total of 41 bioactive compounds in Quyushengxin formula were selected out from the TCMSP database. These bioactive compounds were predicted to target 94 potential proteins by weighted ensemble similarity algorithm. Functional analysis suggested these targets were closely related with inflammatory- and immune-related biological progresses. Furthermore, the results of compound-target network, target-disease network, and target-pathway network indicated that the therapeutic effects of Quyushengxin on UC may be achieved through the synergistic and additive effects.
Conclusion
Quyushengxin may act on immune and inflammation-related targets to suppress UC progression in a synergistic and additive manner.
1. Introduction
Ulcerative colitis (UC) is a chronic and progressive immunologically mediated disease causing consecutive mucosal inflammation of the colon [1, 2]. The onset of UC is most often during young adulthood, which is well characterized by homogeneous and continuous lesions [3]. Although the incidence of UC is increasing in Asia, it is highly diagnosed in the developed countries, especially in Western Europe and North America. Previous reports showed that the overall incidence and prevalence of UC are nearly 1.2/20.3 cases and 7.6/245 per 100,000 persons per year, respectively [4, 5].
UC therapy is aimed to reduce the recurrent rate, as well as improve the life quality and minimize drug-related adverse events. Basic therapies for UC are determined based on the severity of symptoms, which are often thought as step-up approaches. To date, 5-aminosalycilates (5-ASAs) have been the mainstay for treatment of mild-to-moderate UC [6]. Though 5-ASAs are safe and have no dose-related toxicity in short-term use with a dose-response efficacy, long-term use of them might induces adverse events, such as headache, diarrhea, nausea, interstitial nephritis, and hepatitis. In addition, patients with more moderate-to-severe UC after 5-ASAs therapy are typically treated with corticosteroids, and these patients are often followed by transition to a steroid-sparing agent with a thiopurine, adhesion molecule inhibitor, or anti-tumor necrosis factor (TNF) agent [6]. However, these corticosteroid-based therapies also accompany with side effects, such as cataracts, osteopenia, avascular necrosis, insomnia, mood changes, delirium, glaucoma, and adrenal insufficiency [7, 8]. Besides, despite improved medical therapies, it is estimated that about 15% of UC patients still require proctocolectomy [9]. Therefore, it is of great significance to develop more optimized and integrated therapies for UC patients.
To date, an increasing number of traditional Chinese herbal compounds are successfully used for treating UC with less side effects, such as Gegen Qinlian decoction [10], Jianpi Qingchang decoction [11, 12], Zhikang capsule [13], Huangkui Lianchang decoction [14], and Qingchang Wenzhong decoction [15, 16]. Quyushengxin formula is mainly composed of four herbs, Panax ginseng C.A. Mey. (Araliaceae), Astragalus membranaceus (Fisch) Bunge, Pulsatilla chinensis (Bge.) Regel, and Coptis chinensis Franch. Our clinical practice demonstrated Quyushengxin formula could relieve the clinical symptoms in active stage and suppress the inflammatory reaction of UC patients and could be used for treating mild-to-moderate UC [17]. Although the therapeutic effects of Quyushengxin on UC are attractive, molecular mechanisms of its action remain to be further elucidated.
Traditional Chinese medicine- (TCM-) oriented network pharmacology provides us a novel way to unveil the molecular mechanisms of TCM through pharmacokinetic evaluation, network/pathway analysis, and target prediction [18, 19]. In this study, we tried to unveil the molecular mechanisms of Quyushengxin formula acting on UC based on network pharmacology.
2. Materials and Methods
2.1. Screening of Potential Bioactive Compounds in Quyushengxin Formula
Traditional Chinese Medicine Systems Pharmacology Database (TCMSP, http://lsp.nwu.edu.cn) is a systems pharmacology platform of Chinese herbal medicines that captures the relationships between drugs, targets, and diseases [20]. Ingredients along with their molecular weight (MW), water partition coefficient (AlogP), number of hydrogen bond donors (Hodn), number of hydrogen acceptors (Hacc), oral bioavailability (OB), Caco-2 permeability (Caco-2), blood-brain barrier (BBB), drug-likeness (DL), fractional negative accessible surface area (FASA) ,and half-life (HL), of all four herbs in Quyushengxin formula were retrieved from TCMSP. Then, absorption, distribution, metabolism, and excretion (ADME) properties, including OB, DL, and HL, were evaluated for screening out bioactive compounds. The potential bioactive compounds in Quyushengxin were predicted and sifted out via an integrated model including PreOB (for prediction of OL), PreDL (for prediction of DL), and PreHL (for prediction of HL) [21, 22]. In detail, OB value was obtained by OBioavail 1.1, and the compounds with OB ≥ 30% were selected out for further analysis [20, 23]. PreDL was utilized to calculate the DL index of compounds, and compounds with DL ≥ 0.18 were included for further research. The DL evaluation approach was constructed via both Tanimoto coefficient and molecular descriptors, and the formula is listed as follows:
| (1) |
where X was the molecular descriptors of herbal ingredients and Y showed the average molecular properties of all molecules in the DrugBank database (http://www.drugbank.ca/).
Besides, PreHL was estimated by combining multivariable linear regression model and MLR (mixed logistic regression) algorithm [22], as follows:
| (2) |
where R2 was the correlation coefficient of training set and Q2 was the correlation coefficient of external test sets of the model. SEE was the estimated standard deviation of training set. F was the mean square ratio. Besides, Ntraining indicated the number of chemical compounds in the training set, and Ntest indicated the number of chemical compounds in the test set. It was evidenced that there were eight descriptors satisfying the linear regression as follows: nArCO, H7m, D/Dr09, N-070, C-032, JGI6, nRC=N, and Mor02e. Finally, 4 ≤ HL ≤ 8 was defined as appropriate selection criteria for drug HL evaluation.
2.2. Prediction of the Candidate Targets of Bioactive Compounds
Weighted ensemble similarity (WES) algorithm was applied for predicting direct targets of the bioactive compounds via a large scale of drug target relationships [24]. Those targets with likelihood score ≥7 were deemed as direct targets in this study. Thereafter, candidate targets were mapped to Uniprot (http://www.uniprot.org/) for annotation and normalization.
2.3. Functional Enrichment Analyses
Gene Ontology- (GO-) biological processes (BPs) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) pathways of the candidate targets of bioactive compounds were predicted via the Database for Annotation, Visualization, and Integrated Discovery (DAVID) database [25] with P < 0.05 as the criterion for significance.
2.4. Prediction of Target-Related Disease
Target-related diseases were predicted by integrating multisource databases, including Comparative Toxicogenomics Database (CTD, http://ctdbase.org/) [26], Therapeutic Target Database (TTD, http://bidd.nus.edu.sg/group/cjttd/) [27], and PharmGKB database (https://www.pharmgkb.org/) [28].
2.5. Network Construction
Three kinds of networks in this study were established using Cytoscape 3.6.0 software [29]: compound-target network (C-T network), target-disease network (T-D network), and target-pathway network (T-P network). C-T network was composed of bioactive compounds and their potential targets, which was built to reveal the drug-target interactions. T-D network was built based on the potential targets and their related diseases. The pathway information of targets was selected from the results for KEGG pathway enrichment analysis by excluding those pathways with no relevance to UC based the latest pathological information of UC. T-P network was generated based on potential targets and UC-related pathways. In the networks, the nodes represented compounds, targets, diseases, and pathways, and the edges displayed the interactions between two nodes. Furthermore, the significance of each node in the networks was assessed via one crucial topological parameter, namely, “degree,” which was defined as the total of edges related with a node [30, 31]. Degree of all nodes was analyzed using plugin NetworkAnalyzer of Cytoscape 3.6.0.
3. Results
3.1. Screening of Potential Bioactive Compounds from Four Herbs in Quyushengxin Formula
Quyushengxin formula consists of 4 main herbs: Panax ginseng C.A. Mey. (Araliaceae), Astragalus membranaceus (Fisch) Bunge, Pulsatilla chinensis (Bge.) Regel, and Coptis chinensis Franch. After retrieving from TCMSP, 190, 87, 57, and 48 ingredients were obtained for these four herbs, respectively. Based on the criteria of OB ≥ 30%, DL ≥ 0.18, and 4 ≤ HL ≤ 8, 41 potential bioactive compounds, including quercetin, ursolic acid, kaempferol, β-sitosterol, and rutin, were sifted out (Table 1), which accounted for 10.73% of all 382 ingredients in Quyushengxin.
Table 1.
Details of 41 bioactive compounds and their biological parameters.
| ID | Compounds | Structure | OB (%) | DL | HL | Degree | Herb |
|---|---|---|---|---|---|---|---|
| mol01 | Quercetin |

|
46.43 | 0.28 | 14.40 | 73 |
Coptis chinensis Franch Astragalus membranaceus (Fisch) Bunge |
|
| |||||||
| mol02 | Ferulic acid |

|
39.56 | 0.06 | 2.38 | 7 | Coptis chinensis Franch |
|
| |||||||
| mol03 | Palmatine |

|
64.60 | 0.65 | 2.25 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol04 | Jatrorrizine |

|
19.65 | 0.59 | 4.21 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol05 | Berberine |

|
36.86 | 0.78 | 6.57 | 8 | Coptis chinensis Franch |
| mol06 | Columbamine |

|
26.94 | 0.59 | 5.21 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol07 | Coptisine |

|
30.67 | 0.86 | 9.33 | 8 | Coptis chinensis Franch |
|
| |||||||
| mol08 | Worenine |

|
45.83 | 0.87 | 8.41 | 6 | Coptis chinensis Franch |
|
| |||||||
| mol09 | Magnoflorine |

|
0.48 | 0.55 | 6.22 | 8 | Coptis chinensis Franch |
|
| |||||||
| mol10 | Berberrubine |

|
35.74 | 0.73 | 6.46 | 8 | Coptis chinensis Franch |
| mol11 | Epiberberine |

|
43.09 | 0.78 | 6.10 | 7 | Coptis chinensis Franch |
|
| |||||||
| mol12 | (R)-Canadine |

|
55.37 | 0.77 | 6.41 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol13 | Berlambine |

|
36.68 | 0.82 | 7.33 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol14 | Corchoroside A_qt |

|
104.95 | 0.78 | 6.68 | 2 | Coptis chinensis Franch |
| mol15 | Tetrandrine |

|
26.64 | 0.10 | 4.77 | 9 | Coptis chinensis Franch |
|
| |||||||
| mol16 | β-Sitosterol |

|
36.91 | 0.75 | 5.36 | 15 |
Panax ginseng C.A. Mey. (Araliaceae) Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol17 | Kaempferol |

|
41.88 | 0.24 | 14.74 | 26 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol18 | Stigmasterol |

|
43.83 | 0.76 | 5.57 | 10 |
Panax ginseng C.A. Mey. (Araliaceae)G Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol19 | β-Elemene |

|
25.63 | 0.06 | 6.32 | 8 | Panax ginseng C.A. Mey. (Araliaceae) |
| mol20 | Ginsenoside Ro_qt |

|
17.62 | 0.76 | 7.50 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol21 | Dianthramine |

|
40.45 | 0.20 | 5.14 | 3 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol22 | Arachidonate |

|
45.57 | 0.20 | 7.56 | 5 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol23 | Ginsenoside La_qt |

|
15.70 | 0.78 | 5.20 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
| mol24 | Ginsenoside rh2 |

|
36.32 | 0.56 | 11.08 | 9 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol25 | Ginsenoside-Rh3_qt |

|
13.09 | 0.76 | 6.22 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol26 | Ginsenoside-Rh4_qt |

|
31.11 | 0.78 | 6.97 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol27 | Malkangunin |
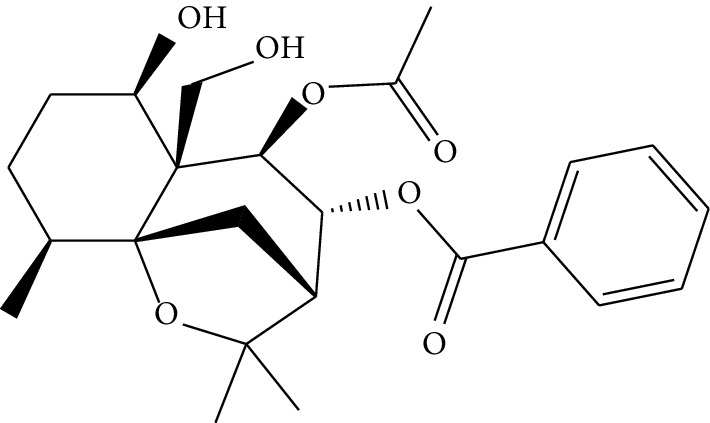
|
57.71 | 0.63 | 4.09 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
| mol28 | Alexandrin_qt |

|
36.91 | 0.75 | 5.53 | 1 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol29 | Ginsenoside rf |

|
17.74 | 0.24 | 4.66 | 5 | Panax ginseng C.A. Mey. (Araliaceae) |
|
| |||||||
| mol30 | Hederagenin |

|
36.91 | 0.75 | 5.35 | 6 | Astragalus membranaceus (Fisch) Bunge |
|
| |||||||
| mol31 | Isorhamnetin |

|
49.60 | 0.31 | 14.34 | 10 |
Astragalus membranaceus (Fisch) Bunge Pulsatilla chinensis (Bge.) Regel |
| mol32 | 7-O-methylisomucronulatol |

|
74.69 | 0.30 | 2.98 | 11 | Astragalus membranaceus (Fisch) Bunge |
|
| |||||||
| mol33 | Rutin |

|
3.20 | 0.68 | 6.22 | 15 | Astragalus membranaceus (Fisch) Bunge |
|
| |||||||
| mol34 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene |

|
39.05 | 0.48 | 7.95 | 5 | Astragalus membranaceus (Fisch) Bunge |
|
| |||||||
| mol35 | Isoferulic acid |

|
50.83 | 0.06 | 2.45 | 7 | Astragalus membranaceus (Fisch) Bunge |
| mol36 | Betulinic acid |

|
55.38 | 0.78 | 8.87 | 1 | Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol37 | Oleanolic acid |

|
29.02 | 0.76 | 5.56 | 6 | Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol38 | Sitosteryl acetate |

|
40.39 | 0.85 | 6.34 | 1 | Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol39 | Lanosterol |

|
42.12 | 0.75 | 5.84 | 1 | Pulsatilla chinensis (Bge.) Regel |
| mol40 | 3-beta,23-Dihydroxy-lup-20(29)-ene-28-O-alpha-L-rhamnopyranosyl-(1-4)-beta-D-glucopyranosyl(1-6)-beta-D-glucopyranoside_qt |

|
37.59 | 0.79 | 6.70 | 1 | Pulsatilla chinensis (Bge.) Regel |
|
| |||||||
| mol41 | Ursolic acid |

|
16.77 | 0.75 | 5.28 | 35 | Pulsatilla chinensis (Bge.) Regel |
3.2. Establishment of C-T Network
Candidate targets of the 41 bioactive compounds were predicted via WES algorithm. A total of 367 potential targets for these 41 bioactive compounds were obtained. After removing the overlapping targets, 94 candidate proteins were reserved. Then, C-T network was built by Cytoscape 3.6.0 which contains 367 connections between 41 compounds and corresponding 94 candidate targets (Figure 1). The degrees of the 41 bioactive compounds in the C-T network were calculated and are displayed in Table 1. The average degree of targets per compound was 4.7, indicating multitarget functions of Quyushengxin formula. Among the 41 bioactive compounds, 8 of them showed a high degree (degree > 10). Quercetin possessed the highest degree of targets (degree = 73), followed by ursolic acid (degree = 35), kaempferol (degree = 26), β-sitosterol (degree = 15), rutin (degree = 15), 7-O-methylisomucronulatol (degree = 11), stigmasterol (degree = 10), and isorhamnetin (degree = 10).
Figure 1.
Compound-target network. A compound node and a target node are connected.
The degree of the candidate targets was also calculated and displayed in Table 2. Eight out of the 94 compounds possessed a degree larger than 10, including ESR1 (estrogen receptor 1, degree = 34), PTGS2 (prostaglandin-endoperoxide synthase 2, degree = 27), NOS2 (nitric oxide synthase 2, degree = 25), PTGS1 (degree = 23), PPARG (peroxisome proliferator-activated receptor gamma, degree = 21), NOS3 (degree = 21), ESR2 (degree = 17), and KCNH2 (Potassium Voltage-Gated Channel Subfamily H Member 2, degree = 13).
Table 2.
Details of 94 UC-related targets of herbs via UniProt.
| ID | UniProt | Protein names | Gene names | Degree | Organism |
|---|---|---|---|---|---|
| 1 | P35228 | Nitric oxide synthase, inducible | NOS2 | 25 | Homosapiens |
| 2 | P23219 | Prostaglandin G/H synthase 1 | PTGS1 | 23 | Homosapiens |
| 3 | P03372 | Estrogen receptor | ESR1 | 34 | Homosapiens |
| 4 | P37231 | Peroxisome proliferator-activated receptor gamma | PPARG | 21 | Homosapiens |
| 5 | P35354 | Prostaglandin G/H synthase 2 | PTGS2 | 27 | Homosapiens |
| 6 | Q92731 | Estrogen receptor beta | ESR2 | 17 | Homosapiens |
| 7 | P11388 | DNA topoisomerase 2-alpha | TOP2A | 5 | Homosapiens |
| 8 | P16389 | Potassium voltage-gated channel subfamily H member 2 | KCNH2 | 13 | Homosapiens |
| 9 | P08709 | Coagulation factor VII | F7 | 6 | Homosapiens |
| 10 | P29474 | Nitric-oxide synthase, endothelial | NOS3 | 21 | Homosapiens |
| 11 | P27338 | Amine oxidase [flavin-containing] B | MAOB | 5 | Homosapiens |
| 12 | Q04206 | Transcription factor p65 | RELA | 6 | Homosapiens |
| 13 | P00533 | Epidermal growth factor receptor | EGFR | 1 | Homosapiens |
| 14 | P31749 | RAC-alpha serine/threonine-protein kinase | AKT1 | 2 | Homosapiens |
| 15 | P15692 | Vascular endothelial growth factor A | VEGFA | 2 | Homosapiens |
| 16 | P24385 | G1/S-specific cyclin-D1 | CCND1 | 3 | Homosapiens |
| 17 | P10415 | Apoptosis regulator Bcl-2 | BCL2 | 5 | Homosapiens |
| 18 | P01100 | Proto-oncogene c-Fos | FOS | 3 | Homosapiens |
| 19 | P38936 | Cyclin-dependent kinase inhibitor 1 | CDKN1A | 4 | Homosapiens |
| 20 | P55211 | Caspase-9 | CASP9 | 4 | Homosapiens |
| 21 | P00749 | Urokinase-type plasminogen activator | PLAU | 4 | Homosapiens |
| 22 | P08253 | 72 kDa type IV collagenase | MMP2 | 2 | Homosapiens |
| 23 | P14780 | Matrix metalloproteinase-9 | MMP9 | 2 | Homosapiens |
| 24 | P22301 | Interleukin-10 | IL10 | 1 | Homosapiens |
| 25 | P01133 | Proepidermal growth factor | EGF | 1 | Homosapiens |
| 26 | P06400 | Retinoblastoma-associated protein | RB1 | 2 | Homosapiens |
| 27 | P01375 | Tumor necrosis factor | TNF | 6 | Homosapiens |
| 28 | P05412 | Transcription factor AP-1 | JUN | 4 | Homosapiens |
| 29 | P05231 | Interleukin-6 | IL-6 | 3 | Homosapiens |
| 30 | P42574 | Caspase-3 | CASP3 | 7 | Homosapiens |
| 31 | P04637 | Cellular tumor antigen p53 | TP53 | 4 | Homosapiens |
| 32 | P11926 | Ornithine decarboxylase | ODC1 | 1 | Homosapiens |
| 33 | Q14790 | Caspase-8 | CASP8 | 3 | Homosapiens |
| 34 | P00441 | Superoxide dismutase [Cu-Zn] | SOD1 | 2 | Homosapiens |
| 35 | P17252 | Protein kinase C alpha type | PRKCA | 2 | Homosapiens |
| 36 | P03956 | Interstitial collagenase | MMP1 | 3 | Homosapiens |
| 37 | P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1 | 2 | Homosapiens |
| 38 | P04626 | Receptor tyrosine-protein kinase erbB-2 | ERBB2 | 1 | Homosapiens |
| 39 | P09601 | Heme oxygenase 1 | HMOX1 | 3 | Homosapiens |
| 40 | P05177 | Cytochrome P450 1A2 | CYP1A2 | 2 | Homosapiens |
| 41 | P01106 | Myc proto-oncogene protein | MYC | 1 | Homosapiens |
| 42 | P05362 | Intercellular adhesion molecule 1 | ICAM1 | 4 | Homosapiens |
| 43 | P01584 | Interleukin-1 beta | IL1B | 5 | Homosapiens |
| 44 | P13500 | C-C motif chemokine 2 | CCL2 | 1 | Homosapiens |
| 45 | P19320 | Vascular cell adhesion protein 1 | VCAM1 | 2 | Homosapiens |
| 46 | P10145 | Interleukin-8 | CXCL8 | 2 | Homosapiens |
| 47 | P05771 | Protein kinase C beta type | PRKCB | 2 | Homosapiens |
| 48 | O15392 | Baculoviral IAP repeat-containing protein 5 | BIRC5 | 2 | Homosapiens |
| 49 | P04792 | Heat shock protein beta-1 | HSPB1 | 1 | Homosapiens |
| 50 | P01137 | Transforming growth factor beta-1 | TGFB1 | 3 | Homosapiens |
| 51 | P60568 | Interleukin-2 | IL2 | 2 | Homosapiens |
| 52 | Q16678 | Cytochrome P450 1B1 | CYP1B1 | 2 | Homosapiens |
| 53 | P00750 | Tissue-type plasminogen activator | PLAT | 1 | Homosapiens |
| 54 | P01579 | Interferon gamma | IFNG | 4 | Homosapiens |
| 55 | P09917 | Arachidonate 5-lipoxygenase | ALOX5 | 3 | Homosapiens |
| 56 | P60484 | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | PTEN | 1 | Homosapiens |
| 57 | P05164 | Myeloperoxidase | MPO | 1 | Homosapiens |
| 58 | Q9UNQ0 | ATP-binding cassette subfamily G member 2 | ABCG2 | 1 | Homosapiens |
| 59 | P09211 | Glutathione S-transferase P | GSTP1 | 3 | Homosapiens |
| 60 | Q16236 | Nuclear factor erythroid 2-related factor 2 | NFE2L2 | 1 | Homosapiens |
| 61 | P15559 | NAD(P)H dehydrogenase [quinone] 1 | NQO1 | 2 | Homosapiens |
| 62 | P09874 | Poly [ADP-ribose] polymerase 1 | PARP1 | 1 | Homosapiens |
| 63 | P35869 | Aryl hydrocarbon receptor | AHR | 2 | Homosapiens |
| 64 | P19875 | C-X-C motif chemokine 2 | CXCL2 | 1 | Homosapiens |
| 65 | O96017 | Serine/threonine-protein kinase Chk2 | CHEK2 | 1 | Homosapiens |
| 66 | Q07869 | Peroxisome proliferator-activated receptor alpha | PPARA | 1 | Homosapiens |
| 67 | P02741 | C-reactive protein | CRP | 1 | Homosapiens |
| 68 | P02778 | C-X-C motif chemokine 10 | CXCL10 | 1 | Homosapiens |
| 69 | Q9NS23 | Ras association domain-containing protein 1 | RASSF1 | 1 | Homosapiens |
| 70 | P17936 | Insulin-like growth factor-binding protein 3 | IGFBP3 | 1 | Homosapiens |
| 71 | P01344 | Insulin-like growth factor II | IGF2 | 1 | Homosapiens |
| 72 | P21860 | Receptor tyrosine-protein kinase erbB-3 | ERBB3 | 1 | Homosapiens |
| 73 | P09488 | Glutathione S-transferase Mu 1 | GSTM1 | 2 | Homosapiens |
| 74 | P28223 | 5-Hydroxytryptamine 2A receptor | HTR2A | 4 | Homosapiens |
| 75 | P84022 | Mothers against decapentaplegic homolog 3 | SMAD3 | 1 | Homosapiens |
| 76 | P08588 | Beta-1 adrenergic receptor | ADRB1 | 3 | Homosapiens |
| 77 | P29466 | Caspase-1 | CASP1 | 2 | Homosapiens |
| 78 | P18509 | Pituitary adenylate cyclase-activating polypeptide | ADCYAP1 | 1 | Homosapiens |
| 79 | O95456 | Proteasome assembly chaperone 1 | PSMG1 | 1 | Homosapiens |
| 80 | P05112 | Interleukin-4 | IL-4 | 1 | Homosapiens |
| 81 | P00325 | Alcohol dehydrogenase 1B | ADH1B | 1 | Homosapiens |
| 82 | P28702 | Retinoic acid receptor RXR-beta | RXRB | 1 | Homosapiens |
| 83 | P04040 | Catalase | CAT | 1 | Homosapiens |
| 84 | P01308 | Insulin | INS | 1 | Homosapiens |
| 85 | P21731 | Thromboxane A2 receptor | TBXA2R | 1 | Homosapiens |
| 86 | P07858 | Cathepsin B | CTSB | 1 | Homosapiens |
| 87 | P40763 | Signal transducer and activator of transcription 3 | STAT3 | 1 | Homosapiens |
| 88 | Q00534 | Cell division protein kinase 6 | CDK6 | 1 | Homosapiens |
| 89 | P09038 | Heparin-binding growth factor 2 | FGF2 | 1 | Homosapiens |
| 90 | P15336 | Cyclic AMP-dependent transcription factor ATF-2 | ATF2 | 1 | Homosapiens |
| 91 | P04141 | Granulocyte-macrophage colony-stimulating factor | CSF2 | 1 | Homosapiens |
| 92 | P17677 | Neuromodulin | GAP43 | 1 | Homosapiens |
| 93 | P18031 | Tyrosine-protein phosphatase nonreceptor type 1 | PTPN1 | 1 | Homosapiens |
| 94 | P30279 | G1/S-specific cyclin-D2 | CCND2 | 1 | Homosapiens |
3.3. GO-BP Analysis
To further validate whether biological processes enriched by candidate targets as mentioned above were correlated with UC, GO-BP enrichment analysis was performed via DAVID. The top 20 significant GO-BP terms are shown in Figure 2. Most of them were strongly associated with inflammatory- and immune-related BPs such as “positive regulation of interleukin-6 biosynthetic process,” “regulation of inflammatory response,” “immune response,” and “positive regulation of T-cell proliferation.” In short, the 41 bioactive compounds in Quyushengxin formula may act on 94 candidate targets with inflammatory- and immune-related effects to affect UC pathogenesis.
Figure 2.
Gene Ontology biological process analysis. The y-axis shows significantly enriched “Biological Processes” categories, and the x-axis shows the enrichment scores of these terms.
3.4. Establishment of T-D Network
Target-related diseases were predicted by mapping them to integrating multisource databases, including CTD, TTD, and PharmGKB. A T-D network consisting of 90 targets and 4 kinds of diseases was built (Figure 3). The four diseases were digestive system disease (degree = 60), pathology (degree = 49), cancer (degree = 23), and signs and symptoms (degree = 14).
Figure 3.
Target-disease network. Red square represents disease and green circle represents target.
3.5. T-P Network Evaluation
KEGG pathway enrichment analysis was performed for the 94 targets, and T-P network was built. Results displayed that 79 targets could be further mapped to 78 pathways, including “mTOR signaling pathway,” “T-cell receptor signaling pathway,” “JAK-STAT signaling pathway,” and “FOXO signaling pathway” (Figure 4). The average degree of targets was 6.85, and the average degree of pathway was 2.8. In addition, 71 candidate targets could be mapped to several pathways (≥5), suggesting that these targets might mediate the cross-talk and interactions between different pathways. Besides, those pathways (70/78) mapped by multiple targets (≥8) might be the main factors for UC development and progression. These pathways were further divided into five function modules, including inflammatory regulation, immune regulation, metabolic regulation, bacterial infection or mycosis and other function.
Figure 4.
Target-pathway network. Red square represents pathway and purple circle represents targets.
3.6. Establishment of Compound-Target-Function Module Network
By combing the networks above, a compound-target-function module network was built, which included 140 nodes (5 function modules, 41 compounds and 95 targets) and 653 edges (Figure 5).
Figure 5.
Compound-target-function module network. Green circle represents compound, blue square represents target, and red hexagon represents function module.
3.7. Details of 4 UC-Related Pathways from T-P Network Analysis
To further unveil the multi-targets mechanisms of Quyushengxin formula in the treatment of UC, an integrated “UC-related pathway” was established according to the key pathways from the T-P network analysis. UC-related pathways as shown in Figure 6 were composed of four pathways, including “T cell receptor signaling pathway” (hsa04660), “FOXO signaling pathway” (hsa04068), “JAK-STAT signaling pathway” (hsa04630) and “mTOR signaling pathway” (hsa04150). Those targets of the integrated “UC-related pathways” displayed the functional relationship with the UC-related proteins. UC-related pathways can be divided into three modules: immunology module, metabolism module and cell apoptosis-related module. Immunology module consisted of “T cell receptor signaling pathway” (hsa04660), and metabolism module consisted of “FOXO signaling pathway” (hsa04068). Cell apoptosis-related module was comprised of “JAK-STAT signaling pathway” (hsa04630) and “mTOR signaling pathway” (hsa04150). Taken together, Quyushengxin formula may well regulate immunology progress, metabolism progress and cell apoptosis progress to suppress UC progression.
Figure 6.
Distribution of targets of Quyushengxin formula in the “UC-related pathway.” Arrow shows activation effect; T-shaped arrow shows inhibition effect, and dotted arrow represents indirect activation effect or inhibition effect.
4. Discussion
TCM has the advantages of high treatment efficacy and low treatment cost and side effect in the treatment of several diseases, including UC in China for several thousands of years [32–34]. After preliminary screening based on ADME properties, 41 potential bioactive compounds of Quyushengxin were screened out. Thereafter, 94 candidate targets of these 41 bioactive compounds were predicted for further analysis. Functional enrichment analyses suggested that these targets were closely related with inflammatory- and immune-related biological processes. Besides, a C-T network, a T-D network, a T-P network, and a compound-target-function module network were built. These networks indicated that the therapeutic effects of Quyushengxin on UC may be achieved through the synergistic and additive effects on multiple molecules and multiple pathways with immune and inflammatory effects to treat UC.
Previous reports showed that the TCMSP-based method was reliable for screening out bioactive compounds of TCM for treatment of thrombosis [35], gastric precancerous lesions [36], cardiocerebrovascular disease [37], and rheumatoid arthritis [38]. In this study, 41 bioactive compounds of Quyushengxin formula were selected out by using TCMSP database in combination with ADME properties. Most of the 41 compounds have been reported to have anti-inflammatory and immune-regulatory effects. For example, quercetin (mol01, OB = 46.43%, DL = 0.28, HL = 14.40) could inhibit lipopolysaccharide- (LPS-) induced interleukin- (IL-) 6 production [39], TNF-α production, and IL-8 production [40, 41] to exert anti-inflammatory effect. Besides, ursolic acid (mol17, OB = 16.77%, DL = 0.75, HL = 5.28) was reported to have human neutrophil elastase inhibitory effect both in vitro and in vivo [42]. Kaempferol (mol17, OB = 41.88%, DL = 0.24, HL = 14.74) was reported to significantly reduce the overproduction of TNF-α, IL-1β, IL-6, intercellular adhesion molecule- (ICAM-) 1, and vascular cell adhesion molecule- (VCAM-) 1 induced by LPS [43]. In addition, β-sitosterol (mol16, OB = 36.91%, DL = 0.75, HL = 5.36) and rutin (mol33, OB = 3.2%, DL = 0.68, HL = 6.22) were shared with significant anti-inflammatory activity [44, 45]. Above all, TCMSP-based systems pharmacology sifted out 41 potential bioactive compounds in Quyushengxin formula for treatment of UC.
Eight of the 94 targets have degree larger than 10 in the C-T network, including ESR1, PTGS2, NOS2, PTGS1, PPARG, NOS3, ESR2, and KCNH2. ESR1 was targeted by 34 compounds, which contributed to T-cell-mediated autoimmune inflammation by promoting T-cell activation and proliferation [46]. Besides, PTGS2 with the second highest degree played a critical role in the pathogenesis of gut inflammation [47, 48]. Moreover, PPARG was demonstrated to be able to downregulate proinflammatory cytokines production, such as IL-4, -5, and -6. In addition, PPARG could also enable to interfere with profibrotic molecules, such as platelet-derived growth factor (PDGF), IL-1, and transforming growth factor beta (TGF-β) [49]. These results suggested that Quyushengxin formula could probably treat UC by regulating anti-inflammatory action and the immune system.
In this study, 94 targets were utilized to perform T-P network analysis, and the results showed that 79 targets could be further mapped to 78 pathways. Meanwhile, numerous pathways mapped by multiple targets might be the main factors for UC progression. Four pathways including “T-cell receptor signaling pathway,” “FOXO signaling pathway,” “JAK-STAT signaling pathway,” and “mTOR signaling pathway” were closely associated with immune and inflammatory effects. T-cell receptors play significant role in function of T cells and formation of the immunological synapse, and they connected T cells and the antigen-presenting cells [50]. T-cell receptor pathway was reported to be important in regulation of UC [51, 52]. FOXO pathway plays a key role in regulating the expression of genes related to cell function such as apoptosis, cell cycle, oxidative stress, and differentiation [53–55]. FOXO3a was shown to control the inflammatory response and help maintain the homeostasis of the intestinal mucosa, which may also be a protective factor in the gut, and maintain a balance between the mucosal immune hemostasis against intravascular bacteria and inflammatory cytokines [56]. Besides, JAK-STAT pathway is the fulcrum for many important cellular processes, including cell survival, differentiation, proliferation, and regulation of immune function [57]. The mTOR pathway plays an important role in regulation of cell metabolism, proliferation, and autophagy. It is reported that mTOR signaling pathway was activated in bacteria-induced colitis in mice [58]. Inhibitors of mTOR signaling pathway are effective as anti-inflammatory drugs in treating colitis [59–61]. Therefore, Quyushengxin might suppress UC progression through targeting these anti-inflammation, autophagy, and immunoregulation pathways.
Nevertheless, limitations in this study could never be neglected. First, results in this study were mainly based on known chemical components in Quyushengxin, related targets, and pathways in UC. With the development of science and technology, new components in Quyushengxin, as well as new targets and pathways in UC will be further discovered, which will supply us with more theoretical evidences for further elucidation of underlying mechanisms of UC pathology. Second, the interaction relationships of the nodes in the networks, such as the action type, e.g., activation, inhibition, binding, and catalysis, and the action effect, e.g., positive, negative, and unspecified, are not investigated due to lack of these data. Third, due to the complex interaction between TCM and the human body, many of its acting mechanisms still needed to be further elucidated via pharmacokinetic test and other experiments.
5. Conclusion
In short, network pharmacology analysis of Quyushengxin showed that 41 bioactive components of Quyushengxin may act on 94 immune and inflammation-related targets to suppress UC progression in a synergistic and additive manner, which may provide us with a new starting point for a more detailed knowledge of mechanisms of UC pathogenesis.
Acknowledgments
The authors would like to thank Ms. Huaping Liu in assistance with data analysis. This work was supported by the National Natural Science Foundation of China project (nos. 81603633, 81874468, and 81403399), Shanghai Committee of Science and Technology project (no. 16401971400), and Peak Research Team Project in Shanghai University of Traditional Chinese Medicine.
Contributor Information
Jiong Wu, Email: 12491947@qq.com.
Zhenyi Wang, Email: yyyygangchangke@163.com.
Data Availability
The datasets used and analyzed during the current study are available by sending email to the corresponding author.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Haojie Yang, Ying Li, and Sichen Shen contributed equally to this study.
References
- 1.Abraham C., Cho J. H. Inflammatory bowel disease. New England Journal of Medicine. 2009;361(21):2066–2078. doi: 10.1056/nejmra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosnes J., Gower–Rousseau C., Seksik P., Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 3.Morsy M. A., Gupta S., Nair A. B., Venugopala K. N., Greish K., El-Daly M. Protective effect of spirulina platensis extract against dextran-sulfate-sodium-induced ulcerative colitis in rats. Nutrients. 2019;11(10):p. 2309. doi: 10.3390/nu11102309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danese S., Fiocchi C. Ulcerative colitis. New England Journal of Medicine. 2011;365(18):1713–1725. doi: 10.1056/nejmra1102942. [DOI] [PubMed] [Google Scholar]
- 5.Loftus E. V., Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126(6):1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Horin S., Andrews J. M., Katsanos K. H., et al. Combination of corticosteroids and 5-aminosalicylates or corticosteroids alone for patients with moderate-severe active ulcerative colitis: a global survey of physicians’ practice. World Journal of Gastroenterology. 2017;23(16):2995–3002. doi: 10.3748/wjg.v23.i16.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreiro-Alonso E., Saro-Gismera C., Sánchez M. Outcomes and prediction of corticosteroid therapy after successive courses of ulcerative colitis treatments. Expert Review of Gastroenterology & Hepatology. 2018;12(7):733–741. doi: 10.1080/17474124.2018.1489231. [DOI] [PubMed] [Google Scholar]
- 8.Salice M., Rizzello F., Calabrese C., Calandrini L., Gionchetti P. A current overview of corticosteroid use in active ulcerative colitis. Expert Review of Gastroenterology & Hepatology. 2019;13(6):557–561. doi: 10.1080/17474124.2019.1604219. [DOI] [PubMed] [Google Scholar]
- 9.Feuerstein J. D., Cheifetz A. S. Ulcerative Colitis. Mayo Clinic Proceedings. 2014;89(11):1553–1563. doi: 10.1016/j.mayocp.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Li R., Chen Y., Shi M., et al. Gegen Qinlian decoction alleviates experimental colitis via suppressing TLR4/NF-κB signaling and enhancing antioxidant effect. Phytomedicine. 2016;23(10):1012–1020. doi: 10.1016/j.phymed.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L., Zhang Y.-L., Dai Y.-C., et al. Jianpi Qingchang decoction alleviates ulcerative colitis by inhibiting nuclear factor-κB activation. World Journal of Gastroenterology. 2017;23(7):1180–1188. doi: 10.3748/wjg.v23.i7.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y.-L., Zheng Y.-Y., Dai Y.-C., Zhang Y.-L., Tang Z.-P. Systems pharmacology approach reveals protective mechanisms of Jian-Pi Qing-Chang decoction on ulcerative colitis. World Journal of Gastroenterology. 2019;25(21):2603–2622. doi: 10.3748/wjg.v25.i21.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei L., Xu K. Zhikang Capsule ameliorates dextran sodium sulfate-induced colitis by inhibition of inflammation, apoptosis, oxidative stress and MyD88-dependent TLR4 signaling pathway. Journal of Ethnopharmacology. 2016;192:236–247. doi: 10.1016/j.jep.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 14.He Z., Zhou Q., Wen K., et al. Huangkui Lianchang decoction ameliorates DSS-induced ulcerative colitis in mice by inhibiting the NF-κB signaling pathway. Evidence-Based Complementary and Alternative Medicine. 2019;2019 doi: 10.1155/2019/1040847.1040847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z., Pei W., Guo Y., et al. Gut microbiota-mediated NLRP12 expression drives the attenuation of dextran sulphate sodium-induced ulcerative colitis by Qingchang Wenzhong decoction. Evidence-Based Complementary and Alternative Medicine. 2019;2019:12. doi: 10.1155/2019/9839474.9839474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao T., Li J., Liu L., et al. Qingchang Wenzhong decoction attenuates DSS-induced colitis in rats by reducing inflammation and improving intestinal barrier function via upregulating the MSP/RON signalling pathway. Evidence-Based Complementary and Alternative Medicine. 2017;2017 doi: 10.1155/2017/4846876.4846876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan D., Han C., Feng Z., et al. Clinical research of Quyu Shengxin formula combined with mesalazine in treating mild to moderate ulcerative colitis. Shanghai Journal of Traditional Chinese Medicine. 2017;8:54–57. [Google Scholar]
- 18.Berger S. I., Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C., Zheng C., Li Y., Wang Y., Lu A., Yang L. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Briefings in Bioinformatics. 2014;15(5):710–733. doi: 10.1093/bib/bbt035. [DOI] [PubMed] [Google Scholar]
- 20.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei T., Zheng C., Huang C., et al. Systematic understanding the mechanisms of vitiligo pathogenesis and its treatment by Qubaibabuqi formula. Journal of Ethnopharmacology. 2016;190:272–287. doi: 10.1016/j.jep.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Li B., Tao W., Zheng C., et al. Systems pharmacology-based approach for dissecting the addition and subtraction theory of traditional Chinese medicine: an example using Xiao-Chaihu-Decoction and Da-Chaihu-Decoction. Computers in Biology and Medicine. 2014;53:19–29. doi: 10.1016/j.compbiomed.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Xu X., Li Y., et al. Systems pharmacology uncovers Janus functions of botanical drugs: activation of host defense system and inhibition of influenza virus replication. Integrative Biology. 2013;5(2):351–371. doi: 10.1039/c2ib20204b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng C., Guo Z., Huang C., et al. Large-scale direct targeting for drug repositioning and discovery. Scientific Reports. 2015;5:p. 11970. doi: 10.1038/srep11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang D. W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Grondin C. J., Davis A. P., Wiegers T. C., Wiegers J. A., Mattingly C. J. Accessing an expanded exposure science module at the comparative Toxicogenomics database. Environmental Health Perspectives. 2018;126(1) doi: 10.1289/ehp2873.014501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y. H., Yu C. Y., Li X. X., et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Research. 2018;46(D1):D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whirl-Carrillo M., McDonagh E. M., Hebert J. M., et al. Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smoot M. E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azuaje F. J., Zhang L., Devaux Y., Wagner D. R. Drug-target network in myocardial infarction reveals multiple side effects of unrelated drugs. Scientific Reports. 2011;1:p. 52. doi: 10.1038/srep00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindman A. B., Cox D. F. Changes in health care costs and mortality associated with transitional care management services after a discharge among medicare beneficiaries. JAMA Internal Medicine. 2018;178(9):1165–1171. doi: 10.1001/jamainternmed.2018.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng X., Xing X., Sun G., et al. Guanxin danshen formulation protects against myocardial ischemia reperfusion injury-induced left ventricular remodeling by upregulating estrogen receptor beta. Frontiers in Pharmacology. 2017;8:p. 777. doi: 10.3389/fphar.2017.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J., Guo F., Zhang M., et al. Signature-oriented investigation of the efficacy of multicomponent drugs against heart failure. The FASEB Journal. 2019;33(2):2187–2198. doi: 10.1096/fj.201800673rr. [DOI] [PubMed] [Google Scholar]
- 35.Yi F., Sun L., Xu L. J., et al. In silico approach for anti-thrombosis drug discovery: P2Y1R structure-based TCMs screening. Frontiers in Pharmacology. 2017;7:p. 531. doi: 10.3389/fphar.2016.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L., Liu W., Hu Z., et al. A systems pharmacology approach for identifying the multiple mechanisms of action of the wei pi xiao decoction for the treatment of gastric precancerous lesions. Evidence-Based Complementary and Alternative Medicine. 2019;2019:15. doi: 10.1155/2019/1562707.1562707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Li Y., Wang J., et al. Systematic investigation of ginkgo biloba leaves for treating cardio-cerebrovascular diseases in an animal model. ACS Chemical Biology. 2017;12(5):1363–1372. doi: 10.1021/acschembio.6b00762. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Li Y., Yang Y., et al. Systems pharmacology dissection of multiscale mechanisms of action for herbal medicines in treating rheumatoid arthritis. Molecular Pharmaceutics. 2017;14(9):3201–3217. doi: 10.1021/acs.molpharmaceut.7b00505. [DOI] [PubMed] [Google Scholar]
- 39.Liu J., Li X., Yue Y., Li J., He T., He Y. The inhibitory effect of quercetin on IL-6 production by LPS-stimulated neutrophils. Cellular & Molecular Immunology. 2005;2(6):455–460. [PubMed] [Google Scholar]
- 40.Geraets L., Moonen H. J. J., Brauers K., Wouters E. F. M., Bast A., Hageman G. J. Dietary flavones and flavonoles are inhibitors of poly(ADP-ribose)polymerase-1 in pulmonary epithelial cells. The Journal of Nutrition. 2007;137(10):2190–2195. doi: 10.1093/jn/137.10.2190. [DOI] [PubMed] [Google Scholar]
- 41.Rao M. K., Ghosh B. Quercetin inhibits LPS-induced nitric oxide and tumor necrosis factor-α production in murine macrophages. International Journal of Immunopharmacology. 1999;21(7):435–443. doi: 10.1016/s0192-0561(99)00024-7. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Xu H. Y., Wu Y. J., Zhang X., Zhang L. Q., Li Y. M. Neutrophil elastase inhibitory effects of pentacyclic triterpenoids from Eriobotrya japonica (loquat leaves) Journal of Ethnopharmacology. 2019;242 doi: 10.1016/j.jep.2019.01.037.111713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bian Y., Liu P., Zhong J., et al. Kaempferol inhibits multiple pathways involved in the secretion of inflammatory mediators from LPSinduced rat intestinal microvascular endothelial cells. Molecular Medicine Reports. 2019;19(3):1958–1964. doi: 10.3892/mmr.2018.9777. [DOI] [PubMed] [Google Scholar]
- 44.Al-Harbi N. O., Imam F., Al-Harbi M. M., et al. Rutin inhibits carfilzomib-induced oxidative stress and inflammation via the NOS-mediated NF-κB signaling pathway. Inflammopharmacology. 2019;27(4):817–827. doi: 10.1007/s10787-018-0550-5. [DOI] [PubMed] [Google Scholar]
- 45.Ding K., Tan Y. Y., Ding Y., et al. β-Sitosterol improves experimental colitis in mice with a target against pathogenic bacteria. Journal of Cellular Biochemistry. 2019;120(4):5687–5694. doi: 10.1002/jcb.27853. [DOI] [PubMed] [Google Scholar]
- 46.Mohammad I., Starskaia I., Nagy T., et al. Estrogen receptor α contributes to T cell-mediated autoimmune inflammation by promoting T cell activation and proliferation. Science Signaling. 2018;11(526) doi: 10.1126/scisignal.aap9415. [DOI] [PubMed] [Google Scholar]
- 47.Bento A. F., Leite D. F. P., Marcon R., et al. Evaluation of chemical mediators and cellular response during acute and chronic gut inflammatory response induced by dextran sodium sulfate in mice. Biochemical Pharmacology. 2012;84(11):1459–1469. doi: 10.1016/j.bcp.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Vong L., Ferraz J. G. P., Panaccione R., Beck P. L., Wallace J. L. A pro-resolution mediator, prostaglandin D2, is specifically up-regulated in individuals in long-term remission from ulcerative colitis. Proceedings of the National Academy of Sciences. 2010;107(26):12023–12027. doi: 10.1073/pnas.1004982107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vetuschi A., Pompili S., Gaudio E., Latella G., Sferra R. PPAR-gamma with its anti-inflammatory and anti-fibrotic action could be an effective therapeutic target in IBD. European Review for Medical and Pharmacological Sciences. 2018;22(24):8839–8848. doi: 10.26355/eurrev_201812_16652. [DOI] [PubMed] [Google Scholar]
- 50.Yokosuka T., Saito T. The immunological synapse, TCR microclusters, and T cell activation. Current Topics in Microbiology and Immunology. 2010;340:81–107. doi: 10.1007/978-3-642-03858-7_5. [DOI] [PubMed] [Google Scholar]
- 51.Lee J. C., Lyons P. A., McKinney E. F., et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. Journal of Clinical Investigation. 2011;121(10):4170–4179. doi: 10.1172/jci59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seidelin J. B., Coskun M., Kvist P. H., Holm T. L., Holgersen K., Nielsen O. H. IL-33 promotes GATA-3 polarization of gut-derived T cells in experimental and ulcerative colitis. Journal of Gastroenterology. 2015;50(2):180–190. doi: 10.1007/s00535-014-0982-7. [DOI] [PubMed] [Google Scholar]
- 53.Accili D., Arden K. C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117(4):421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 54.Nakashima K., Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Bioscience, Biotechnology, and Biochemistry. 2007;71(7):1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 55.Rathbone C. R., Booth F. W., Lees S. J. FoxO3a preferentially induces p27Kip1 expression while impairing muscle precursor cell-cycle progression. Muscle & Nerve. 2008;37(1):84–89. doi: 10.1002/mus.20897. [DOI] [PubMed] [Google Scholar]
- 56.He Z., He X., Chen Z., et al. Activation of the mTORC1 and STAT3 pathways promotes the malignant transformation of colitis in mice. Oncology Reports. 2014;32(5):1873–1880. doi: 10.3892/or.2014.3421. [DOI] [PubMed] [Google Scholar]
- 57.Coskun M., Salem M., Pedersen J., Nielsen O. H. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacological Research. 2013;76:1–8. doi: 10.1016/j.phrs.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Sun X., Threadgill D., Jobin C. Campylobacter jejuni induces colitis through activation of mammalian target of rapamycin signaling. Gastroenterology. 2012;142(1):86–95. doi: 10.1053/j.gastro.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhonde M. R., Gupte R. D., Dadarkar S. D., et al. A novel mTOR inhibitor is efficacious in a murine model of colitis. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;295(6):G1237–G1245. doi: 10.1152/ajpgi.90537.2008. [DOI] [PubMed] [Google Scholar]
- 60.Farkas S., Hornung M., Sattler C., et al. Rapamycin decreases leukocyte migration in vivo and effectively reduces experimentally induced chronic colitis. International Journal of Colorectal Disease. 2006;21(8):747–753. doi: 10.1007/s00384-005-0793-7. [DOI] [PubMed] [Google Scholar]
- 61.Kim H., Banerjee N., Barnes R. C., et al. Mango polyphenolics reduce inflammation in intestinal colitis-involvement of the miR-126/PI3K/AKT/mTOR axis in vitro and in vivo. Molecular Carcinogenesis. 2017;56(1):197–207. doi: 10.1002/mc.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available by sending email to the corresponding author.








