Abstract
FAT1 is a mutant gene found frequently in human cervical cancer (CC), but its expression and relevance in CC proliferation, invasion, and migration are still unknown. We aimed to explore the role and novel mechanism of FAT1 in CC progression. The expression of FAT1 in CC and adjacent normal tissues was analysed, and we investigated the proliferation, migration, and invasion of HeLa and C33A cells treated with wild-type FAT1 plasmid or FAT1 siRNA. Meanwhile, we evaluated the effect of FAT1 on the epithelial-mesenchymal transition (EMT) and the β-catenin-mediated transcription of target genes. Here, we showed that FAT1 expression was significantly lower in CC tissues than in adjacent tissues. FAT1 overexpression significantly dysregulated CC cell proliferation, invasion, and migration, whereas FAT1 knockdown had the opposite effect. FAT1 overexpression promoted the expression of phosphorylated β-catenin and E-cadherin protein and inhibited the expression of vimentin, TWIST, and several downstream targets of β-catenin, namely, c-MYC, TCF-4 and MMP14. In contrast, FAT1 silencing notably increased the expression c-MYC, TCF-4, and MMP14 and promoted the EMT in HeLa and C33A cells. Endogenous and exogenous FAT1 was confirmed to interact with β-catenin, and the overexpression of β-catenin could partially block the effect of FAT1 on the proliferation, migration, and invasion of HeLa and C33A cells. Conclusion: FAT1 acts as a tumor suppressor by inhibiting β-catenin-mediated transcription and might be used as a novel anti-metastatic agent in targeted CC therapy.
Keywords: Cervical cancer, FAT1, β-catenin, metastasis, epithelial-mesenchymal transition
Introduction
Cervical cancer (CC) is one of the most common gynecologic cancers and is the fourth leading cause of mortality in women worldwide. The survival rates for International Federation of Gynecology and Obstetrics (FIGO) stage I patients are reported as 80-98% but decrease markedly to 50% when patients also have metastasis [4]. Although many patients have greatly benefitted from surgery and chemoradiotherapy, the recurrence and metastasis rates remain high (approximately 29% to 38%) and are the main causes of death [32]. Thus, the molecular pathogenesis underlying CC progression must be identified to elucidate new therapeutic targets.
FAT1, a member of the FAT cadherin superfamily, is a ~500-kD transmembrane protein that regulates actin dynamics, cell-cell adhesion, and cell polarity [13,24,26]. FAT1 is expressed differently in human cancer cells. Our preliminary work showed that the mutation rate of FAT1 was 8.8%. Notably, FAT1 mutations, including termination (44%) and missense (56%) mutations, occurred in the functional area. A previous study showed that the expression of FAT1 was downregulated in intrahepatic cholangiocarcinoma tissue [22], oral cancer cell lines, primary oral cancers (Nakaya et al. 2005) and astrocytic tumors [6]. In contrast, Fat1 protein is expressed in leukemia cell lines, such as Jurkat and THP-1 cells, whereas no visible expression of FAT1 is detected in normal peripheral blood and bone marrow cells [7]. Mechanically, FAT1 plays a dual role in difference cancers by the Hippo, Wnt, and MAPK/ERK signalling pathways [1,11,17]. It is conventionally considered to be a tumor suppressor gene in oral cancer [6], head and neck squamous cell carcinoma [9], and astrocytic tumors [6], among others [18]. Conversely, FAT1 is also considered an oncogene that promotes acute myeloid leukaemia and acute lymphoblastic leukaemia proliferation [7], modulates epithelial-mesenchymal transition (EMT) and the expression of stemness-related genes in hypoxic glioblastoma [25]. To date, the role and mechanisms of FAT1 in CC remain unclear.
The Wnt signalling pathway is highly conserved and critical for cell proliferation, differentiation, invasion and migration [5,29,31]. As a canonical mediator of the Wnt pathway, β-catenin plays a key role in Wnt signal transduction cascades and the activation of the transcription of downstream target genes, such as T cell factor/lymphoid enhancer factor (TCF/LEF) and c-Myc, after its stabilization in the cytoplasm and translocation into the nucleus [21]. MacDonald et al. showed that aberrant activation of the Wnt/β-catenin signalling pathway contributes to many cancers [16]. Shinohara and colleagues revealed that β-catenin is notably accumulated in the cytoplasm and nucleus in 47% of cervical cancer tissues [23]. Moreover, nuclear expression of β-catenin is related to poor prognosis and chemo/radioresistance in human CC [34]. Other studies have demonstrated that the inhibition of β-catenin could suppress the proliferation, migration, and invasion of CC cells [12,27,28,30]. These data hint that β-catenin plays a critical role in controlling the fate of CC. However, the mechanism underlying the aberrant activation of the β-catenin signaling pathway and the regulation of β-catenin in CC cells remains unclear.
In this study, we aimed to determine the expression and function of FAT1 and evaluated the relationship between FAT1 and β-catenin in CC cells as well as the underlying molecular mechanism. Taken together, the data demonstrate that FAT1 severs as a tumor suppressor in CC by promoting the phosphorylation of β-catenin.
Materials and methods
Materials and samples
40 CC patients’ tumor and adjacent tissues were collected in the Department of Obstetrics & Gynecology, Southwest Hospital, Army Medical University. Patients who were enrolled provided written informed consent, and the Ethical Committee of Southwest Hospital of the Third Military Medical University approved the study (approval number: KY201336). TRI Reagent, CCK-8 and anti-FAT1 primary antibody were purchased from Sigma-Aldrich (Shanghai, China). Other primary antibodies, including E-cadherin, vimentin, TWIST, total β-catenin and phospho-β-catenin (p-β-catenin), transcription factor 4 (TCF-4), matrix metalloproteinase-14 (MMP-14), c-Myc and β-actin antibodies, were purchased from CST Co., Ltd. (Shanghai, China). A RevertAidTM First Strand cDNA Synthesis Kit and Protein Extraction Reagent for Mammalian, RPMI 1640 Medium, Minimum Essential Media (MEM), fetal bovine serum (FBS), Opti-MEM and 0.25% trypsin were ordered from Thermo Fisher Scientific Co., Ltd. (Shanghai, China). PCR Mix for real-time PCR was purchased from Roche (Basel, Switzerland). Transwell® inserts were purchased from Corning Co., Ltd. (New York, USA). Protein beads (A/G PLUS-Agarose) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated secondary antibodies were purchased from Beijing Golden Bridge Biotech (Beijing, China). Immobilon Western Chemiluminescent HRP Substrate was purchased from Merck KGaA (Darmstadt, Germany). ViaFect™ transfection reagent was ordered from Promega Co., Ltd. (Madison, WI, USA). pcDNA3.1-FAT1, pcDNA3.1(+)-β-catenin and pcDNA3.1(+) (vector) plasmids were constructed by Sangon Biotech Co., Ltd. (Shanghai, China). Other reagents were from Sigma-Aldrich and used as received (Shanghai, China).
RNA isolation and Real-time PCR
Total RNA was isolated from CC tissues, adjacent nontumor tissues, and harvested cells, and cDNA was synthesized according to a previous report [14]. The following primers: FAT1 forward: 5’-CCTTCCAACAGCCACATCCACTAC-3’, reverse: 5’-TTGAACCGTGAGCGTGTAACCTG-3’; β-actin forward: 5’-GAGCACAGAGCCTCGCCTTT-3’, reverse: 5’-AGAGGCGTACAGGGATAGCA-3’. FAT1 gene expression was normalized to that of β-actin.
Immunohistochemistry (IHC)
IHC assay was carried out as previously described [33]. Briefly, tissue sections were deparaffinized in xylene and rehydrated. After antigen retrieval, the samples blocked with 10% BSA. The sections incubated with the primary antibody for FAT1. Finally, the bound antibodies were stained with diaminobenzidine (DAB), and the sections were counterstained with hematoxylin. Each experiment was repeated 3 times.
Cell cultures
HeLa (HPV18+) cells were purchased from a typical Cell Culture Collection Committee of the Chinese Academy of Sciences Library, and C33A (HPV-) CC cells were obtained from ATCC. These cells had been identified by STR. Hela and C-33A cells were identified again by Genechem and Fuheng Biology Co. Ltd in October 2017 and November 2018, respectively. These cells were cultured in RPMI 1640 or MEM containing 10% FBS at 37°C in 5% CO2.
Silencing and overexpression of FAT1 in HeLa and C33A cells
Three FAT1 siRNA oligos and a negative control (NC) were synthesized by RiboBio Co., Ltd. (Guangzhou, China). Cells were seeded into 6-cm dishes overnight. The cells were transfected with three FAT1 siRNA oligos and the NC using ViaFect™ transfection reagent in Opti-MEM for 6 h. The medium was removed, and fresh complete medium was added for another 48 h of culturing. Silencing efficiency at the mRNA level was evaluated by RT-PCR. ViaFect™ transfection reagent was used to transfect HeLa and C33A cells with pcDNA3.1-FAT1 or the vector for 12 h. The Opti-MEM was then removed, and fresh complete medium was added for further study.
Co-immunoprecipitation and western blot
To analyse whether exogenous flagged FAT1 binds endogenous β-catenin in these cells, they were infected with 3 × Flag-FAT1 adenovirus for 24 h then harvested and lysed with lysis buffer. Either β-catenin monoclonal antibody or control IgG was incubated with the cell lysate at 4°C for overnight. Protein A/G PLUS-agarose was added to that cell lysate and incubated for 2 h. The agarose beads were then washed 3 times with lysis buffer. The precipitates were analysed by western blot as previously described [14]. A similar protocol was used to analyze whether endogenous FAT1 bound the endogenous β-catenin.
Cell viability assays
The viability of HeLa and C33A cell was detected by CCK-8. Briefly, cells (5000 per well) were seeded into 96-well plates, allowed to attach for 12 h, then treated as indicated for an additional 48 h. After removing the medium, 100 μL of fresh medium with 10 μL of CCK-8 reagent was added to each well, and then incubated at 37°C for 4 h. Finally, each well’s absorbance at 450 nm was detected.
Migration and invasion assays
The ability of cell migration and invasion was determined by transwell assay according to manufacturer’s instructions. 5 × 103 cells in 100 μL of serum-free medium were seeded into the upper chamber for the migration assay. For the invasion assay, the upper chamber was coated with Matrigel, and the bottom chamber was coated with 700 μL of complete culture medium. Cells on the upper surface of the upper chamber were removed after incubating at 37°C for 24 h. Invaded cells on the lower surface of the upper chamber were fixed with 4% PFA then stained with 1% crystal violet for 10 min.
Statistical analysis
The data are presented as the means ± SDs from at least three independent experiments. These data were analysed using SPSS 17.0 (SPSS, Chicago, IL, USA). The data were compared between two group using two-tailed Student’s t-tests and one-way analysis of variance (ANOVA) was carried out to compare data among three and more groups. P < 0.05 was considered significant.
Results
Analysis of FAT1 expression in CC tissues
In our study, we evaluated the expression of FAT1 mRNA in 40 CC tissue samples and matched adjacent non-tumor tissues by qRT-PCR and immunohistochemistry (IHC). As shown in Figure 1A, the relative expression of FAT1 mRNA was 0.42 ± 0.05 and 1.07 ± 0.12 in CC tissues and adjacent non-tumor tissues, respectively (Figure 1A). The expression of FAT1 mRNA in CC tissues was lower than that in adjacent non-tumor tissues (P < 0.001). Additionally, FAT1 protein was weakly stained in CC tissues, whereas obvious staining of FAT1 protein was observed in adjacent non-tumour tissues (Figure 1B). These data suggest that FAT1 exhibited lower expression in CC tissues.
Figure 1.
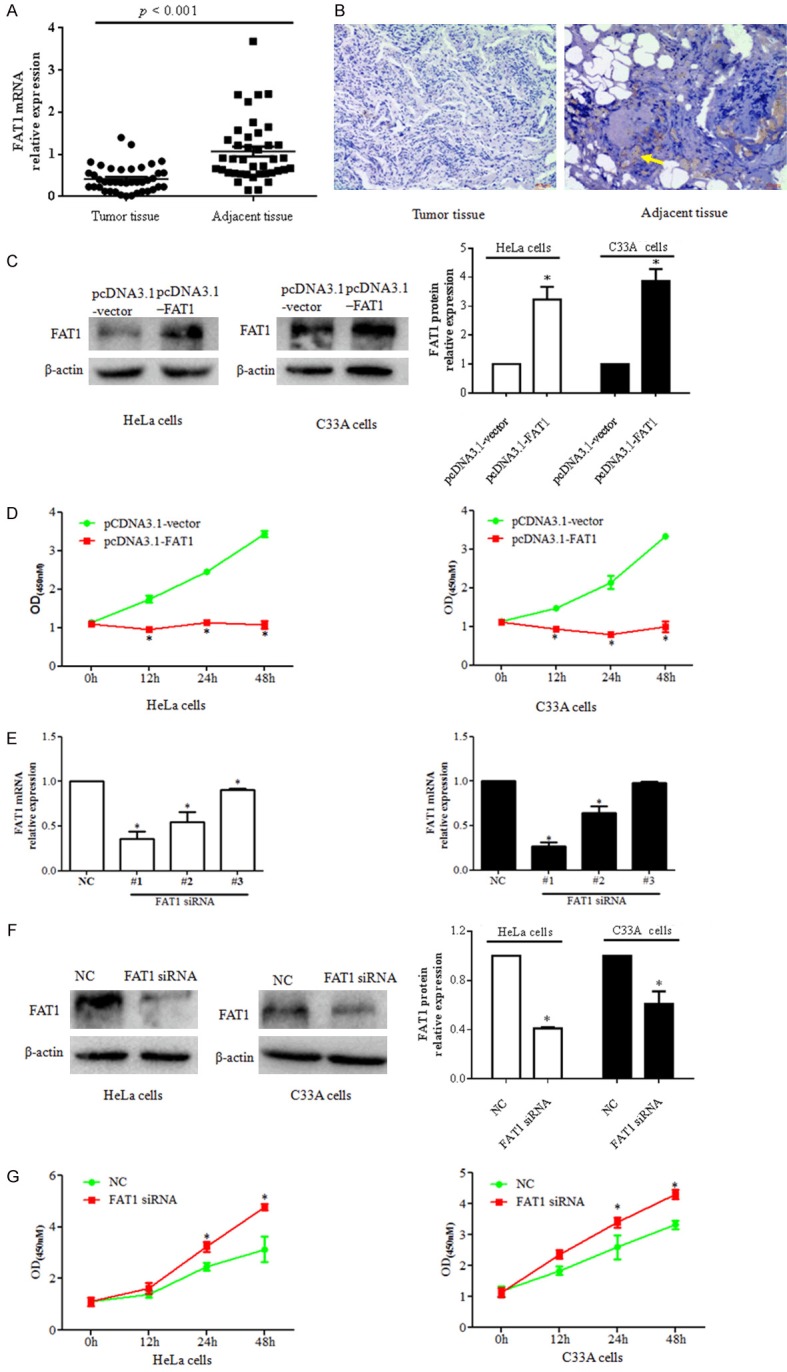
FAT1 regulates cervical cancer cell proliferation. A. Relative expression of FAT1 mRNA was evaluated by qPCR. B. Immunohistochemical analysis of FAT1 protein expression and location in cervical cancer (right panel) and adjacent tissue (left panel). Immunoreactivity appears brown, with a blue hematoxylin counterstain. Scale bar, 40 μm. C. FAT1 expression in HeLa (right panel) and C33A cells (middle panel) transfected with the pcDNA3.1-FAT1 plasmid and pcDNA3.1-vector was evaluated by western blot. The histograms in the left panel illustrate the quantitative analysis of the FAT1 protein levels, which were normalized to the β-actin levels. *P < 0.05 vs. the pcDNA3.1-vector group. D. CCK-8 was used to analyse HeLa (right panel) and C33A cell (left panel) viability at the indicated time points. *P < 0.05 vs. pcDNA3.1-vector group. HeLa and C33A cells were transfected with pcDNA3. 1-FAT1 plasmid and pcDNA3.1-vector for 48 h, respectively. E. The relative expression of FAT1 was detected by qPCR. FAT1 mRNA was silenced by three FAT1 siRNA oligos in HeLa (left histogram) and C33A cells (right histogram) after three FAT1 siRNA sequences and NC were transfected into HeLa and C33A for 48 h, respectively. *P < 0.05 vs. the NC group. F. Western blot was used to analyse the relative expression levels of FAT1 proteins after treating cells with FAT1 siRNA for 48 h in HeLa (left panel) and C33A (middle panel) cells. The histograms in the left panel illustrate the quantitative analysis of the FAT1 protein levels, which were normalized to the β-actin levels. *P < 0.05 vs. NC group. G. CCK-8 was used to analyze the viability of HeLa (left panel) and C33A cells (right panel) at the indicated time points after transfection with FAT1 siRNA. *P < 0.05 vs. NC group.
FAT1 regulated the proliferation of CC cells in vitro
We investigated the effect of FAT1 on the proliferation of CC cells. First, compared with the pcDNA3.1 vector group, the expression of FAT1 protein was significantly elevated in HeLa and C33A cells after transfection of the pcDNA3.1-FAT1 plasmid (P < 0.05) (Figure 1C). A CCK-8 assay showed that the viability of HeLa and C33A cells at 12 h, 24 h, and 48 h was notably suppressed in the pcDNA3.1-FAT1 group compared with the pcDNA3.1 vector group (P < 0.05) (Figure 1D). Second, we silenced FAT1 mRNA using three FAT1 siRNAs (#1, #2 and #3 oligo sequences), and a real-time PCR assay showed that the expression of FAT1 mRNA in both HeLa and C33A cells was lower in the three FAT1 siRNA groups than in the NC group (P < 0.05). Moreover, FAT1 siRNA #1 oligo more effectively silenced FAT1 expression than the other siRNA oligos (Figure 1E), and this oligo was selected for the subsequent studies. Consistent with the above result, a western blot assay showed that the expression of FAT1 protein was significantly decreased in the FAT1 siRNA group compared with the NC group (P < 0.05) (Figure 1F). A CCK-8 assay revealed that the viability of HeLa and C33A cells at 24 h and 48 h was higher in the FAT1 siRNA groups than in the NC groups (P < 0.05) (Figure 1G). These data demonstrate that FAT1 regulates the proliferation of CC cells.
FAT1 overexpression inhibits the migration, invasion, and EMT of CC cells
Metastasis is a main factor responsible for CC progression and induces the death of CC proteins [4]. We explored the effect of FAT1 on the EMT, migration, and invasion of CC cells. The expression of EMT markers, including E-cadherin, vimentin and TWIST, in HeLa and C33A cells 48 h after transfection with pcDNA3.1-FAT1 plasmid was evaluated by western blot. The data showed that the level of E-cadherin protein was higher and those of vimentin and TWIST protein were lower in the pcDNA3.1-FAT1 group than in the pcDNA3.1-vector group (P < 0.05) (Figure 2A and 2B). In contrast, the silencing of FAT1 downregulated E-cadherin and upregulated TWIST and vimentin in HeLa and C33A cells (Figure 2C and 2D).
Figure 2.
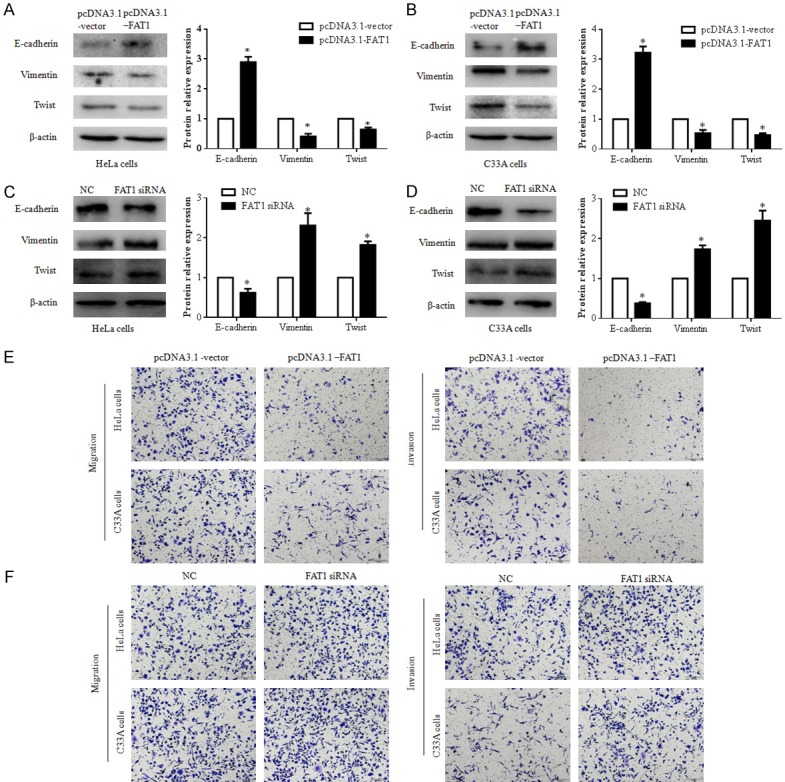
FAT1 suppressed EMT, migration, and invasion of HeLa and C33A cells. (A and B) FAT1 supressed EMT of HeLa and C33A cells. The expression of E-cadherin, vimentin, and TWIST in HeLa (A) and C33A cells (B) were detected by western blot after these cells were treated with pcDNA3.1-FAT1 or pcDNA3.1-vector plasmid for 48 h, respectively. The histograms illustrate quantitative analysis of the E-cadherin, vimentin, and TWIST protein levels, which were normalized to the β-actin levels. *P < 0.05 vs. the pcDNA3.1-vector group. (C and D) Silenced FAT1 promotes the EMT of HeLa and C33A cells. The expression of E-cadherin, vimentin, and TWIST in HeLa (A) and C33A cells (B) were detected by western blot after these cells were treated with NC or FAT1 siRNA for 48 h, respectively. The histograms illustrate the quantitative analysis of the E-cadherin, vimentin, and TWIST protein levels, which were normalized to β-actin levels. *P < 0.05 vs. the NC group. (E and F) The effect of FAT1 on HeLa and C33A cell migration (E) and invasion (F) capacity was analyzed using a transwell assay. These cells were transfected with the pcDNA3.1-FAT1/pcDNA3.1-vector plasmid or NC/FAT1 siRNA for 24 h, respectively.
We subsequently assessed the influence of FAT1 on CC cell migration and invasion using a classic transwell assay. The results showed that the overexpression of FAT1 significantly suppressed the migration and invasion of HeLa and C33A cells (Figure 2E and 2F). FAT1 knockdown promoted cell migration and invasion in HeLa and C33A cells, respectively, compared with the control cells (Figure 2E and 2F). Thus, FAT1 inhibited the EMT, migration and invasion of CC cells in vitro.
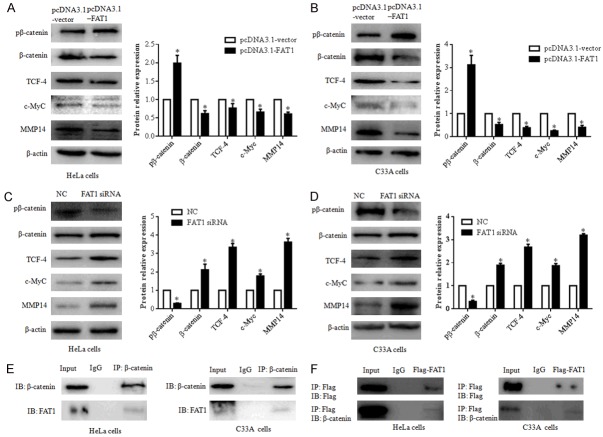
FAT1 suppresses the β-catenin mediated signalling pathway
To explore the effect of FAT1 on the β-catenin signalling pathway, we evaluated the expression of phospho-β-catenin (pβ-catenin), β-catenin, and its downstream targets TCF-4, c-Myc, and MMP14. A western blot showed that the p-β-catenin protein level was significantly increased and the β-catenin, TCF-4, c-Myc and MMP14 protein levels were decreased in the pcDNA3.1-FAT1 group compared with the pcDNA3.1-vector group (P < 0.05) (Figure 3A and 3B). Conversely, in FAT1 siRNA-transfected cells, the pβ-catenin protein level was significantly downregulated compared with that in the NC group, whereas β-catenin was upregulated. The downstream targets of β-catenin, including TCF-4, c-Myc, and MMP-14, were significantly upregulated (Figure 3C and 3D).
Figure 3.
FAT1 interacts with β-catenin and regulates β-catenin-mediated transcription. (A and B) Overexpression of FAT1 inhibited β-catenin-mediated transcription by phospho-β-catenin (pβ-catenin). pβ-catenin, β-catenin, TCF-4, c-Myc and MMP14 expression levels were evaluated by western blot after transfecting HeLa (A) and C33A (B) cells with the pcDNA3.1-FAT1 plasmid and pcDNA3.1-vector for 48 h, respectively. The histograms illustrate the quantitative analysis of the pβ-catenin, β-catenin, TCF-4, c-Myc, and MMP14 protein levels, which were normalized to the β-actin levels. *P < 0.05, vs. the pcDNA3.1-vector group. (C and D) FAT1 knockdown promoted β-catenin-mediated transcription of HeLa and C33A. pβ-catenin, β-catenin, TCF-4, c-Myc and MMP14 expression levels were evaluated by western blot after transfecting HeLa (C) and C33A (D) cells with NC and FAT1 siRNA for 48 h, respectively. The histograms illustrate the quantitative analysis of the pβ-catenin, β-catenin, TCF-4, c-Myc and MMP14 protein levels, which were normalized to the β-actin levels. *P < 0.05 vs. the NC group. (E) Immunoprecipitation assays detected that endogenous FAT1 bound endogenous β-catenin in HeLa and C33A cells. (F) Immunoprecipitation assays detected that exogenous FAT1-flag bound endogenous β-catenin in HeLa and C33A cells. HeLa and C33A cells were infected with Adv-flag-FAT1 for 48 h. Immunoprecipitation (IP) antibody or immunoglobulin G (IgG) negative controls were used as indicated. β-actin was used as a loading control.
To further investigate β-catenin regulation by FAT1, we performed a co-immunoprecipitation assay with HeLa and C33A cells and found that endogenous FAT1 interacted with β-catenin. Furthermore, we transfected FAT1 plasmids into both CC cell types and found a similar interaction between FAT1 and β-catenin (Figure 3E and 3F).
Overexpression of β-catenin attenuates the effect of FAT1 on CC cells
The abovementioned data showed that the protein expression levels of β-catenin, c-Myc, TCF-4, MMP-14, TWIST, and vimentin were higher in the pcDNA3.1-β-catenin/Adv.Flag-FAT1 group than in the vector/Adv.Flag-FAT1 group (P < 0.05). Furthermore, the E-cadherin protein level was decreased in the pcDNA3.1-β-catenin/Adv.Flag-FAT1 group compared with that in the vector/Adv.Flag-FAT1 group (P < 0.05) (Figure 4A and 4B). The viability of HeLa and C33A cells was also analyzed by CCK-8 assay (Figure 4C), and the data showed that cell viability at 24 and 48 h was significantly increased in the β-catenin group compared with the vector group (P < 0.05). Compared with the vector/Adv.Flag-FAT1 group, cell viability was increased at 12, 24, and 48 h in the pcDNA3.1-β-catenin/Adv.Flag-FAT1 group (P < 0.05) (Figure 4C). Additionally, we analyzed the migration and invasion of HeLa and C33A cells after pretreatment with a β-catenin plasmid with or without Adv.Flag-FAT1 by transwell assay (Figure 4D and 4E). The results demonstrated that HeLa and C33A cell migration and invasion were significantly increased in the pcDNA3.1-β-catenin group compared with those in the vector group. Moreover, cell migration and invasion were restored in the pcDNA3.1-β-catenin/Adv.Flag-FAT1 group compared with the vector/Adv.Flag-FAT1 group. These data suggested that β-catenin upregulation could partially reverse the FAT1-mediated inhibitory effect on the proliferation, migration, and invasion of HeLa and C33A cells.
Figure 4.
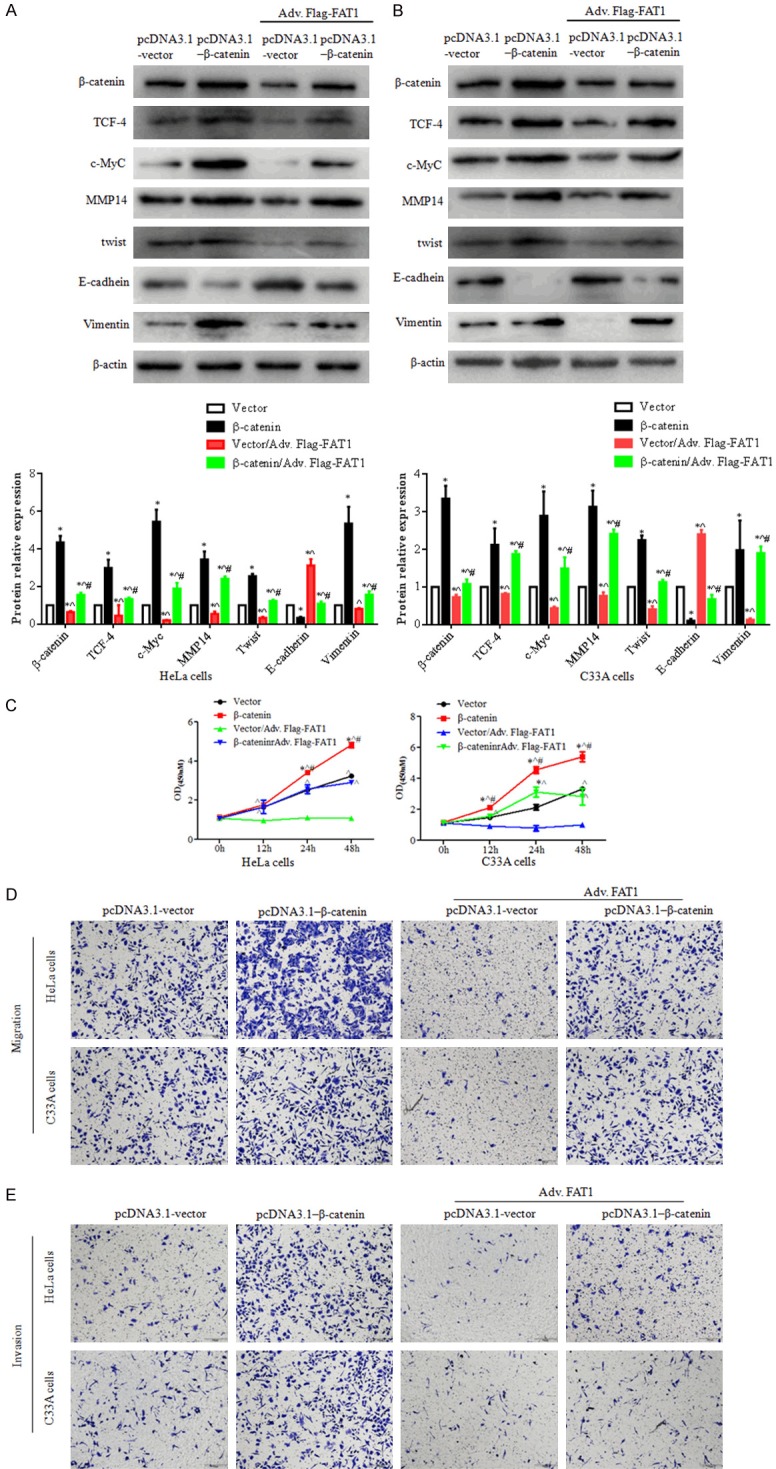
Upregulation of β-catenin partly reversed FAT1’s effect on proliferation, migration and invasion. HeLa and C33A cells were transfected with the vector and β-catenin plasmid for 12 h, then infected with adv. Flag-FAT1. (A and B) Western blotting showed the expression levels of β-catenin, TCF-4, c-Myc, MMP14, TWIST, E-cadherin, and vimentin in HeLa and C33A cells. The histograms illustrate the quantitative analysis of the pβ-catenin, β-catenin, TCF-4, c-Myc and MMP14 protein levels, which were normalized to the β-actin levels. *P < 0.05, vs. the pcDNA3.1-vector group. ^P < 0.05, vs. the β-catenin group. #P < 0.05, vs. the Vector/Adv.Flag-FAT1 group. (C) CCK-8 assays were used to analyze HeLa (left panel) and C33A (right panel) cell viability. *P < 0.05 vs. the vector group. ^P < 0.05 vs. the vector/adv. Flag-FAT1 group. #P < 0.05 vs. the β-catenin/adv. Flag-FAT1 group. (D and E) A transwell assay was used to evaluate HeLa and C33A cell migration (D) and invasion (E) capacity with co-expression of β-catenin and FAT1. Treated cells were seeded in upper chamber for 24 h, respectively.
Discussion
We found that FAT1 mRNA expression is downregulated in CC tissues, and the upregulation of FAT1 suppressed proliferation, EMT, migration, and invasion in HeLa and C33A cells. In contrast, the silencing of FAT1 promoted cell proliferation, EMT, and migration. Further study found that FAT1 could interact with β-catenin, promoting the phosphorylation and degradation of β-catenin, and thereby inactivate its downstream targets, namely, c-MYC, TCF4 and MMP14. These data hinted that FAT1 could regulate cellular proliferation and metastasis through the phosphorylation and degradation of β-catenin.
FAT1 is regarded as a key regulator of cell proliferation, migration, and polarity and plays critical roles in tumor progression. Due to its regulation of the Salvador/Warts/Hippo signalling pathway, FAT1 was first thought to have tumor suppressor function in Drosophila melanogaster [21]. Morris et al. identified a FAT1 mutation in glioblastoma (20.5%), colorectal cancer (7.7%), and head and neck cancer (6.7%), and this mutation resulted in FAT1 inactivation and loss of function and thus served as a tumor suppressor [18]. Martin’s research demonstrated that inactivating mutations of FAT1 resulted in YAP1 activation by the Hippo signalling pathway, leading to the progression of head and neck squamous cell carcinoma [17]. Another study showed that FAT1 is downregulated and considered a tumor suppressor in oral cancer, glioblastoma multiforme, and invasive breast cancer [13 18]. These research data hinted that FAT1 abolishes its tumour suppressor function in several cancers by downregulating its expression or by inducing an inactivating mutation. In our study, we found that the expression of FAT1 mRNA was lower in CC tissues than in matched adjacent non-tumor tissues. Moreover, our previous study also found that a FAT1 mutation was present in 8.8% of CC tissues. The forced expression of FAT1 through a FAT1 plasmid suppressed the proliferation of CC cells, whereas the knockdown of FAT1 promoted the proliferation of these cells. These data suggested that FAT1 is a tumor suppressor in CC.
CC metastasis is one of major causes of patient death. It is closely related to EMT, which is characterized by the loss of epithelial features and the gain of mesenchymal and migratory features [5]. Hu et al. showed that FAT1 suppresses the EMT, cell migration, and invasion in ESCC by disruption of the MAPK/ERK pathway [11,12]. Srivastava and colleagues showed that FAT1 regulates the EMT of glioblastoma both through or independent of HIF-1α [25]. Another study revealed that the knockdown of FAT1 using siRNA inhibited cell migration and invasion in oral squamous cell by disorganized localization of β-catenin [20]. Our data showed that FAT1 overexpression significantly elevated the expression of E-cadherin, suppressed the level of vimentin and Twist, and suppressed the migration and invasion of CC cells, indicating that FAT1 plays an important role in EMT, migration, and invasion in CC.
Aberrant activation of the Wnt/β-catenin pathway contributes to CC carcinogenesis and progression [3,8]. β-catenin, the central factor in canonical Wnt signalling, promotes the transcription of various genes, such as c-Myc, TCF-1, MMP14 and Cyclin D1, and thus controls the proliferation, migration, and invasion of CC when located in the nucleus [2]. A previous study showed that the FAT1 intracellular domain (Fat1IC) could interact with β-catenin to prevent the nuclear localization of β-catenin and inhibit its transcriptional activity, resulting in decreased VSMC proliferation and vessel remodelling [10]. Further study revealed that mutated FAT1 is unable to bind β-catenin, whereas upregulated wild-type FAT1 could interact with β-catenin and subsequently suppress the β-catenin-mediated signal pathway [18]. In the present study, we demonstrated that FAT1 could bind to β-catenin, promote its phosphorylation, and thus inactivate its downstream targets c-Myc, TCF4 and MMP14. Conversely, the silencing of FAT1 elevated the levels of total β-catenin and its target genes. Moreover, β-catenin upregulation partly reversed the FAT1-mediated inhibitory effect on proliferation, migration/invasion, and EMT progression in vitro. These results indicated that FAT1 interacts with β-catenin, induces the phosphorylation of β-catenin in the cytoplasm, and thereby prevents β-catenin nuclear localization and inactivates its transcription [18].
Conclusions
In conclusion, FAT1 suppresses CC cell proliferation, migration, and invasion by promoting β-catenin phosphorylation and inactivating Wnt/β-catenin-mediated transcription. These findings suggest that FAT1 might act as a tumor suppressor in CC and is a potential target in new molecular therapeutic strategies to improve CC prognosis.
Acknowledgements
This work was supported by the National Natural Youthful Science Foundation of China (No. 81602287). We thank Prof. Zhiqing Liang and Dr. Xiaolong Liang for their advice.
Disclosure of conflict of interest
None.
References
- 1.Alamoud KA, Kukuruzinska MA. Emerging insights into Wnt/β-catenin signaling in head and neck cancer. J Dent Res. 2018;97:665–673. doi: 10.1177/0022034518771923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asem MS, Buechler S, Wates RB, Miller DL, Stack MS. Wnt5a signaling in cancer. Cancers. 2016;8:79. doi: 10.3390/cancers8090079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahrami A, Hasanzadeh M, ShahidSales S, Yousefi Z, Kadkhodayan S, Farazestanian M, Joudi Mashhad M, Gharib M, Mahdi Hassanian S, Avan A. Clinical significance and prognosis value of Wnt signaling pathway in cervical cancer. J Cell Biochem. 2017;118:3028–3033. doi: 10.1002/jcb.25992. [DOI] [PubMed] [Google Scholar]
- 4.Benedetti Panici P, Basile S, Angioli R. Pelvic and aortic lymphadenectomy in cervical cancer: the standardization of surgical procedure and its clinical impact. Gynecol Oncol. 2009;113:284–90. doi: 10.1016/j.ygyno.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Borcherding N, Kusner D, Kolb R, Xie Q, Li W, Yuan F, Velez G, Askeland R, Weigel RJ, Zhang W. Paracrine WNT5A signaling inhibits expansion of tumor-initiating cells. Cancer Res. 2015;75:1972–82. doi: 10.1158/0008-5472.CAN-14-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chosdol K, Misra A, Puri S, Srivastava T, Chattopadhyay P, Sarkar C, Mahapatra AK, Sinha S. Frequent loss of heterozygosity and altered expression of the candidate tumor suppressor gene ‘FAT’ in human astrocytic tumors. BMC Cancer. 2009;9:5. doi: 10.1186/1471-2407-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bock CE, Ardjmand A, Molloy TJ, Bone SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J, Spanevello MD, Nelmes G, Catchpoole DR, Lincz LF, Boyd AW, Burns GF, Thorne RF. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26:918–26. doi: 10.1038/leu.2011.319. [DOI] [PubMed] [Google Scholar]
- 8.Ford CE, Henry C, Llamosas E, Djordjevic A, Hacker N. Wnt signalling in gynaecological cancers: a future target for personalised medicine? Gynecol Oncol. 2016;140:345–51. doi: 10.1016/j.ygyno.2015.09.085. [DOI] [PubMed] [Google Scholar]
- 9.Hayes TF, Benaich N, Goldie SJ, Sipilä K, Ames-Draycott A, Cai W, Yin G, Watt FM. Integrative genomic and functional analysis of human oral squamous cell carcinoma cell lines reveals synergistic effects of FAT1 and CASP8 inactivation. Cancer Lett. 2016;383:106–114. doi: 10.1016/j.canlet.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE. The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J Cell Biol. 2006;173:417–29. doi: 10.1083/jcb.200508121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang J, Qian Y, Ma Y, Wang F, Li H, Cheng C, Zhang L, Jia Z, Li Y, Yang B, Xu E, Wang J, Yang J, Bi Y, Chang L, Wang Y, Zhang Y, Song B, Li G, Shi R, Liu J, Zhang M, Cheng X, Cui Y. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93. doi: 10.1016/j.canlet.2017.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Zhai Y, Shi R, Qian Y, Cui H, Yang J, Bi Y, Yan T, Yang J, Ma Y, Zhang L, Liu Y, Li G, Zhang M, Cui Y, Kong P, Cheng X. FAT1 inhibits cell migration and invasion by affecting cellular mechanical properties in esophageal squamous cell carcinoma. Oncol Rep. 2018;39:2136–2146. doi: 10.3892/or.2018.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18:523–8. [PubMed] [Google Scholar]
- 14.Li H, He B, Liu X, Li J, Liu Q, Dong W, Xu Z, Qian G, Zuo H, Hu C, Qian H, Mao C, Wang G. Regulation on toll-like receptor 4 and cell barrier function by Rab26 siRNA-loaded DNA nanovector in pulmonary microvascular endothelial cells. Theranostics. 2017;7:2537–2554. doi: 10.7150/thno.17584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Yang WT, Zheng PS, Liu XF. SOX17 restrains proliferation and tumor formation by down-regulating activity of the Wnt/β-catenin signaling pathway via trans-suppressing β-catenin in cervical cancer. Cell Death Dis. 2018;9:741. doi: 10.1038/s41419-018-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal V, Moroishi T, Thorne RF, Zaida M, Siegele B, Cheong SC, Molinolo AA, Samuels Y, Tamayo P, Guan KL, Lippman SM, Lyons JG, Gutkind JS. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat Commun. 2018;9:2372. doi: 10.1038/s41467-018-04590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly I, Liau L, Cloughesy TC, Mischel PS, Mellinghoff IK, Chan TA. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–61. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakaya K, Yamagata HD, Arita N, Nakashiro KI, Nose M, Miki T, Hamakawa H. Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene. 2007;26:5300–8. doi: 10.1038/sj.onc.1210330. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa Y, Miyazaki T, Nakashiro K, Yamagata H, Isokane M, Goda H, Tanaka H, Oka R, Hamakawa H. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of β-catenin. Oncol Rep. 2011;26:587–92. doi: 10.3892/or.2011.1324. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran I, Thavathiru E, Ramalingam S, Natarajan G, Mills WK, Benbrook DM, Zuna R, Lightfoot S, Reis A, Anant S, Queimado L. Wnt inhibitory factor 1 induces apoptosis and inhibits cervical cancer growth, invasion and angiogenesis in vivo. Oncogene. 2012;31:2725–37. doi: 10.1038/onc.2011.455. [DOI] [PubMed] [Google Scholar]
- 22.Settakorn J, Kaewpila N, Burns GF, Leong AS. FAT, E-cadherin, beta catenin, HER 2/neu, Ki67 immuno-expression, and histological grade in intrahepatic cholangiocarcinoma. J Clin Pathol. 2005;58:1249–54. doi: 10.1136/jcp.2005.026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinohara A, Yokoyama Y, Wan X, Takahashi Y, Mori Y, Takami T, Shimokawa K, Tamaya T. Cytoplasmic/nuclear expression without mutation of exon 3 of the beta-catenin gene is frequent in the development of the neoplasm of the uterine cervix. Gynecol Oncol. 2001;82:450–5. doi: 10.1006/gyno.2001.6298. [DOI] [PubMed] [Google Scholar]
- 24.Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, Schäfer T, Ramachandran H, Mlodzik M, Huber TB, Kuehn EW, Kim E, Kramer-Zucker A, Walz G. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A. 2009;106:8579–84. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava C, Irshad K, Dikshit B, Chattopadhyay P, Sarkar C, Gupta DK, Sinha S, Chosdol K. FAT1 modulates EMT and stemness genes expression in hypoxic glioblastoma. Int J Cancer. 2018;142:805–812. doi: 10.1002/ijc.31092. [DOI] [PubMed] [Google Scholar]
- 26.Tanoue T, Takeichi M. New insights into fat cadherins. J Cell Sci. 2005;118:2347–53. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- 27.Wang CH, Li YH, Tian HL, Bao XX, Wang ZM. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3002–3009. doi: 10.26355/eurrev_201805_15057. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Qin Q, Song R, Zhao C, Liu H, Yang Y, Gu S, Zhou D, He J. NHERF1 inhibits beta-catenin-mediated proliferation of cervical cancer cells through suppression of alpha-actinin-4 expression. Cell Death Dis. 2018;9:668. doi: 10.1038/s41419-018-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Fang J, Yang Z, Chen F, Liu J, Wang Y. Wnt inhibitory factor-1 regulates glioblastoma cell cycle and proliferation. J Clin Neurosci. 2012;19:1428–32. doi: 10.1016/j.jocn.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Wang M, Li X, Xie Y, Xia X, Tian J, Zhang K, Tang A. Wnt signaling in cervical cancer. J Cancer. 2018;9:1277–1286. doi: 10.7150/jca.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao H, Ashihara E, Maekawa T. Targeting the Wnt/β-catenin signaling pathway in human cancers. Expert Opin Ther Targets. 2011;15:873–87. doi: 10.1517/14728222.2011.577418. [DOI] [PubMed] [Google Scholar]
- 32.Yessaian A, Magistris A, Burger RA, Monk BJ. Radical hysterectomyfollowed by tailored postoperative therapy in the treatment of stage IB2 cervical cancer: feasibility and indications for adjuvant therapy. Gynecol Oncol. 2004;94:61–6. doi: 10.1016/j.ygyno.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 33.Yu MH, Luo Y, Qin SL, Wang ZS, Mu YF, Zhong M. Up-regulated CKS2 promotes tumor progression and predicts a poor prognosis in human colorectal cancer. Am J Cancer Res. 2015;5:2708–18. [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu B, Zhao Q, Hou T, Huang X. Nuclear localizaiton of β-catenin is associated with poor survival and chemo-/radioresistance in human cervical squamous cell cancer. Int J Clin Exp Pathol. 2014;7:3908–17. [PMC free article] [PubMed] [Google Scholar]