Abstract
This study aimed to investigate the correlation between serum microRNA levels and histological stages of liver fibrosis in patients with chronic hepatitis B (CHB). A total of 28 patients with CHB who received liver biopsy at China Medical University Hospital between October 2012 and April 2013 were included in the study. The patients were divided into four groups according to the histological stages of liver fibrosis by using the METAVIR score. Serum microRNA levels were tested using quantitative real-time PCR after microRNA extraction from patients’ serum. Of all the tested microRNAs, miR-21, miR-29, and miR-221 were expressed in the serum. The expression levels of serum miR-21 were significantly correlated with liver fibrosis stages (r = 0.420, P = 0.026). The expression levels of serum miR-21 were significantly correlated with cirrhosis (METAVIR F4 vs. F1-F3, r = 0.386, P = 0.043). The grades of serum miR-21 showed significant ordered differences among different stages of liver fibrosis (P = 0.019). However, miR-21 exhibited an inferior predictive performance for liver fibrosis F2-F4 (AUROC = 0.69) compared with other noninvasive markers of liver fibrosis, namely the aspartate aminotransferase (AST) to platelet ratio index (APRI) and Fibrosis-4 (FIB-4) score (AUROC = 0.83 and 0.86, respectively). Serum miR-21 correlated with the histological stage of liver fibrosis in patients with CHB. The predictive performance of serum miR-21 for the histological stage of liver fibrosis tended to be inferior to those of the APRI and FIB-4 score.
Keywords: miR-21, liver fibrosis, APRI, FIB-4, chronic hepatitis B
Introduction
Liver fibrosis is a tissue-healing process that occurs to maintain hemostasis in response to chronic liver injury [1-4]. Liver injuries initiate recruitment of inflammatory macrophages into the liver. Chronic activation of inflammatory macrophages induces activation of quiescent hepatic stellate cells (HSCs) into activated HSCs, the myofibroblasts. Myofibroblasts produce the extracellular matrix (ECM), and deposition of the ECM promotes liver scarring [5]. Advanced liver fibrosis leads to liver cirrhosis, which increases the risk of hepatocellular carcinoma and liver failure and requires liver transplantation.
The main causes of liver fibrosis and cirrhosis in developed countries include chronic infection with hepatitis C virus, habitual alcohol consumption, and nonalcoholic fatty liver disease. Hepatitis B infection is endemic and remains a crucial public health problem in the Asia-Pacific region and sub-Saharan Africa [6]. Hepatitis B e antigen (HBeAg)-positive chronic hepatitis B (CHB) leads to cirrhosis at an incidence rate of 1.6%-3.8% per 100 person years, and the cirrhosis incidence rate is 2.8%-9.7% per 100 person years in patients with HBeAg-negative CHB [7]. Hepatitis B-related cirrhosis was traditionally believed to be irreversible. With the advent of the nucleos(t)ide analog (NA) therapy, reversal of liver fibrosis and cirrhosis has been observed in large proportions of CHB patients [8,9]. Nonetheless, patients receiving antiviral therapy are still at a risk of hepatocellular carcinoma and cirrhosis-associated complications, and the risk is associated with the severity of liver fibrosis [10]. Therefore, identifying diagnostic and therapeutic biological targets for liver fibrosis is crucial.
Assessment of liver fibrosis is important because the degree of liver fibrosis is associated with the risk of complications caused by liver fibrosis and cirrhosis, such as hepatic encephalopathy and esophageal variceal bleeding. Notably, reversal of liver fibrosis after antiviral therapy has been observed among patients with CHB [8,9] and chronic hepatitis C (CHC)-related liver fibrosis [11] and cirrhosis and among cirrhotic patients with alcohol abstinence [12]. The gold-standard diagnosis method for liver fibrosis is liver biopsy. Common histological scores include the METAVIR [13,14] and Ishak scores [15]. Liver biopsy is an invasive procedure with a potential risk of complications to patients, especially in cirrhotic patients with thrombocytopenia [16]. Therefore, noninvasive assessment methods of liver fibrosis have been developed and extensively used in the past 20 years. Noninvasive imaging modalities, such as transient elastography (FibroScan®) [17] and acoustic radiation force impulse elastography [18], are commonly used in clinical practice.
Indirect serum markers of liver fibrosis are collection indexes, including markers that reflect liver inflammation and liver function [19-21], such as serum aminotransferase, γ-glutamyl transferase (γ-GT), platelet count, prothrombin time, apolipoprotein A, α2 macroglobulin, α2 globulin, and γ globulin [20]. The aspartate aminotransferase (AST) to platelet ratio index (APRI) and Fibrosis-4 (FIB-4) index are inexpensive, noninvasive tests for assessing liver fibrosis in low- and middle-income countries, as recommended by the recent WHO guideline [22]. APRI and FIB-4 showed high specificity but low sensitivity for hepatitis B-related advanced liver fibrosis and cirrhosis [23,24]. These noninvasive tests are not capable of assessing all stages of liver fibrosis; therefore, diagnosis of advanced liver fibrosis and cirrhosis may be missed by using these noninvasive tests only due to their low sensitivity [22].
Direct serum markers of liver fibrosis are under investigation and offer another method to assess liver fibrosis. Liver fibrogenesis occurs through the activation of HSCs and several inflammatory pathways to produce the ECM [25]. Direct serum markers reflect the dynamics of deposition and removal of the ECM during fibrogenesis [26]. Markers associated with ECM deposition include procollagen type I carboxy terminal peptide (PICP), procollagen type III amino-terminal peptide (PIIINP), and tissue inhibitor of metalloproteinases (TIMPs). Markers associated with ECM degradation include the procollagen IV C peptide (PIVCP), procollagen IV N peptide (PIVNP), type IV collagen, and metalloproteinases (MMPs) [26-29]. Other direct markers include hyaluronic acid (HA), YKL-40 (chitinase-3-like-1 or human cartilage glycoprotein-39), and cytokines that are involved in ECM metabolism, such as transforming growth factor β (TGF-β) and platelet-derived growth factor (PDGF) [27]. PICP and PIIINP are released through proteolytic cleavage of procollagen (a precursor of collagen) into the serum during ECM deposition and remodeling. PICP reflects the severity of alcoholic cirrhosis more accurately than chronic liver disease related to other etiologies [27]. PIIINP predicts both liver fibrosis and liver inflammation. PIIINP exhibits more accurate prediction of liver inflammation than liver fibrosis [27]. Propeptides of type IV procollagen, PIVCP and PIVNP, enhance the basement membrane turnover in active fibrosis [30]. Type IV collagen reflects fibrolysis and remodeling of the interstitial connective tissue [30]. HA is an essential component of the ECM produced by activated HSCs [31]. HA exhibits more accurate predictive performance for liver fibrosis than PIIINP [32]. MMPs and their inhibitors, TIMPs, together play a role in ECM metabolism. MMP-2 is expressed by activated HSCs during liver injuries. MMP-2 induces ECM degradation, and TIMPs inhibit MMP-2-induced ECM degradation [33]. YKL-40 is a chitinase-like protein expressed in multiple tissues including the liver. It functions as a growth factor for fibroblasts and epithelial cells [26]. Moreover, YKL-40 is involved in ECM remodeling. Serum YKL-40 is associated with the liver fibrosis stage in CHB [34]. TGF-β is released from necrotic hepatocytes during chronic liver injury [35]. TGF-β1 activates HSCs, inhibits HSC apoptosis, suppresses MMPs, and upregulates TIMP, and thereby induces ECM production and prevents ECM degradation [35].
HSC activation is pivotal in liver fibrogenesis. It requires epigenetic remodeling of the de novo expression of regulators of the myofibroblast phenotype [36,37]. Mechanisms of epigenetic modulation and post-transcriptional gene regulation of HSCs include DNA methylation, histone modification, and alteration of mRNA degradation and translation repression by microRNAs [38]. MicroRNAs are small noncoding RNA molecules that post-transcriptionally affect mRNA stability and translation by targeting the 3’-untranslated region (3’-UTR) of various transcripts. Dysregulation of microRNAs epigenetically affects some liver fibrogenesis pathways [37]. miR-21 is upregulated in many cancer types [39]. miR-21 is also enhanced after initiation of fibrosis of the heart, kidney, and lung [40,41]. miR-21 acts as a profibrogenic microRNA, activates HSCs through PTEN/Akt signaling [42], and represses the TGF-β inhibitory Smad 7 protein [43]. miR-21 upregulation during liver fibrogenesis is based on TGF-β-induced Smad-2 and Smad-3 phosphorylation. Phosphorylated Smad-3 aggregates with Smad-4 and transcriptionally induces pri-miR-21 synthesis [38,44]. TGF-β also promotes the expression of mature miR-21 by the DROSHA complex. After TGF-β exposure, Smad-2 and Smad-3 interact with RNA helicase p68, a component of the DROSHA microprocessor complex, and further promote the formation of pre-miR-21 [45]. miR-29 is highly expressed in HSCs. miR-29 mediates the regulation of liver fibrosis and is part of a signaling nexus involving TGF-β- and NF-κB-dependent downregulation of miR-29 family members in HSCs with subsequent upregulation of extracellular matrix genes [46]. miR-29 mediates the repression of ECM and growth factors such as PDGF and insulin-like growth factor-1 (IGF-I). Stimulation of TGF-β and PDGF results in miR-29 repression and upregulation of profibrogenic expression of the ECM, PDGF-C, or IGF-I. PDGF and IGF-I stimulate stellate cell proliferation and ECM production [37]. miR-221/222 upregulation correlates with the mRNA expression levels of α1 (I) collagen and α-smooth muscle actin and activation of HSCs [47].
To identify serum microRNAs as the potential biomarkers of liver fibrosis, this study investigated the correlation of serum profibrogenic and antifibrogenic microRNA levels with the histological stages of liver fibrosis in patients with CHB.
Materials and methods
Patients
A total of 28 patients with CHB who received liver biopsy at China Medical University Hospital between October 2012 and April 2013 were included. All patients met the following inclusion criteria: age ≥ 18 years and ≤ 70 years and HBsAg positivity for more than 6 months. Patients were excluded if they had concurrent diagnosis of hepatitis C infection, alcoholic hepatitis, autoimmune hepatitis, Wilson’s disease, hemochromatosis, or hepatocellular carcinoma. All the patients underwent blood tests at the time of liver biopsy. Blood chemistry, complete blood count, HBV DNA, HBeAg, anti-HBe, and α-fetoprotein were measured. Another 10 mL of the blood sample was stored at -80°C until further study of microRNAs.
Liver biopsy and assessment of liver fibrosis
Liver biopsy was performed with real-time guidance of abdominal sonography by puncturing a biopsy needle into the right lobe of the liver through the right intercostal space. A liver tissue piece of length > 1.0 cm was sampled. Formalin-fixed liver tissues were then stained with hematoxylin and eosin and Masson’s trichrome. For histological assessment of liver fibrosis, the METAVIR scoring system was used.
Blood sampling
Serum RNA was extracted using the miRNeasy Serum/Plasma Kit (QIAGEN®, Hilden, Germany) following the manufacturer’s protocol. cDNA was obtained from reverse transcription of total serum RNA by using the miScript II RT Kit (QIAGEN®).
Real-time quantitative PCR analysis of serum microRNAs
The expression of serum microRNAs was quantified through real-time quantitative PCR by using the SYBR green method. Primer sequences of microRNAs are listed in Table S1. Cel-miR-39 was chosen for internal control by using miRNeasy Serum/Plasma Spike-In Control (QIAGEN®). Real-time quantitative PCRs were performed in triplicate in a LightCycler® 480 system (Roche Molecular Systems, Inc. Pleasanton, CA, USA).
Noninvasive fibrosis markers: APRI and FIB-4 score
The APRI was calculated using the patient’s AST level and platelet count. The formula for APRI calculation is as below [48] (Equation 1):
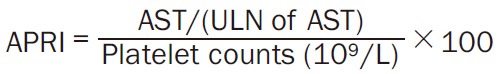 |
ULN of AST: 34 U/L.
The FIB-4 score was calculated using the patient’s AST level, ALT level, platelet count, and age. The formula for FIB-4 score calculation is as below [49] (Equation 2):
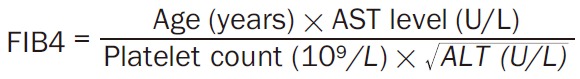 |
Statistical analysis
All variables were first tested for distribution by using normality tests. Continuous variables that fitted with normal distribution are presented as mean ± standard deviation (SD). Continuous variables that did not match normal distribution are presented as medians (25 percentiles, 75 percentiles). Categorical variables are presented as numbers (percentage). The correlation analysis was performed using Pearson’s correlation. To examine the difference in the relative quantifications of serum microRNA levels, we graded each level of microRNA according to its mean ± 1 SD value. Grade 1 was defined as the low microRNA level (0 to mean - 1 SD); Grade 2 was defined as the medium level (between mean - 1 SD and mean + 1 SD); and Grade 3 was defined as the high level (> mean + 1 SD). The Jonckheere-Terpstra trend test was then performed to examine the ordered difference between serum microRNA grades and stages of histological liver fibrosis. All the aforementioned tests were performed with IBM® SPSS® Statistics software, version 23, IBM Corp, Armonk, NY, USA. Receiver operating characteristic (ROC) curve analysis was performed using SigmaPlot software, version 14.0, Systat Software, Inc, San Jose, CA, USA. A p value of < 0.05 was considered significant.
Results
Patient demographics
The demographic data and clinical data of the 28 CHB patients with liver fibrosis are listed in Table 1. Seventeen (60.7%) patients had genotype B infection. Six (21.4%) patients were HBeAg-positive. Twenty-five (89.3%) patients exhibited fatty metamorphosis on histology. The characteristics of CHB patients stratified by liver fibrosis are presented in Table 2.
Table 1.
Baseline characteristics of 28 patients with CHB
| Age (year) | 50.9 ± 6.9 |
| Gender (Male, %) | 21 (75%) |
| BMI (kg/m2) | 24.7 ± 3.7 |
| AST (U/L) | 110.3 ± 223.6, 53 [33.3, 104] |
| ALT (U/L) | 148.5 ± 219.5, 63.5 [38.3, 164] |
| Total bilirubin (mg/dL) | 1.3 ± 1.1 |
| Albumin | 4.1 ± 0.47 |
| PT (INR) | 1.1 ± 0.1 |
| Platelet count (× 103/µL) | 162.3 ± 46.1 |
| AFP (ng/mL) | 14.6 ± 44.8, 4.0 [2.9, 7.5] |
| Genotype (B vs. C), % | 17 (60.7%) |
| Serum HBV DNA (IU/mL) | 2.1 × 1010 ± 1.1 × 1011, 2.5 × 105 [1.6 × 104, 4.27 × 107] |
| HBeAg (positive vs. negative), % | 6 (21.4%) |
| qHBsAg (IU/mL) | 705.6 [213.8, 1945.3] |
| METAVIR score (1 vs. 2 vs. 3 vs. 4) | 7 (25%), 7 (25%), 7 (25%), 7 (25%) |
| Fatty metamorphosis (0 vs. 1 vs. 2 vs. 3) | 3 (10.7%), 3 (0.7%), 21 (75%), 1 (3.6%) |
Median [first quartile, third quartile]. Number (percentage).
Table 2.
Characteristics of patients stratified by liver fibrosis
| Liver fibrosis METAVIR score | Patient Groups | P | |||
|---|---|---|---|---|---|
|
| |||||
| F1 (n = 7) | F2 (n = 7) | F3 (n = 7) | F4 (n = 7) | ||
| Age (year) | 47 ± 6.1 | 49 ± 6.1 | 55.6 ± 9.6 | 51.6 ± 1.9 | 0.266 |
| Gender (Male, %) | 6 (85.7) | 5 (71.4) | 4 (57.1) | 6 (85.7) | |
| BMI (kg/m2) | 21.7 ± 1.5 | 23.9 ± 2.8 | 26.5 ± 2.9 | 26.5 ± 4.9 | 0.041* |
| AST (U/L) | 52.3 ± 56.2 | 207.7 ± 200.4 | 53.7 ± 30.7 | 45 ± 15.6 | 0.016* |
| ALT (U/L) | 64.4 ± 57.4 | 325.6 ± 237.1 | 62.9 ± 36.4 | 45.6 ± 7.5 | 0.031* |
| Total bilirubin (mg/dL) | 0.9 ± 0.1 | 1.8 ± 2.2 | 1.0 ± 0.1 | 1.3 ± 0.4 | 0.372 |
| Albumin | 4.5 ± 0.3 | 3.8 ± 0.5 | 4.0 ± 0.3 | 3.9 ± 0.5 | 0.032* |
| PT (INR) | 1.0 ± 0.03 | 1.1 ± 0.1 | 1.1 ± 0.04 | 1.2 ± 0.1 | 0.010* |
| Platelet count (× 103/µL) | 196.3 ± 27.7 | 166.6 ± 25.8 | 165.3± 46.7 | 121 ± 50.7 | 0.031* |
| AFP (ng/mL) | 2.9 ± 0.6 | 8.7 ± 8.1 | 8.3 ± 4.4 | 37.8 ± 88 | 0.050 |
| Genotype (B vs. C), % | 6 (85.7) | 7 (100) | 7 (100) | 6 (85.7) | |
| HBV DNA (IU/mL) | 1.3 × 108 ± 3.2 × 108 | 8.1 × 1010 ± 2.1 × 1011 | 4.3 × 106 ± 9.1 × 106 | 2.2 × 105 ± 1.8 × 105 | 0.002* |
| HBeAg (positive vs. negative), % | 1 (14.3) | 2 (28.6) | 0 | 3 (42.9) | |
| qHBsAg (IU/mL) | 617 ± 519.6 | 1108.1 ± 1272.3 | 2532.4 ± 3616.0 | 697.5 ± 687.8 | 0.795 |
| Fatty metamorphosis (0 vs. 1 vs. 2 vs. 3) | 0:3:4:0 | 3:0:3:1 | 0:0:7:0 | 0:0:7:0 | |
P < 0.05.
Expression of serum miR-21, miR-29, and miR-221 in CHB patients with liver fibrosis
Of all the tested microRNAs, miR-21, miR-29, and miR-221 were expressed in the serum (Figure 1). By contrast, miR-122, miR-138-2, miR-140, miR-143, and miR-222 were not expressed in the serum of CHB patients with liver fibrosis. cel-miR-39 was expressed in all serum samples (data not shown).
Figure 1.
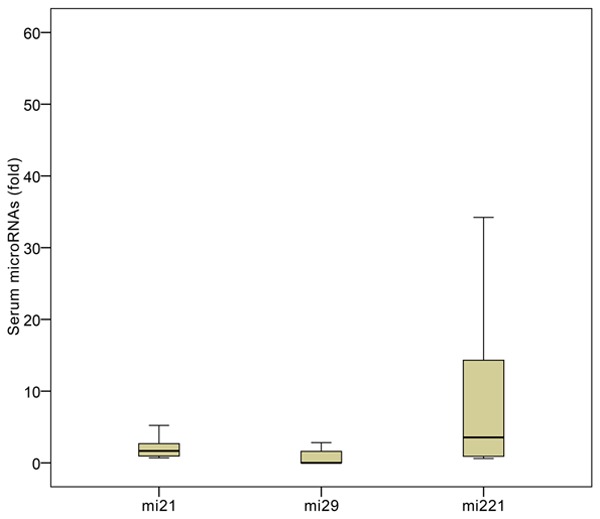
Expression of serum microRNAs in CHB patients with liver fibrosis.
The serum miR-21 level is correlated with the stage of liver fibrosis and cirrhosis
The expression levels of serum miR-21 were significantly correlated with the stage of liver fibrosis (P = 0.026). By contrast, miR-29 and miR-221 were not significantly correlated with the liver fibrosis stage (Table 3).
Table 3.
Correlation of serum miR-21, miR-29, and miR-221 levels with liver fibrosis stage
| MicroRNAs | Correlation coefficient, r | p value |
|---|---|---|
| miR-21 | 0.420 | 0.026* |
| miR-29 | -0.040 | 0.841 |
| miR-221 | 0.192 | 0.328 |
P < 0.05.
The expression levels of serum miR-21 were significantly correlated with cirrhosis (METAVIR F4 vs. F1-F3) (P = 0.043). Serum levels of miR-21 were not significantly correlated with significant fibrosis (METAVIR F2-4 vs. F0-F1) (P = 0.143) or severe fibrosis (METAVIR F3-F4 vs. F1-F2) (P = 0.061) (Table 4).
Table 4.
Correlation of serum miR-21 levels with the early and late stages of liver fibrosis
| Stage of fibrosis | Correlation coefficient, r | p value |
|---|---|---|
| Significant fibrosis (F2-4 vs. F0-F1) | 0.284 | 0.143 |
| Severe fibrosis (F3-F4 vs. F1-F2) | 0.358 | 0.061 |
| Cirrhosis (F4 vs. F1-F3) | 0.386 | 0.043* |
P < 0.05.
Grades of serum miR-21 show ordered differences among different stages of liver fibrosis
The serum levels of miR-21, miR-29, and miR-221 showed no ordered differences among different stages of liver fibrosis (Figure 2). The distribution of serum miR-21 grades in each stage of liver fibrosis is shown in Table 5. The grades of serum miR-21 showed significant ordered differences among different stages of liver fibrosis (P = 0.019) (Figure 3). The grades of miR-29 (P = 0.626) and miR-221 (P = 0.849) did not show significant differences among different stages of liver fibrosis (data not shown).
Figure 2.
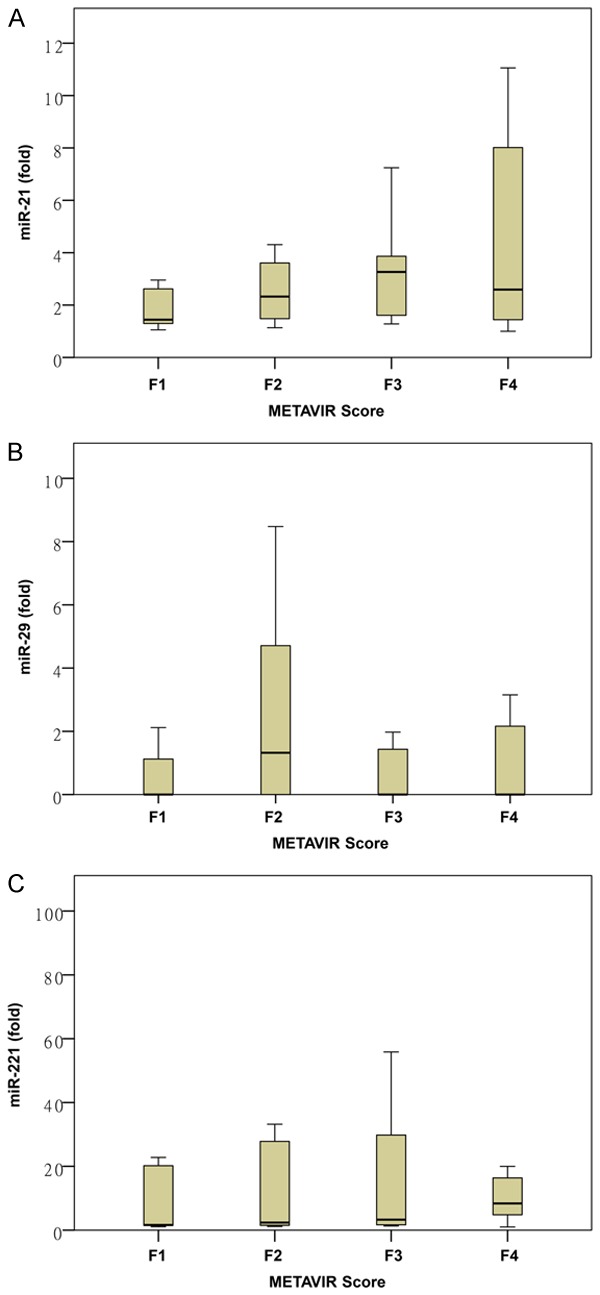
Expression of (A) serum miR-21, (B) miR-29, and (C) miR-221 among different stages of liver fibrosis: miR-21 (P = 0.120), miR-29 (P = 0.964), and miR-221 (P = 0.512) showed no ordered differences among different stages of liver fibrosis.
Table 5.
Grades of serum miR-21 exhibit significant correlation with the liver fibrosis stage
| METAVIR score | Grades of miR-21 | p value | ||
|---|---|---|---|---|
|
| ||||
| Low (n = 19) | Medium (n = 5) | High (n = 4) | ||
| F1 | 7 (36.8) | 0 | 0 | 0.019* |
| F2 | 5 (26.3) | 2 (40) | 0 | |
| F3 | 3 (15.8) | 3 (60) | 1 (25) | |
| F4 | 4 (21.1) | 0 | 3 (75) | |
Cutoffs for low, medium, and high serum miR-21 levels are defined by mean ± 1 standard deviation.
P < 0.05.
Figure 3.
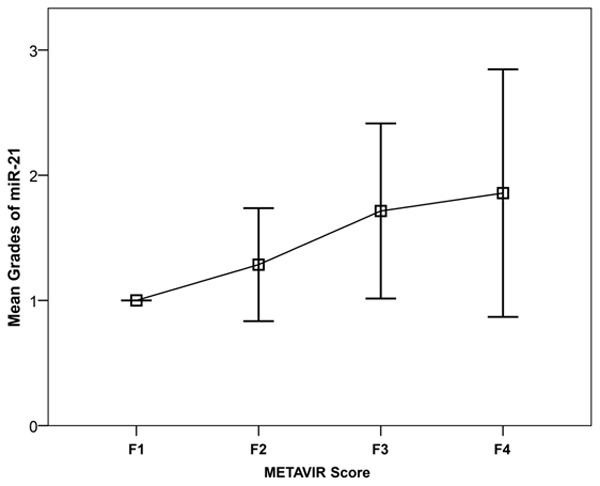
Correlation of miR-21 grades with the liver fibrosis stage.
Comparison of the predictive performances of miR-21, APRI, and FIB-4 for liver fibrosis stage
The APRI and FIB-4 score were not significantly correlated with the liver fibrosis stage (P = 0.818 and p = 0.106, respectively) (Table 6). The AUROC of miR-21 was lower than those of the APRI and FIB-4 score in predicting METAVIR F1 vs. METAVIR F2-F4, and the differences were not significant (miR-21 vs. APRI, P = 0.4413, miR-21 vs. FIB-4, P = 0.335). Similar results were noted in the prediction of F1-F2 vs. F3-F4 (miR-21 vs. APRI, P = 0.3641, miR-21 vs. FIB-4, P = 0.8835) and F1-F3 vs. F4 (miR-21 vs. APRI, P = 0.8871, miR-21 vs. FIB-4, P = 0.6757). The ROC curves are shown in Figure 4.
Table 6.
Correlation of APRI and FIB-4 with the liver fibrosis stage
| Correlation coefficient, r | p value | |
|---|---|---|
| APRI | -0.046 | 0.818 |
| FIB-4 | 0.312 | 0.106 |
Figure 4.
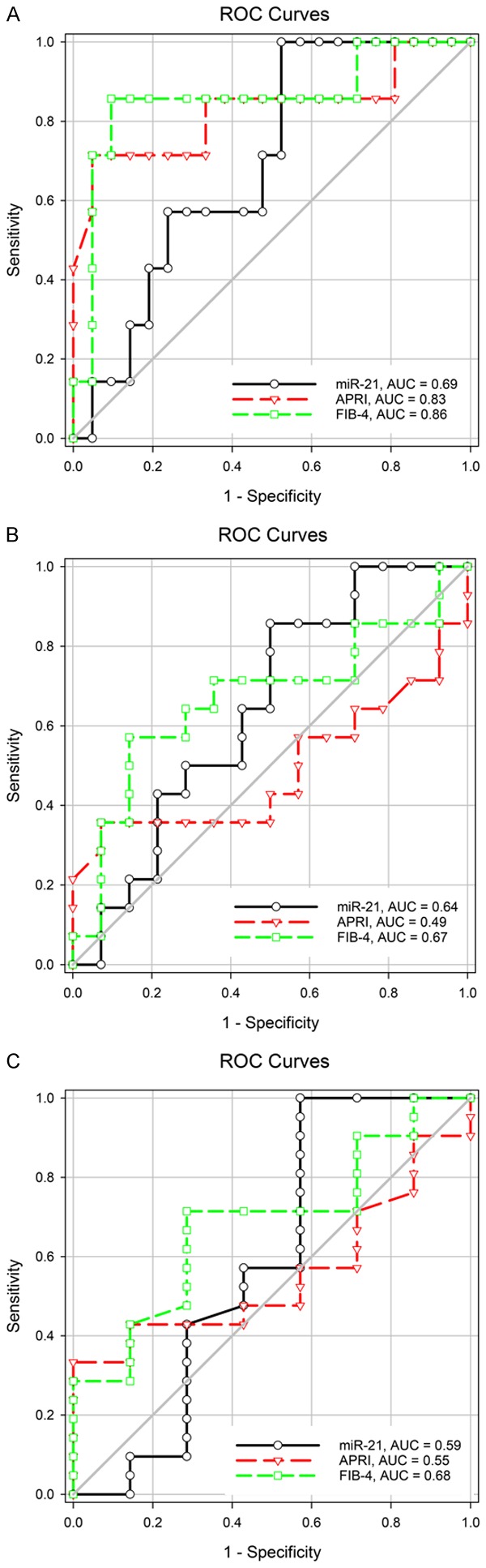
The predictive performance of miR-21 for the liver fibrosis stage was lower than that of APRI and FIB-4. A. METAVIR F1 vs. F2-F4. B. METAVIR F1-F2 vs. F3-F4. C. METAVIR F1-F3 vs. F4.
Discussion
According to our results, miR-21, miR-29, and miR-221 were expressed in the serum of CHB patients with liver fibrosis. Of these microRNAs, serum miR-21 was correlated with the histological stage of liver fibrosis. Studies have shown a correlation between miR-21 and liver fibrosis. miR-21 activates HSCs through PTEN/Akt signaling [42]. miR-21 promotes α-SMA and collagen I expression in HSCs through the Smad 7 signaling pathway [43]. miR-21 overexpression enhances TGF-β1-induced fibrogenic epithelial-to-mesenchymal transition by targeting RECK in hepatic oval cells [50]. microRNA-21 along with programmed cell death protein 4 and activation protein-1 is associated with the development of hepatic fibrosis [51]. Although a recent study showed that miR-21 and Dicer are responsible for HSC activation and the development of toxic liver fibrosis caused by CCl4 [52], our results showed a correlation of miR-21 with HBV-associated liver fibrosis. Different contexts of chronic liver injury may induce different fibrogenic pathways; therefore, an inconsistent correlation between microRNAs and the liver fibrosis stage may be observed.
In our study, the APRI and FIB-4 score were not significantly correlated with the liver fibrosis stage (P = 0.818 and P = 0.106, respectively). However, studies have demonstrated that both APRI and FIB-4 are reliable predictors of liver fibrosis [53,54]. We further compared the predictive performance of miR-21, APRI, and FIB-4 for liver fibrosis. We demonstrated that FIB-4 and APRI showed good predictive performance for liver fibrosis F2-F4 (both AUROCs > 0.8). miR-21 exhibited inferior predictive performance for liver fibrosis F2-F4 (AUROC = 0.69), with no significant differences among different stages of liver fibrosis. Although the predictive performance of miR-21 for cirrhosis was also inferior to that of FIB-4 (METAVIR F4 vs. F1-F3, AUROC = 0.59 vs. 0.68, respectively), miR-21 correlated with cirrhosis (METAVIR F4 vs. F1-F3, r = 0.386, P = 0.043). In our study, APRI exhibited more accurate predictive performance for significant fibrosis (METAVIR F2-F4 vs. F1, AUROC = 0.83) than cirrhosis (METAVIR F4 vs. F1-F3, AUROC = 0.55). This result was consistent with those of previous studies [23,54]. A meta-analysis on APRI in hepatitis B-related fibrosis and cirrhosis revealed AUROC = 0.79 for significant fibrosis and AUROC = 0.75 for cirrhosis [23]. In another meta-analysis, APRI exhibited AUROC = 0.7407 for significant fibrosis detection (METAVIR F2-F4 vs. F1), 0.7347 for advanced fibrosis detection (METAVIR F3-F4 vs. F1-F2), and 0.728 for cirrhosis detection (METAVIR F4 vs. F1-F3) [54]. Furthermore, our results were consistent with those of previous meta-analysis [54] and suggested that APRI had a lower accuracy for predicting HBV-related significant fibrosis, severe fibrosis, and cirrhosis compared with FIB-4 (AUROC = 0.83, 0.49, and 0.55 and AUROC = 0.86, 0.76, and 0.68, respectively).
In our study, the predictive performance of FIB-4 for cirrhosis (METAVIR F4 vs. F0-F3, AUROC = 0.68) was lower than that for significant fibrosis (METAVIR F2-F4 vs. F0-F1, AUROC = 0.86). This result differed from those of the previous studies [23,53,54]. In a previous meta-analysis, FIB-4 exhibited higher diagnostic accuracy in predicting hepatitis B-related liver cirrhosis (METAVIR F4 vs. F0-F3, AUROC = 0.89) than significant fibrosis (METAVIR F2-F4 vs. F0-F1, AUROC = 0.78) and severe fibrosis (METAVIR F3-F4 vs. F0-F2, AUROC = 0.79) [53]. Another recent meta-analysis also revealed that FIB-4 showed higher predictive performance for cirrhosis (METAVIR F4 vs. F1-F3, AUROC = 0.8448) than for advanced fibrosis (METAVIR F3-F4 vs. F1-F2, AUROC = 0.8165) and significant fibrosis (METAVIR F2-F4 vs. F1, AUROC = 0.7844) [54]. The small numbers of cases of patients with F3 and F4 might have accounted for the lower AUROCs of FIB-4 in our study.
The present study had some limitations. First, our study had a small sample size. Thus, the statistical power is insufficient to generate a conclusion for the general population. Nonetheless, our preliminary result indicated that three specific microRNAs were expressed in the serum of patients with HBV-related liver fibrosis. Further research should be conducted on these microRNAs to elucidate their roles in liver fibrogenesis in the context of liver tissues. Second, we were unable to determine the expression levels of microRNAs in the liver tissues due to the limited amount of available biopsy specimens. The relationships between these microRNAs in the serum and fibrotic liver tissues were not explored in this study. Some published reports have evidenced the expression of these microRNAs in the liver tissues, including the expression of miR-21 through miRNA array hybridization [55], miR-29 through microarray analysis [46], and miR-221 through microarray analysis [47]. A correlative study of microRNAs between these two sources is warranted to evaluate the feasibility of using serum microRNAs as surrogate biomarkers for liver fibrosis. Third, demographic parameters (BMI, AST, ALT, and HBV DNA level) were not matched in the four liver fibrosis groups. Studies have suggested that miR-21 is involved in nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH). Liver miR-21 is overexpressed in NASH in mouse models [56]. Serum levels of miR-21, miR-34a, miR-122, and miR-451 were higher in patients with NAFLD [57]. In addition, HBV virologic factors may influence the expression of miR-21. HBV X protein induced miR-21 expression in cell culture models [58,59]. NAFLD and HBV virologic factors may confound the correlation between microRNAs and the liver fibrosis stage [56-58].
In conclusion, serum miR-21 correlated with the histological stage of liver fibrosis in patients with CHB. The predictive value of serum miR-21 for the histological stage of liver fibrosis tended to be inferior to those of the APRI and FIB-4 scores.
Acknowledgements
This manuscript was edited by Wallace Academic Editing.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Su TH, Kao JH, Liu CJ. Molecular mechanism and treatment of viral hepatitis-related liver fibrosis. Int J Mol Sci. 2014;15:10578–10604. doi: 10.3390/ijms150610578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman SL. Liver fibrosis - from bench to bedside. J Hepatol. 2003;38(Suppl 1):S38–53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 4.Wynn T. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trautwein C, Friedman SL, Schuppan D, Pinzani M. Hepatic fibrosis: concept to treatment. J Hepatol. 2015;62:S15–S24. doi: 10.1016/j.jhep.2015.02.039. [DOI] [PubMed] [Google Scholar]
- 6.Tsochatzis EA, Bosch J, Burroughs AK. Liver cirrhosis. Lancet. 2014;383:1749–1761. doi: 10.1016/S0140-6736(14)60121-5. [DOI] [PubMed] [Google Scholar]
- 7.Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Chang TT, Liaw YF, Wu SS, Schiff E, Han KH, Lai CL, Safadi R, Lee SS, Halota W, Goodman Z, Chi YC, Zhang H, Hindes R, Iloeje U, Beebe S, Kreter B. Long-term entecavir therapy results in the reversal of fibrosis/cirrhosis and continued histological improvement in patients with chronic hepatitis B. Hepatology. 2010;52:886–893. doi: 10.1002/hep.23785. [DOI] [PubMed] [Google Scholar]
- 9.Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Aguilar Schall R, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet. 2013;381:468–475. doi: 10.1016/S0140-6736(12)61425-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang HW, Lai HC, Hu TH, Su WP, Lu SN, Lin CH, Hung CH, Chuang PH, Wang JH, Lee MH, Chen CH, Peng CY. Stratification of hepatocellular carcinoma risk through modified FIB-4 index in chronic hepatitis B patients on entecavir therapy: modified FIB-4 index and hepatocellular carcinoma. J Gastroenterol Hepatol. 2019;34:442–449. doi: 10.1111/jgh.14372. [DOI] [PubMed] [Google Scholar]
- 11.George SL, Bacon BR, Brunt EM, Mihindukulasuriya KL, Hoffmann J, Di Bisceglie AM. Clinical, virologic, histologic, and biochemical outcomes after successful HCV therapy: a 5-year follow-up of 150 patients. Hepatology. 2009;49:729–738. doi: 10.1002/hep.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi H, Shigefuku R, Maeyama S, Suzuki M. Cirrhosis improvement to alcoholic liver fibrosis after passive abstinence. BMJ Case Rep. 2014;2014:bcr2013201618. doi: 10.1136/bcr-2013-201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR cooperative study group. Hepatology. 1996;24:289–93. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 14.Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- 15.Ishak K, Baptista A, Bianchic L, Callead F, De Grootee J, Gudat F, Denkg H, Desmeth V, Korbi G, MacSweenj RNM, Phillipsk MJ, Portmann BG, Poulsen H, Scheuer PJ, Schmid M, Thaler H. PlumX Metrics histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 16.Seeff LB, Everson GT, Morgan TR, Curto TM, Lee WM, Ghany MG, Shiffman ML, Fontana RJ, Di Bisceglie AM, Bonkovsky HL, Dienstag JL HALT-C Trial Group. Complication rate of percutaneous liver biopsies among persons with advanced chronic liver disease in the HALT-C Trial. Clin Gastroenterol Hepatol. 2010;8:877–883. doi: 10.1016/j.cgh.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel K, Wilder J. The clinical utility of FibroScan® as a noninvasive diagnostic test for liver disease. Med Devices Evid Res. 2014;7:107–114. doi: 10.2147/MDER.S46943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Qiu L, Liu D, Qian L. Acoustic radiation force impulse (ARFI) elastography for non-invasive evaluation of hepatic fibrosis in chronic hepatitis B and C patients: a systematic review and meta-analysis. Med Ultrason. 2017;19:23. doi: 10.11152/mu-942. [DOI] [PubMed] [Google Scholar]
- 19.Schuppan D. Liver fibrosis: common mechanisms and antifibrotic therapies. Clin Res Hepatol Gastroenterol. 2015;39:S51–S59. doi: 10.1016/j.clinre.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Manning DS, Afdhal NH. Diagnosis and quantitation of fibrosis. Gastroenterology. 2008;134:1670–1681. doi: 10.1053/j.gastro.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Schuppan D, Kim YO. Evolving therapies for liver fibrosis. J Clin Invest. 2013;123:1887–1901. doi: 10.1172/JCI66028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization: guidelines for the prevention care and treatment of persons with chronic hepatitis B virus infection. Geneva, Switzerland: WHO Press; 2015. [PubMed] [Google Scholar]
- 23.Jin W, Lin Z, Xin Y, Jiang X, Dong Q, Xuan S. Diagnostic accuracy of the aspartate aminotransferase-to-platelet ratio index for the prediction of hepatitis B-related fibrosis: a leading meta-analysis. BMC Gastroenterol. 2012;12:14. doi: 10.1186/1471-230X-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Xu MY, Zheng RD, Xian JC, Xu HT, Shi JP, Li SB, Qu Y, Dong YW, Lu LG. Prediction of significant fibrosis and cirrhosis in hepatitis B e-antigen negative patients with chronic hepatitis B using routine parameters: liver fibrosis prediction using routine data. Hepatol Res. 2013;43:441–451. doi: 10.1111/j.1872-034X.2012.01094.x. [DOI] [PubMed] [Google Scholar]
- 25.Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afdhal NH, Nunes D. Evaluation of liver fibrosis: a concise review. Am J Gastroenterol. 2004;99:1160–1174. doi: 10.1111/j.1572-0241.2004.30110.x. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad W, Ijaz B, Gull S, Asad S, Khaliq S, Jahan S, Sarwar MT, Kausar H, Sumrin A, Shahid I, Hassan S. A brief review on molecular, genetic and imaging techniques for HCV fibrosis evaluation. Virol J. 2011;8:53. doi: 10.1186/1743-422X-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Wang X, Karsdal MA, Leeming DJ, Genovese F. Molecular serum markers of liver fibrosis. Biomark Insights. 2012;7:105–117. doi: 10.4137/BMI.S10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lurie Y. Non-invasive diagnosis of liver fibrosis and cirrhosis. World J Gastroenterol. 2015;21:11567. doi: 10.3748/wjg.v21.i41.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuppan D. Connective tissue polypeptides in serum as parameters to monitor antifibrotic treatment in hepatic fibrogenesis. J Hepatol. 1991;13(Suppl 3):S17–25. doi: 10.1016/0168-8278(91)90004-u. [DOI] [PubMed] [Google Scholar]
- 31.Neuman MG, Cohen LB, Nanau RM. Hyaluronic acid as a non-invasive biomarker of liver fibrosis. Clin Biochem. 2016;49:302–315. doi: 10.1016/j.clinbiochem.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Guéchot J, Laudat A, Loria A, Serfaty L, Poupon R, Giboudeau J. Diagnostic accuracy of hyaluronan and type III procollagen amino-terminal peptide serum assays as markers of liver fibrosis in chronic viral hepatitis C evaluated by ROC curve analysis. Clin Chem. 1996;42:558. [PubMed] [Google Scholar]
- 33.Hemmann S, Graf J, Roderfeld M, Roeb E. Expression of MMPs and TIMPs in liver fibrosis - a systematic review with special emphasis on anti-fibrotic strategies. J Hepatol. 2007;46:955–975. doi: 10.1016/j.jhep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Yan L, Deng Y, Zhou J, Zhao H, Wang G China HepB-Related Fibrosis Assessment Research Group. Serum YKL-40 as a biomarker for liver fibrosis in chronic hepatitis B patients with normal and mildly elevated ALT. Infection. 2018;46:385–393. doi: 10.1007/s15010-018-1136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J Histochem Cytochem. 2016;64:157–167. doi: 10.1369/0022155415627681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann DA. Epigenetics in liver disease. Hepatology. 2014;60:1418–1425. doi: 10.1002/hep.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front Physiol. 2012;3:49. doi: 10.3389/fphys.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YA, Wallace MC, Friedman SL. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2008;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 42.Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387–392. doi: 10.1016/j.biopha.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Fu RQ, Hu DP, Hu YB, Hong L, Sun QF, Ding JG. miR-21 promotes α-SMA and collagen I expression in hepatic stellate cells via the Smad7 signaling pathway. Mol Med Rep. 2017;16:4327–4333. doi: 10.3892/mmr.2017.7054. [DOI] [PubMed] [Google Scholar]
- 44.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol. 2011;22:1668–1681. doi: 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, Tacke F, Trautwein C, Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 48.Wai CA. Simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2005;38:518–526. doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- 49.Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, S Sulkowski M, Torriani FJ, Dieterich DT, Thomas DL, Messinger D, Nelson M APRICOT Clinical Investigators. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 50.Gao S, Chen D, Zou L, Huang L, Dai R, Zheng J, Mo Z, Hu W, Shan Y. MiR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target RECK in hepatic oval cells. Int J Clin Exp Pathol. 2016;9:4779–4785. [Google Scholar]
- 51.Zhang Z, Zha Y, Hu W, Huang Z, Gao Z, Zang Y, Chen J, Dong L, Zhang J. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem. 2013;288:37082–37093. doi: 10.1074/jbc.M113.517953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caviglia JM, Yan J, Jang MK, Gwak GY, Affo S, Yu L, Olinga P, Friedman RA, Chen X, Schwabe RF. MicroRNA-21 and dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology. 2018;67:2414–2429. doi: 10.1002/hep.29627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Chen Y, Zhao Y. The diagnostic value of the FIB-4 index for staging hepatitis B-related fibrosis: a meta-analysis. PLoS One. 2014;9:e105728. doi: 10.1371/journal.pone.0105728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao G, Yang J, Yan L. Comparison of diagnostic accuracy of aspartate aminotransferase to platelet ratio index and fibrosis-4 index for detecting liver fibrosis in adult patients with chronic hepatitis B virus infection: a systemic review and meta-analysis. Hepatology. 2015;61:292–302. doi: 10.1002/hep.27382. [DOI] [PubMed] [Google Scholar]
- 55.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loyer X, Paradis V, Hénique C, Vion AC, Colnot N, Guerin CL, Devue C, On S, Scetbun J, Romain M, Paul JL, Rothenberg ME, Marcellin P, Durand F, Bedossa P, Prip-Buus C, Baugé E, Staels B, Boulanger CM, Tedgui A, Rautou PE. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. 2016;65:1882. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, Hamajima N, Hashimoto S. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99–103. doi: 10.1016/j.cca.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 58.Lamontagne J. Hepatitis B virus and microRNAs: complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol. 2015;21:7375. doi: 10.3748/wjg.v21.i24.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Damania P, Sen B, Dar SB, Kumar S, Kumari A, Gupta E, Sarin SK, Venugopal SK. Hepatitis B virus induces cell proliferation via HBX-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN) PLoS One. 2014;9:e91745. doi: 10.1371/journal.pone.0091745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


