Abstract
Objective: To investigate the association between SNP in the BTNL2 gene region and the susceptibility to osteoarthritis of the knee. Methods: The blood samples of 103 knee osteoarthritis and 134 healthy subjects were collected. Four SNP in the BTNL2 gene region were selected, whole DNA was extracted using the QIAamp blood DNA purification mini reagent, the BTNL2 gene fragment was amplified and sequenced, and the genotype and corresponding frequency were counted. The results were statistically analyzed. Results: The four SNP (rs41521946, rs28362677, rs28362678, rs28362675) in the BTNL2 gene region were analyzed using a chi-square test (mutation heterozygote, homozygous, and normal homozygote), and the genotypes of the four mutation points were found to be statistically significant (P=0.003, 0.013, 0.005, and 0.045, respectively). Among the four SNP, the first three SNP were in Hardy-Weinberg equilibrium, and a multivariate logistic regression analysis was used to correlate them with knee osteoarthritis (P=0.003, 0.013, 0.005, respectively). rs28362675 was not in Hardy-Weinberg equilibrium but was associated with knee osteoarthritis (P=0.045), which might be smaller samples or an ethnicity differential allelic variation. The P values of the statistical analysis of age and height in the baseline data of both groups were less than 0.05. Considering the possible impact on the results, they were used as covariates in the analysis. The SNP of rs41521946 and rs28362677 showed a significant change in their associations with mutations, and the genotype P values of rs41521946 (AC+AA)/CC and rs2836267 (AG+AA)/GG were 0.002, 0.006, respectively. Conclusion: Four SNP (rs41521946, rs28362677, rs28362678, rs28362675) in the BTNL2 gene region were significantly associated with knee osteoarthritis, and the target population might be significantly affected by rs28362675.
Keywords: Osteoarthritis, BTNL2 gene, single nucleotide polymorphism (SNP), knee, association study
Introduction
Osteoarthritis (OA) is a common kind of articular cartilage degenerative joint disease caused by multiple factors, and its main characteristics are joint cartilage damage, bone hyperplasia, limited movement, and even joint deformity [1]. With the increasing incidence of knee osteoarthritis [2,3], More and more attention has been paid to it. The treatments of osteoarthritis are limited, which places a heavy burden on society and families [4], so it is particularly important to prevent the disease [5,6]. Some studies on the etiology and pathological mechanisms of osteoarthritis have shown that inflammatory factors may aggravate the process of articular cartilage damage during the evolution of osteoarthritis [5]. There are many risk factors for osteoarthritis of the knee [7], and epidemiological studies have proved that osteoarthritis is a complex disease due to both environmental and genetic interactions [8]. The important role of genetic factors has been demonstrated in twin and family studies [9], especially in genome-wide association studies to further confirm the influence of genetic factors on knee osteoarthritis and analyze SNP in racial and regional heterogeneity [9-11]. Genetic variation may influence OA risk factors such as obesity, skeletal shape, bone mass and synovitis and could also affect the risk of genetic sensitivity to pain and the risk of disease progression [9]. Some studies have reported the association of rs7775228 and rs10947262 SNP on BTNL2 gene regions with knee osteoarthritis in different populations is different [11,12]. In order to further clarify the association between BTNL2 gene polymorphisms and the Chinese knee osteoarthritis population, we investigated the association between BTNL2 gene polymorphisms and osteoarthritis in the Han population.
Materials and methods
Study population
This study was approved by the Medical Ethics Committee of Beijing Bo’ai Hospital, China Rehabilitation Research Center. Patients and healthy controls admitted to Beijing Bo’ai Hospital of China Rehabilitation Research Center and Beijing Chaoyang Hospital Affiliated to Capital Medical University from January 1, 2017 to August 31, 2018 were selected, and we recorded detailed records of their names, sex, age, height, weight and body mass index (BMI), which was calculated as weight in kg divided by the square of height in meters (kg/m2). In our study, blood samples of 237 subjects (103 OA patients and 134 healthy controls) were collected.
Diagnostic criteria
The diagnostic criteria of osteoarthritis of the knee of the American College of Rheumatology were adopted [13]. The diagnostic criteria for osteoarthritis of the knee-include knee pain plus at least 3 out of the 7 following criteria: ① age ≥50 years old, ② stiffness <30 minutes, ③ crepitus, ④ bony enlargement, ⑤ bony tenderness, ⑥ no palpable warmth, and ⑦ osteophyte formation on radiological examination. Inclusion criteria of the patient group: ① the Han people of northern China, living and working in this region for more than 20 years, ② aged between 40 and 70 years old, ③ body mass index <30 Kg/m2 [9], ④ corresponding to the diagnostic criteria of osteoarthritis of the knee, ⑤ volunteered to participate in this study. Exclusion criteria of the patient group: ① the transient population, or less than 20 years of life in this region, ② age >70 or <40 years of age, ③ body mass index ≥30 Kg/m2, ④ incompatible diagnostic criteria of knee joint osteoarthritis, ⑤ secondary OA, inflammatory arthritis (rheumatoid arthritis, systemic lupus erythematosus, hemophilic arthritis, etc.), the above diagnosis must be accompanied by a certificate of diagnosis issued by a Third Level 1st Class Hospital, ⑥ diabetes, hyperuricemia, heart disease, infectious diseases, severe mental disorders.
Sample processing
According to the inclusion and exclusion criteria, the samples meeting the requirements were strictly screened, and 2 ml EDTA (ethylene diamine tetraethylene) anticoagulant blood was collected and the samples were divided into 300 microliters and stored at -80°C.
DNA amplification and genotyping
Genomic DNA was extracted from peripheral blood leukocytes (according to the QIAamp DNA Mini Kit operation protocol). The extracted DNA was stored at 4°C for future analysis. The genotyping of the four SNP in the BTNL2 gene was performed using the pairs of primers: sense primer 5’-TGCTCCAGCCAGCCTCAT-3’, anti-sense primer 5’-GGTCTTCATATTTATTTTCC-3’ (Primer PREMIER Version 5.0, Primer 5). Each 25.0 μL PCR reaction mixture consisted of 5.0 μL of genomic DNA, 1.0 μL of each primer (10 pmol/L), 12.5 μL of 2 × Taq PCR Mastermix (50 mM Tris-HCl, pH 8.7, 100 mM KCl, 4 mM MgCl2, 0.1 U Taq Platinum polymerase/μL, 500 μM dNTP each), and 5.5 μL of ddH2O (DNase/RNase-free). The reaction mixture was subjected to denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 45 s, 52.3°C for 45 s, 72°C for 45 s, then a final extension at 72°C for 5 min. Agarose gel electrophoresis was performed on the products of PCR. With DL 2000 DNA marker as the reference, amplified bands appeared on the horizontal line of the target sequence and were relatively obvious, which were sequenced in the second generation (Shanghai Majorbio Bio-pharm Technology Co., LTD). We used the NCBI blast database (http://www.ncbi.nlm.nih.gov/BLAST/) in the detection after the sequencing of sequence, and the matching degree between the target sequences and BTNL2 sequences was greater than or equal to 98.9%. Alleles and genotypes of BTNL2 were determined by peak mapping using chromas software (version 1.62 graph 1).
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) test was applied to confirm the independent segregation of the alleles. Qualitative variables were expressed as a raw count and a percentage. Mean ± SD was used for the presentation of quantitative variables. Genotypic and allelic frequencies were calculated by direct counting. Pearson’s χ2 test was used to evaluate the difference in genotype distribution and sex ratio between the groups. The differences in the general characteristics between the patient group and the control group were evaluated using the independent-Sample T-test. A multivariate logistics regression was used to assess the association between SNP and osteoarthritis of the knee. A two sided P<0.05 was considered significant. We estimated the interactions between the SNPs and age and height, which were regarded as covariates to perform a multiple logistic regression analysis again (Table 5). Odds ratios (ORs) were also calculated. All statistical analyses were carried out using the statistical software package SPSS 23.0 (SPSS Inc., Chicago, Illinois, USA).
Table 5.
The association of polymorphisms with osteoarthritis risk after adjustment for age and height
| Gene | rs number | Genotype | Frequency | OR | P value | |
|---|---|---|---|---|---|---|
|
| ||||||
| OA (103) | Normal (134) | |||||
| BTNL2 | rs41521946 | CC | 76 | 119 | 1 | |
| AC | 24 | 14 | 2.68 | 0.007 | ||
| AA | 3 | 1 | 4.70 | 0.184 | ||
| AC+AA | 27 | 15 | 3.64 | 0.002 | ||
| rs28362677 | GG | 76 | 116 | 1 | ||
| AG | 24 | 16 | 2.29 | 0.020 | ||
| AA | 3 | 2 | 2.29 | 0.370 | ||
| AG+AA | 27 | 18 | 2.98 | 0.006 | ||
| rs28362678 | CC | 75 | 117 | 1 | ||
| CT | 25 | 16 | 3.34 | 0.003 | ||
| TT | 3 | 1 | 2.30 | 0.539 | ||
| CT+TT | 28 | 17 | 3.28 | 0.003 | ||
| rs28362675 | GG | 89 | 126 | 1 | ||
| TG | 12 | 7 | 2.38 | 0.127 | ||
| TT | 2 | 1 | 3.71 | 0.308 | ||
| TG+TT | 14 | 8 | 2.49 | 0.051 | ||
ORs were estimated by multiple logistic regression analyses after adjustment for age and height.
Results
General characteristics
A comparison of the generalized features can be found in Table 1. The values of mean age (67.18±9.53 vs. 60.23±6.69) and height (160.11±6.05 vs. 163.12±4.28) were significant between the groups (P<0.05, respectively), but the statistical analyses adjusted for age and height still showed the same association. There was no significant difference in sex ratio, weight and BMI between the two groups (P>0.05, respectively).
Table 1.
Baseline characteristics of the participants whose DNA was extracted in this study
| Variable | OA (103) | Normal (134) | X2/t | P value |
|---|---|---|---|---|
| Age (SD), year | 67.18±9.53 | 60.23±6.69 | 6.31 | .00 |
| Female, n (%) | 86 (83.50%) | 99 (73.88%) | 3.14 | .08 |
| Heigh (SD), cm | 160.11±6.05 | 163.12±4.28 | -4.30 | .00 |
| Weight (SD), Kg | 69.05±11.05 | 69.57±9.10 | 0.40 | .69 |
| BMI (SD), kg/m2 | 26.91±3.93 | 26.17±3.59 | 1.50 | .14 |
Values were the mean (SD). Individuals were classified as normal if they did not have disease in either knee at both examinations and as having osteoarthritis (OA) if they met the diagnostic criteria.
Electrophoresis, genotyping, and sequencing
The amplification of genomic DNA yielded PCR products of 489 bp. After agarose gel electrophoresis, the PCR products meeting the requirements were sequenced, and their genotypes were identified by peak mapping software (Figure 1), and there were three genotypes in each single nucleotide polymorphism. Thus, XX represented a homozygous genotype, and XY was on half of a heterozygous genotype, and YY stood for the mutated homozygote (Table 2).
Figure 1.
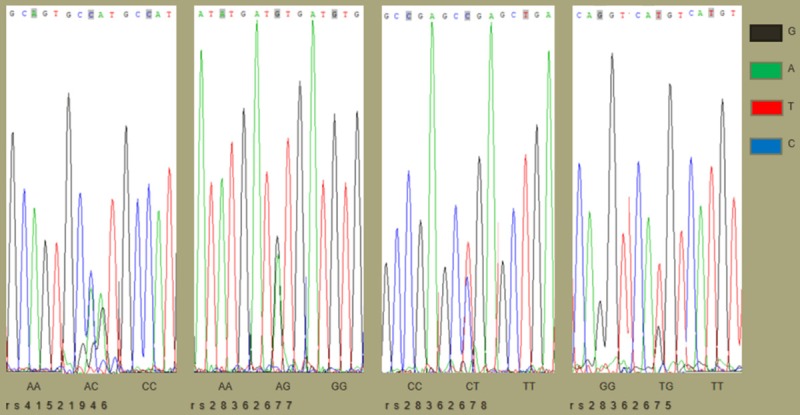
The adjacent sequences and genotypes of four SNP on the BTNL2 gene. The specific fragment of BTNL2 was amplified by the action of restriction endonucleases, and the genotypes of four SNP were determined by chromas peak mapping software after sequencing. The position of each light-colored shadow marker mutation site was judged by the peak map, the single peak was homozygous, and the double peak was heterozygous. The genotype of this locus is indicated below the graph. Each color represents a base, and black, green, red, and blue represent G, A, T, and C, respectively.
Table 2.
Hardy-Weinberg equilibrium
| SNP | XX | XY | YY | X2 | P value |
|---|---|---|---|---|---|
| rs41521946 | 195 | 38 | 4 | 1.72 | 0.41 |
| rs28362677 | 192 | 40 | 5 | 2.65 | 0.27 |
| rs28362678 | 192 | 41 | 4 | 1.06 | 0.59 |
| rs28362675 | 215 | 19 | 3 | 9.26 | 0.01 |
XX, XY, YY represent wild type, homozygous, mutant genotype.
Genotypic and allelic frequencies
In our study, four SNP (rs41521946, rs28362677, rs28362678, rs28362675) in the BTNL2 gene region were evaluated using chi-square tests (mutation heterozygote, homozygous and normal homozygote), and the genotypes of the four mutation points were statistically significant (P=0.003, 0.013, 0.005, and 0.045, respectively) (Table 3). Among the four SNP, the first three SNP were in Hardy-Weinberg equilibrium (Table 2), and a multivariate logistic regression analysis was used to associate them with knee osteoarthritis (P=0.003, 0.013, 0.005, respectively). rs28362675 was not in Hardy-Weinberg equilibrium but was also associated with knee osteoarthritis (P=0.045) (Table 3). The P values of the statistical analysis of age and height in the baseline data of both groups were less than 0.05. Considering the possible impact on the results, they were used in a covariate analysis for the multiple logistic regression analysis. The SNP of rs41521946 and rs28362677 showed a significant change in the association of mutations, and the genotype P values of rs41521946 (AC+AA)/CC and rs2836267 (AG+AA)/GG were 0.002, 0.006, respectively (Tables 4, 5). The corresponding P and OR values were calculated, and the OR values of all four SNP were more than 1, which could prove that they were risk factors.
Table 3.
Results of the genotype frequency analysis of the SNP
| SNP | Genotype | Frequency (n%) | X2 | P Value | |
|---|---|---|---|---|---|
|
| |||||
| OA | Normal | ||||
| rs41521946 | CC | 76 (73.79) | 119 (88.80) | - | - |
| AC | 24 (23.30) | 14 (10.45) | 7.59 | 0.006 | |
| AA | 3 (2.91) | 1 (0.75) | 2.13 | 0.145 | |
| CC/(AC+AA) | 27 (26.21) | 15 (11.2) | 9.01 | 0.003 | |
| rs28362677 | GG | 76 (73.79) | 116 (86.57) | - | - |
| AG | 24 (23.30) | 16 (11.94) | 5.63 | 0.018 | |
| AA | 3 (2.91) | 2 (1.49) | 0.21 | 0.647 | |
| GG (AG+AA) | 27 (26.21) | 18 (13.43) | 6.18 | 0.013 | |
| rs28362678 | CC | 75 (72.82) | 117 (87.31) | - | - |
| CT | 25 (24.27) | 16 (11.94) | 6.62 | 0.010 | |
| TT | 3 (2.91) | 1 (0.75) | 0.88 | 0.349 | |
| CC (CT+TT) | 28 (27.18) | 17 (12.69) | 7.96 | 0.005 | |
| rs28362675 | GG | 89 (86.41) | 126 (94.03) | - | - |
| TG | 12 (11.65) | 7 (5.22) | 3.37 | 0.066 | |
| TT | 2 (1.94) | 1 (0.75) | 0.09 | 0.770 | |
| GG (TG+TT) | 14 (13.59) | 8 (5.97) | 4.02 | 0.045 | |
Table 4.
The association between the polymorphism and the risk of osteoarthritis
| Gene | rs number | Genotype | Frequency | OR | P value | |
|---|---|---|---|---|---|---|
|
| ||||||
| OA (103) | Normal (134) | |||||
| BTNL2 | rs41521946 | CC | 76 | 119 | 1 | |
| AC | 24 | 14 | 2.68 | 0.007 | ||
| AA | 3 | 1 | 4.70 | 0.184 | ||
| AC+AA | 27 | 15 | 2.82 | 0.003 | ||
| rs28362677 | GG | 76 | 116 | 1 | ||
| AG | 24 | 16 | 2.29 | 0.020 | ||
| AA | 3 | 2 | 2.29 | 0.370 | ||
| AG+AA | 27 | 18 | 2.29 | 0.014 | ||
| rs28362678 | CC | 75 | 117 | 1 | ||
| CT | 25 | 16 | 2.44 | 0.012 | ||
| TT | 3 | 1 | 4.68 | 0.185 | ||
| CT+TT | 28 | 17 | 2.57 | 0.006 | ||
| rs28362675 | GG | 89 | 126 | 1 | ||
| TG | 12 | 7 | 2.43 | 0.073 | ||
| TT | 2 | 1 | 2.83 | 0.398 | ||
| TG+TT | 14 | 8 | 2.49 | 0.051 | ||
ORs were estimated by multiple logistic regression analyses.
Discussion
We chose the population that had lived in the north of China for more than 20 years to reduce the environmental impact and to avoid the ethnic and geographical heterogeneity as much as possible. Four SNP (rs41521946, rs28362677, rs28362678, rs28362675) in the BTNL2 gene region were analyzed using a chi-square test (mutation heterozygote, homozygous, and normal homozygote), and the genotypes of the four mutation points were found to be statistically significant (P=0.003, 0.013, 0.005, and 0.045, respectively) (Table 3). Among the four single nucleotide polymorphisms, the first three SNP were in Hardy-Weinberg equilibrium (Table 2), and a multivariate logistic regression analysis was used to correlate them with knee osteoarthritis (P=0.003, 0.014, 0.006, respectively). The single nucleotide polymorphism rs28362675 was not in Hardy-Weinberg equilibrium and seemed to be associated with knee osteoarthritis (P=0.051) (Table 4). On the one hand, this might be caused by a small sample size or an allelic variation based on ethnic differences and could possibly be due to genotyping errors; on the other hand, it might be affected by multiple factors, such as sex, age, BMI, and so on. The P values of the statistical analysis of age and height in the baseline data of both groups were less than 0.05. We considered the possible impact on the results, which were regarded as covariates to perform a multiple logistic regression analysis again (Table 5). The SNP of rs41521946 and rs28362677 showed a significant change in the association of mutations, The P value of rs41521946 genotype (AC+AA)/CC and the rs28362677 genotype (AG+AA)/GG were 0.002, 0.006, respectively (Table 5). We considered that these changes might be affected by age, height, weight, and body mass index.
Genome-wide association studies demonstrated that there were many SNPs in the BTNL2 gene regions [10-12], but their biological significance were not clear. A study turned out that SNP of rs10947262 and rs7775228 on BTL2 gene regions were significantly associated with knee osteoarthritis in the Japanese population (P=3.07 × 10-3, OR=1.53, 5.74 × 10-3, OR=1.47, respectively) [12], but not in European descent (P=0.42, 0.28, respectively) [11]. All of these might demonstrate racial and geographic heterogeneity in the SNP located in the BTNL2 gene region. Our research also supported this view. The three SNP in the BTNL2 gene region (rs41521946, rs28362677, rs28362678) were correlated with the susceptibility to rheumatoid arthritis [14], and Spanish scholars also confirmed that the BTNL2 gene was associated with the onset of rheumatoid arthritis and could be an independent risk factor for rheumatoid osteoarthritis [15]. However, their association with knee osteoarthritis had not been reported, which was verified by our study. From the perspective of OR, four single nucleotide polymorphisms might be risk factors for osteoarthritis of the knee. Some reports had found that rs7775228 and rs10947262 had significant association with SNP located in the 340 kb region of the HLA locus, including BTNL2, HLA-DRA, HLA-DRB5, HLA-DRB1, HLA-DQA1, and HLA-DQB1. Only SNPs in the 340 kb region had strong correlations with OA, but SNPs outside this region have no significant correlation with OA [12]. Our results showed that the four SNP in this region were consistent with this view, and it was not known whether other SNP outside this region were associated with knee osteoarthritis.
Some scholars have reported changes in pathophysiological mechanisms following mutations in the BTNL2 gene. In a linkage study between HLA-DPB1 and other HLA loci, five apparent recombinations between DPB1 and DR/DQ loci were observed, which changed the protein expression [16], and genes encoding structural extracellular matrix components (such as DVWA) and molecules involved in prostaglandin metabolism (such as DQB1 and BTNL2) might be present in OA [17]. Body adipose tissue had been shown to be related to the development and progression of knee OA [18], which could change our body center of gravity and exercise patterns. Meanwhile, most scholars have confirmed the correlation between BTNL2 and sarcoidosis [19-21] and have reported an association of sarcoidosis and HLA class I and especially class II alleles in different populations [22].
The current treatments for OA are limited and insufficient to prevent the occurrence and development of the disease. Genetic studies of OA patients can elucidate the molecular mechanisms that lead to the progression of specific diseases, including joint damage, pain, and chronic pain and can provide new insights into treatment.
Conclusion
Four SNP (rs41521946, rs28362677, rs28362678, rs28362675) in the BTNL2 gene region are significantly associated with knee osteoarthritis, and the target population might be significantly affected by rs28362675.
Limitations
There were some limitations in this study. First, the number of patients in this study and the healthy controls were still not large enough and might not reveal the potential link between the mutation site and OA. Second, the patients in this study were from Beijing Bo’ai Hospital and Beijing Chaoyang hospital, so the study might be unable to represent all OA targets. Finally, all normal controls in this study were OA-free, but it is possible that a small number of the control patients who had mild asymptomatic and radiological OA were selected as normal controls. Radiological examination was not possible due to objective conditions, including economic reasons. Therefore, it is necessary to conduct further research in a wider population and even in other ethnic groups. This study also had its own advantages. Strict laboratory procedures ensured a high reliability of the genotyping results. Secondly, logistic regression analysis was used to remove the interference of baseline data on the experimental results, which could make the results more credible.
Acknowledgements
The study was approved by Medical Ethics Committee of China Rehabilitation Research Center (No. 2017-075-1). Source: This work was funded by the Fundamental Research Funds for Central Public Welfare Research Institutes, China Rehabilitation Research Center (No. 2016CZ-2 and 2018CZ-6).
Disclosure of conflict of interest
None.
References
- 1.Pereira D, Ramos E, Branco J. Osteoarthritis. Acta Med Port. 2015;28:99–106. doi: 10.20344/amp.5477. [DOI] [PubMed] [Google Scholar]
- 2.Wallace IJ, Worthington S, Felson DT, Jurmain RD, Wren KT, Maijanen H, Woods RJ, Lieberman DE. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc Natl Acad Sci U S A. 2017;114:9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q, Niu J, Huang J, Ke Y, Tang X, Wu X, Li R, Li H, Zhi X, Wang K, Zhang Y, Lin J. Knee osteoarthritis and all-cause mortality: the wuchuan osteoarthritis study. Osteoarthritis Cartilage. 2015;23:1154–7. doi: 10.1016/j.joca.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Cross M, Smith E, Hoy D, Nolte S, Ackerman I, Fransen M, Bridgett L, Williams S, Guillemin F, Hill CL, Laslett LL, Jones G, Cicuttini F, Osborne R, Vos T, Buchbinder R, Woolf A, March L. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:1323–30. doi: 10.1136/annrheumdis-2013-204763. [DOI] [PubMed] [Google Scholar]
- 5.Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!) Osteoarthritis Cartilage. 2013;21:16–21. doi: 10.1016/j.joca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Bitton R. The economic burden of osteoarthritis. Am J Manag Care. 2009;15(Suppl):S230–5. [PubMed] [Google Scholar]
- 7.Roos EM, Arden NK. Strategies for the prevention of knee osteoarthritis. Nat Rev Rheumatol. 2016;12:92–101. doi: 10.1038/nrrheum.2015.135. [DOI] [PubMed] [Google Scholar]
- 8.Chapman K, Valdes AM. Genetic factors in OA pathogenesis. Bone. 2012;51:258–64. doi: 10.1016/j.bone.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Valdes AM, Spector TD. Genetic epidemiology of hip and knee osteoarthritis. Nat Rev Rheumatol. 2011;7:23–32. doi: 10.1038/nrrheum.2010.191. [DOI] [PubMed] [Google Scholar]
- 10.Yang HC, Liang YJ, Chung CM, Chen JW, Pan WH. Genome-wide gene-based association study. BMC Proc. 2009;3(Suppl 7):S135. doi: 10.1186/1753-6561-3-s7-s135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdes AM, Styrkarsdottir U, Doherty M, Morris DL, Mangino M, Tamm A, Doherty SA, Kisand K, Kerna I, Tamm A, Wheeler M, Maciewicz RA, Zhang W, Muir KR, Dennison EM, Hart DJ, Metrustry S, Jonsdottir I, Jonsson GF, Jonsson H, Ingvarsson T, Cooper C, Vyse TJ, Spector TD, Stefansson K, Arden NK. Large scale replication study of the association between HLA class II/BTNL2 variants and osteoarthritis of the knee in european-descent populations. PLoS One. 2011;6:e23371. doi: 10.1371/journal.pone.0023371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima M, Takahashi A, Kou I, Rodriguez-Fontenla C, Gomez-Reino JJ, Furuichi T, Dai J, Sudo A, Uchida A, Fukui N, Kubo M, Kamatani N, Tsunoda T, Malizos KN, Tsezou A, Gonzalez A, Nakamura Y, Ikegawa S. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010;5:e9723. doi: 10.1371/journal.pone.0009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, Howell D, Kaplan D, Koopman W, Longley S, Mankin H, McShane DJ, Medsger T, Meenan R, Mikkelsen W, Moskowitz R, Murphy W, Rothschild B, Segel M, Sokoloff L, Wolfe F. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and therapeutic criteria committee of the american rheumatism association. Arthritis Rheum. 1986;29:1039–49. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 14.Mitsunaga S, Hosomichi K, Okudaira Y, Nakaoka H, Kunii N, Suzuki Y, Kuwana M, Sato S, Kaneko Y, Homma Y, Kashiwase K, Azuma F, Kulski JK, Inoue I, Inoko H. Exome sequencing identifies novel rheumatoid arthritis-susceptible variants in the BTNL2. J Hum Genet. 2013;58:210–5. doi: 10.1038/jhg.2013.2. [DOI] [PubMed] [Google Scholar]
- 15.López Herráez D, Martínez-Bueno M, Riba L, García de la Torre I, Sacnún M, Goñi M, Berbotto GA, Paira S, Musuruana JL, Graf CE, Alvarellos AJ, Messina OD, Babini AM, Strusberg I, Marcos JC, Scherbarth H, Spindler AJ, Quinteros A, Toloza SM, Moreno JL, Catoggio LJ, Tate G, Eimon A, Citera G, Catalán Pellet A, Nasswetter GG, Cardiel MH, Miranda P, Ballesteros F, Esquivel-Valerio JA, Maradiaga-Ceceña MA, Acevedo-Vásquez EM, García García C, Pons-Estel BA, Alarcón-Riquelme ME. Rheumatoid arthritis in latin Americans enriched for Amerindian ancestry is associated with loci in chromosomes 1, 12, and 13, and the HLA class II region. Arthritis Rheum. 2013;65:1457–67. doi: 10.1002/art.37923. [DOI] [PubMed] [Google Scholar]
- 16.Mitsunaga S, Kuwata S, Tokunaga K, Uchikawa C, Takahashi K, Akaza T, Mitomi Y, Juji T. Family study on HLA-DPB1 polymorphism: linkage analysis with HLA-DR/DQ and two “new” alleles. Hum Immunol. 1992;34:203–11. doi: 10.1016/0198-8859(92)90113-2. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima M, Kizawa H, Saitoh M, Kou I, Miyazono K, Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J Biol Chem. 2007;282:32185–92. doi: 10.1074/jbc.M700522200. [DOI] [PubMed] [Google Scholar]
- 18.Belluzzi E, El Hadi H, Granzotto M, Rossato M, Ramonda R, Macchi V, De Caro R, Vettor R, Favero M. Systemic and local adipose tissue in knee osteoarthritis. J Cell Physiol. 2017;232:1971–1978. doi: 10.1002/jcp.25716. [DOI] [PubMed] [Google Scholar]
- 19.Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R, Adler A, Kelly JA, Kaufman KM, Lessard CJ, Moser KL, Kimberly RP, Harley JB, Iannuzzi MC, Rybicki BA, Montgomery CG. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One. 2012;7:e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Wei J, Fan L, Cheng D. BTNL2 gene polymorphism and sarcoidosis susceptibility: a meta-analysis. PLoS One. 2015;10:e0122639. doi: 10.1371/journal.pone.0122639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morais A, Lima B, Peixoto MJ, Alves H, Marques A, Delgado L. BTNL2 gene polymorphism associations with susceptibility and phenotype expression in sarcoidosis. Respir Med. 2012;106:1771–7. doi: 10.1016/j.rmed.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Morais A, Alves H, Lima B, Delgado L, Gonçalves R, Tafulo S. HLA class I and II and TNF-alpha gene polymorphisms in sarcoidosis patients. Rev Port Pneumol. 2008;14:727–46. [PubMed] [Google Scholar]


