Abstract
Excessive proteinase inhibitor 9 (PI9) levels predict a poor outcome for patients with several tumor types. We compared the expression of PI9 in HepG2 cells and in 21 pairs of tumor and peritumor tissue samples using western blotting. Immunohistochemical staining was used to detect PI9 expression in 105 cases of hepatocellular carcinoma (HCC) and in 20 adjacent normal liver tissues. Changes in the degree of apoptosis and proliferation were determined before and after transfection with pcDNA3.1-PI9 and small interfering (si)RNA-PI9 using MTT analysis, colony formation assay, and flow cytometry. The correlation between PI9 expression and prognosis was determined in a large HCC patient cohort (n=105). Western blotting showed that PI9 expression was significantly higher in tumor tissues than in adjacent tissues. PI9 immunohistochemical staining was positive in 78.1% of HCC tissues, which was significantly higher than that seen in adjacent normal liver tissue (35%). PI9 expression in HCC correlated closely with the extent of tumor differentiation, tumor-node-metastasis staging, and tumor size. Cox regression analysis demonstrated that PI9 is an independent predictor of prognosis in patients with HCC, and is related to overall survival. The apoptosis of HepG2 cells was significantly increased following PI9 up-regulation and significantly decreased after siRNA interference against PI9 expression. Cell proliferation was significantly decreased following PI9 up-regulation and significantly increased after siRNA interference of PI9 expression. PI9 appears to contribute to the apoptosis of HCC, and could be an independent predictor of recurrence and a suitable pharmaceutical target in patients with HCC.
Keywords: PI9, immune escape, hepatocellular carcinoma, apoptosis, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer death worldwide and the fifth most common type of cancer. The 5-year survival rate for HCC after surgical resection remains low [1]. In the past few decades, great progress has been made in understanding the molecular biology of liver cancer; however, our knowledge on the occurrence and development of malignant liver tumors involving multiple factors and genes is still insufficient.
The gene encoding serine protease inhibitor 9 (PI9) consists of seven exons and six introns and is located on chromosome 6p25 [2]. In healthy humans, PI9 is mainly found in lymphoid tissues, immune-specific sites, and T lymphocytes. It is expressed most abundantly in the placenta and lung, but is also expressed in cytotoxic T lymphocytes (CTLs) - where it inhibits apoptosis, natural killer (NK) cells - the ovary, and the spleen [3]. This expression suggests that PI9 protects CTLs from GrB damage. Recent studies have also found that PI9 is expressed in cancers of the lung, prostate, colon, and other tumors [4]. PI9 has been associated with poor clinical prognosis in lymphoma and nasopharyngeal carcinoma [5].
However, its function is not fully understood. Recent evidence suggests that PI9 inactivates the protease granzyme B (GrB) efficiently and irreversibly in vitro, while PI9 overexpression prevented CTL-mediated cytotoxicity through the GrB pathway. GrB’s natural inhibitor is PI9 (serpin B9), a 42 kD clade B serpin which inhibit serine proteases intracelluarly. Serpins irreversibly inhibit their target protease, which can be detected by the formation of an SDS-stable complex with the target [6]. Therefore, PI9 may be involved in the formation of an immune escape mechanism, which is closely related to immune surveillance and the occurrence, development, and prognosis of tumors [7].
No previous studies have investigated the expression of PI9 in liver cancer. Therefore, in this study, we determined the expression of PI9 in tumor samples by western blotting and immunohistochemistry, and explored the clinical significance of PI9 expression changes in the progression of HCC. We observed that PI9 expression affected the extent of HCC cell apoptosis and proliferation. These results suggest that PI9 may be a tumor marker for poor prognosis in HCC.
Materials and methods
Patients and specimens
We collected 21 random HCC tissue specimens and paired adjacent tissue samples from patients who had undergone surgical resection for HCC in Nantong University Affiliated Hospital (Nantong City, People’s Republic of China) between December 2014 and July 2015 for western blot analysis. Tumor specimens were randomly retrieved from 105 patients with HCC from the same sample bank between January 2009 and May 2011 for use in tissue microarrays. Clinicopathologic characteristics of all patients were recorded. None of the patients had received any preoperative anticancer treatment. The study protocol, including tumor specimens and the use of human cell lines, was approved by patients and the Ethics Review Board of Nantong University Affiliated Hospital.
Tissue microarray and immunohistochemistry
Tissue microarrays and immunohistochemistry were performed as described previously [8]. Briefly, after neutralizing endogenous peroxidase and antigen retrieval by microwave irradiation, we pre-incubated the slides with blocking serum and incubated them overnight with mouse antihuman PI9 polyclonal antibody. Sections were then incubated with a secondary antibody and exposed to horseradish peroxidase-conjugated streptavidin. Using a Leica CCD camera (DFC420), we captured photographs of three representative fields using Leica QWin Plus v3 software. Immunohistochemical staining was assessed by two independent pathologists with no knowledge of patient characteristics. A scoring system was applied as previously described [9]. The staining intensity of PI9 was scored first (0: negative; 1: weak; 2: high), followed by the percentage of positive cells (1: 0%-25%; 2: 26%-50%; 3: 51%-75%; 4: ≥ 75%). The final score for each sample was calculated by multiplying the staining intensity score by the percentage score. Tumors with a staining index ≥ 2 were defined as high-expression tumors.
Western blot analysis
Western blot analysis was carried out as described previously. Briefly, cell or tissue lysates were generated and total proteins were separated using standard sodium dodecyl sulfate polyacrylamide gel electrophoresis, then transferred to polyvinylidenedifluoride membranes. The membranes were washed and blocked before being incubated with polyclonal antibody PI9 (Abcam, USA) followed by incubation with horseradish peroxidase-conjugated secondary antibodies. The reactions were detected using an enhanced chemiluminescence assay. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. The relative intensity of each band was determined using Image Lab 3.0 imaging methods (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell culture
The human HCC cell line HepG2 was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA) and penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA, USA).
Transfection
We cultured HepG2 cells in 24-well plates or 6 cm dishes to 60%-70% confluence, then transfected them with small interfering (si)RNA targeting the human PI9 (siRNA sequence: 5’-CAGUUCCUCUCAACGUUUAUU-3’) using Lipofectamine2000 following the manufacturer’s protocol (Invitrogen). After 48 h post-transfection, the cells were harvested for western blot analysis.
The PI9 construct sub-cloned into the pcDNA3.1 vector was linearized with Pvu I restriction enzyme digestion. Following purification, PI9 was transfected into HepG2 cells using Lipofectamine2000 (Invitrogen). Approximately 48 h post-transfection, cells were selected using medium containing 500 μg/ml G418 (Invitrogen). At this concentration of G418, non-transfected cells were killed in approximately 10 days. Cells that survived G418 selection were expanded and used for all subsequent assays.
MTT assay
The proliferation and viability of HepG2 cells were measured using the MTT assay. In brief, cells were seeded in 96-well plates at densities of 5 × 103 cells/well and treated with 30 nmol/L siRNA-PI9 and pcDNA3.1-PI9 for 48 h. After transfection, cells were treated with MTT dye (5 mg/mL) for 4 h, then the medium was replaced by 150 µL dimethyl sulfoxide. The absorbance values were measured using a microplate reader (Model 550, BIO-RAD, Shanghai, China) at a wavelength of 490 nm. Each experiment was repeated at least three times.
Flow cytometric analysis
Cell apoptosis was evaluated by Annexin-V staining according to the manufacturer’s protocol (MR Biotech, China). Briefly, 5 μL of Annexin V-FITC were incubated with 3 × 105 cells for 15 min in the dark. After staining, the cells were resuspended in 400 μL binding buffer. Two ml of HepG2 cells (5 × 105 cells) were seeded into a 6-well plate and incubated for 4 h, followed by incubation with 20 μM caspase inhibitor Z-VAD-FMK (Beyotime, China) for 1 h. Different concentrations of PPP were added into respective wells and incubated for 24 h. HepG2 cells were then collected, rinsed twice in pre-cold PBS, and centrifuged at 1,000 × g for 5 min. The supernatant was removed and the cells were resuspended in 100 μL of binding buffer. Then, the cells were stained with 5 μL of Annexin V-FITC and 10 μL of PI for 15 min in the dark followed by the addition of 400 μL of binding buffer. The apoptosis rate was analyzed by a FACS-Calibur flow cytometer.
Statistical analysis
Statistical analyses were performed with SPSS 20.0 and Graphpad Prism 7.0 software. Quantitative data were expressed as mean ± SD. Differences between groups were analyzed by two-tailed Student’s t-test or analysis of variance between groups (ANOVA), followed by Bonferroni’s post hoc test. Categorical data were evaluated by the χ2 test. P-values < 0.05 was considered significant.
Results
Expression of PI9 in HCC tissues and adjacent tissues
To assess the role of PI9 in HCC tissues, we performed western blotting to measure the expression of PI9 in 21 freshly collected HCC tissues and adjacent non-tumorous liver tissues. PI9 protein was found to be significantly upregulated in 15 of 21 cases compared with corresponding adjacent non-tumorous tissues (P < 0.05), and we randomly selected and demonstrated 5 pairs of 15 obvious experimental results as follows (Figure 1A, 1B).
Figure 1.
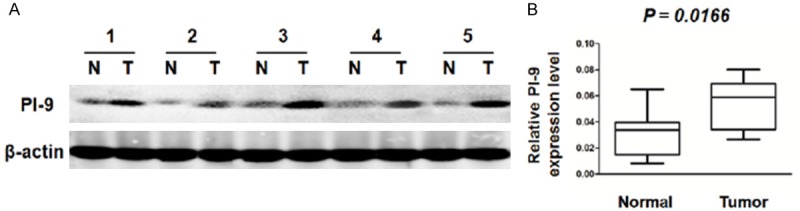
A, B. PI9 protein expression was examined in HCC tissues and normal tissue using western blot analysis. HCC: hepatocellular carcinoma; Normal: matched adjacent normal tissue.
Immunohistochemical analysis of PI9 in HCC tissues
Immunohistochemical analysis of PI9 revealed brown staining that was present mainly in the cytoplasm (Figure 2). Of the 105 patients with HCC, 82 (78.1%) were positive for PI9, with 47 (44.8%) considered high-expression patients and 35 (33.3%) considered low-expression patients. 23 (21.9%) patients were negative for PI9 expression, while the positive rate of adjacent normal tissues was 35% (7/20) (P < 0.05). In Table 1, we show that high PI9 expression in HCC was significantly associated with the extent of differentiation, tumor-node-metastasis stage, and tumor size (χ2 = 9.272, P = 0.002*; χ2 = 10.202, P = 0.001*; χ2 = 8.219, P = 0.004*). There was no significant correlation with gender, age, tumor number, hepatitis B infection, or tumor metastasis in patients with HCC.
Figure 2.
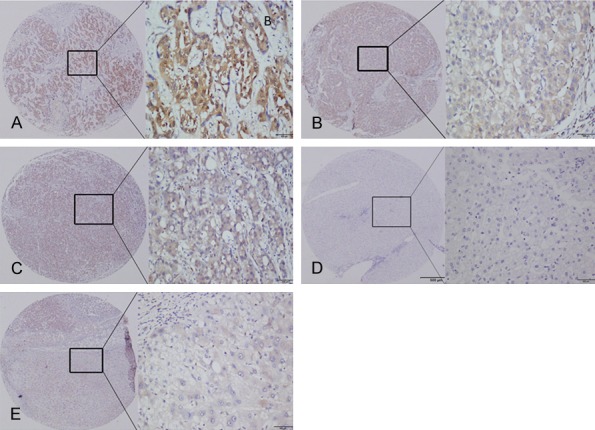
PI9 expression was detected in (A) poorly differentiated tissues, (B) moderately differentiated tissues, and (C) highly differentiated tissues. (D) PI9 was not expressed in normal liver tissues. (E) PI9 was expressed in normal tissues.
Table 1.
Relationship between PI9 expression and clinicopathological parameters in liver tissue
| Group | n | PI9 (n) | χ2 | P | |
|---|---|---|---|---|---|
|
| |||||
| High | Low | ||||
| Age | |||||
| ≥ 45 | 56 | 31 | 25 | 0.056 | 0.814 |
| < 45 | 49 | 26 | 23 | ||
| Gender | |||||
| Male | 80 | 43 | 37 | 0.039 | 0.844 |
| Female | 25 | 14 | 11 | ||
| Tumor size (cm) | |||||
| < 4.5 | 54 | 22 | 32 | 8.219 | 0.004** |
| ≥ 4.5 | 51 | 35 | 16 | ||
| Tumor number | |||||
| Single | 72 | 39 | 33 | 0.001 | 0.917 |
| Multiple | 33 | 18 | 15 | ||
| TNM | |||||
| I/II | 50 | 19 | 31 | 10.202 | 0.001** |
| III/IV | 55 | 38 | 17 | ||
| HBsAg | |||||
| Negative | 18 | 8 | 10 | 2.075 | 0.150 |
| Positive | 87 | 49 | 38 | ||
| Differentiation degree | |||||
| Well/Moderate | 53 | 46 | 7 | 9.272 | 0.002** |
| Poor | 52 | 42 | 10 | ||
| Neoplasm metastasis | |||||
| No | 89 | 48 | 41 | 0.395 | 0.529 |
| Yes | 16 | 9 | 7 | ||
P < 0.01.
Relationship between PI9 protein expression and prognosis in HCC
To study the prognostic value of PI9 expression, we determined the overall survival of the 105 patients with primary liver cancer. We found that the survival status of patients was significantly associated with multiple clinicopathologic parameters including tumor size, TNM, differentiation, and PI9 expression (Table 2). We further investigated the survival curves of patients with HCC with high and low expression of PI9. The survival time of patients with high PI9 expression was significantly shorter than that of the low expression group (Figure 3).
Table 2.
Multivariate analyses of prognostic factors associated with survival
| Group | n | PI9 (n) | χ2 | P | |
|---|---|---|---|---|---|
|
| |||||
| High | Low | ||||
| Age | |||||
| ≥ 45 | 56 | 42 | 14 | 0.170 | 0.680 |
| < 45 | 49 | 35 | 14 | ||
| Gender | |||||
| Male | 80 | 59 | 21 | 0.030 | 0.863 |
| Female | 25 | 18 | 7 | ||
| Tumor size (cm) | |||||
| < 4.5 | 54 | 34 | 20 | 6.664 | 0.013* |
| ≥ 4.5 | 51 | 43 | 8 | ||
| Tumor number | |||||
| Single | 72 | 52 | 20 | 0.253 | 0.615 |
| Multiple | 33 | 25 | 8 | ||
| TNM | |||||
| I/II | 50 | 28 | 22 | 4.665 | 0.000*** |
| III/IV | 55 | 49 | 6 | ||
| HBsAg | |||||
| Negative | 18 | 12 | 6 | 0.482 | 0.494 |
| Positive | 87 | 65 | 22 | ||
| Differentiation degree | |||||
| Well/Moderate | 66 | 44 | 22 | 4.038 | 0.044* |
| Poor | 39 | 33 | 6 | ||
| Neoplasm metastasis | |||||
| No | 89 | 66 | 23 | 0.203 | 0.652 |
| Yes | 16 | 11 | 5 | ||
| PI9 expression | |||||
| High | 57 | 47 | 10 | 4.420 | 0.036* |
| Low | 48 | 30 | 18 | ||
P < 0.05;
P < 0.001.
Figure 3.
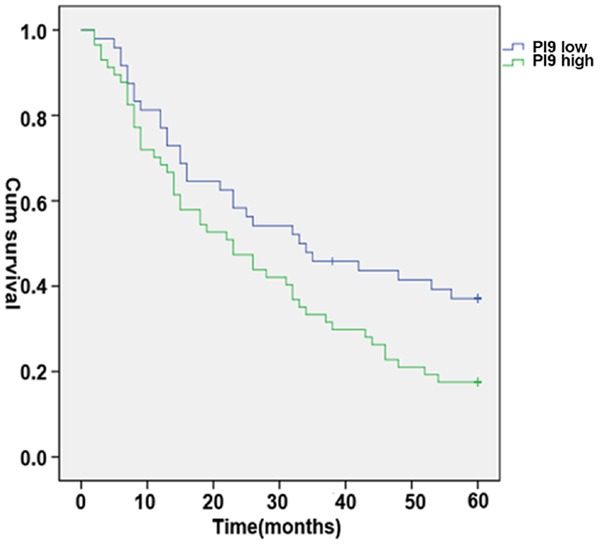
Kaplan-Meier survival curves of HCC patients positive and negative for expression of PI9. The 5-year overall survival rate of HCC patients with high PI9 expression correlated significantly with shorter overall survival times (P < 0.05).
Transfection of HepG2 cells with pcDNA3.1-PI9 and siRNA-PI9
First, we used siRNA1-PI9, siRNA2-PI9, and siRNA3-PI9 to interfere with the expression of PI9 in HepG2 cells by western blot analysis compared with control levels. PI9 protein levels were significantly lower in HepG2 cells transfected with siRNA1-PI9 than siRNA2-PI9 and siRNA3-PI9 (P < 0.05) (Figure 4A). Transfection of HepG2 cells with pcDNA3. 1-PI9 was shown to significantly increase protein levels of PI9 by western blot analysis compared with control levels (P < 0.05). Conversely, PI9 protein levels were significantly lower in HepG2 cells transfected with siRNA-PI9 than in the control group, indicating that siRNA-PI9 inhibits the expression of PI9 protein (Figure 4B).
Figure 4.
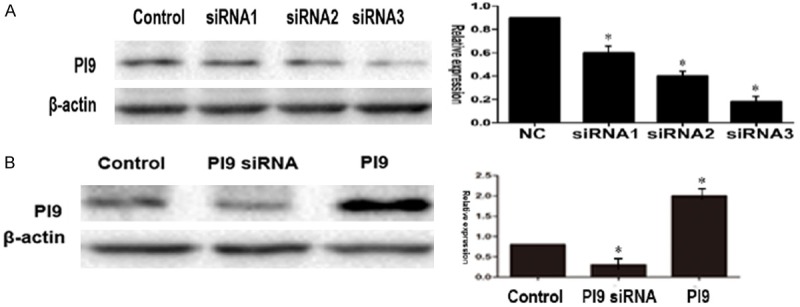
A. PI9 protein levels were significantly lower in HepG2 cells transfected with siRNA1-PI9 than siRNA2-PI9 and siRNA3-PI9 (P < 0.05). B. Compared with the control group, PI9 protein levels were significantly increased in HepG2 cells transfected with pcDNA3.1-PI9 (P < 0.05). The level of PI9 protein in the siRNA-PI9 group was significantly lower than in the control group (P < 0.05). (The PI9 siRNA was designed by Dharmacon (Lafayette, CO, USA), and has the sequence siRNA1: 5’-GAGAAAAUAGACCAUAAUGAU-3’; siRNA2: 5’-GGCCAAAUAAAAUUGTAUUAU-3’; siRNA3: 5’-CAGUUCCUCUCAACGUUUAUU-3’.
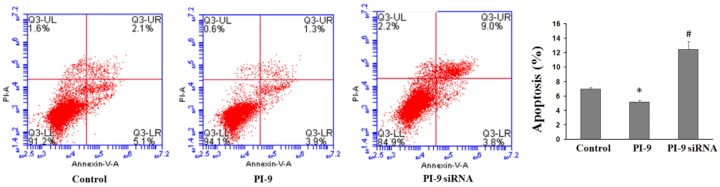
Effects of PI9 gene expression on the apoptosis of HCC cells
HepG2 cells transfected with siRNA-PI9 or pcDNA3.1-PI9 were assessed for apoptosis by flow cytometry (Figure 5). Compared with the control group, apoptosis was significantly enhanced in HepG2 cells transfected with siRNA-PI9 (#P < 0.05), while apoptosis was significantly decreased in HepG2 cells transfected with pcDNA3.1-PI9 (*P < 0.05).
Figure 5.
Flow cytometry results revealing significant differences in apoptosis between the siRNA-PI9 group, pcDNA3.1-PI9 group, and control group of HepG2 cells.
Effects of PI9 gene expression on the proliferation of HCC cells
HepG2 cells transfected with siRNA-PI9 or pcDNA3.1-PI9 were assessed for proliferation by MTT and colony formation assay. Compared with the control group, proliferation was significantly enhanced in HepG2 cells transfected with pcDNA3.1-PI9 (*P < 0.05), while proliferation was significantly decreased in HepG2 cells transfected with siRNA-PI9 (#P < 0.05) (Figures 6, 7).
Figure 6.
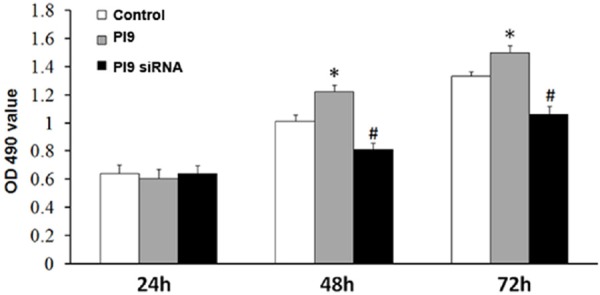
MTT assay analysis of cell proliferation. Quantitative data represent the mean ± SD of three independent experiments performed in triplicate. *P < 0.05, #P < 0.05.
Figure 7.
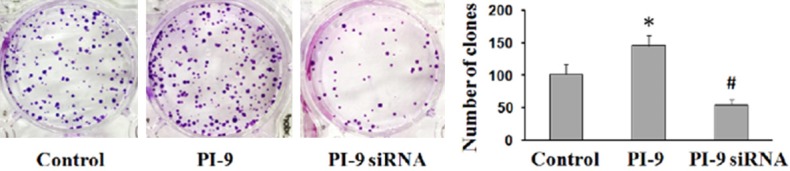
The capacity of colony formation was determined by colony formation assay. Quantitative data represent the mean ± SD of three independent experiments performed in triplicate. *P < 0.05, #P < 0.05.
Discussion
HCC is the most common primary liver cancer and the third leading cause of death from cancer worldwide, accounting for 50% of cancer-related deaths. Compared with chemotherapy, surgical resection is an effective treatment for liver cancer, but the 5 year survival rate is still low. However, in recent years, the monitoring and destruction of viral infections or cancer cells using the immune system has become a promising new method of treatment [10]. Therefore, the discovery of novel factors involved in the immune escape mechanism of HCC will provide effective benefits for treatments and prognosis of HCC patients. In this study, we find PI9 could take part in the immune escape mechanism and it could be a prognostic factor for HCC. However, this study collected HCC samples from a single clinical center and in the future we intend to collect samples from a large number of clinical research centers to verify our results.
PI9 is a novel serine protease inhibitor from the human placenta cDNA library. The protein contains around 374 amino acid residues and has a molecular mass of ~42 kD [11]. Our study showed for the first time that PI9 was significantly higher in HCC tissues than in the adjacent liver tissues by IHC analyses. PI9 expression has been detected in breast cancer, prostate cancer, and other tumors, and appears to be closely related to the occurrence, development, and prognosis of tumors [12,13]. For example, previous studies have significantly correlated the high expression of PI9 in lung cancer cells with tumor stage. Increasing the expression of PI9 was also found to effectively prevent GrB-mediated cytotoxicity by lung cancer cells. Van Houdt et al. [4] reported that PI9 is expressed in melanoma cells, and that high PI9 expression occurs in metastatic melanoma cells, and is closely related to overall survival and disease-free survival. They also observed that first diagnosis or patients or patients with recurrence with low activated lymphocyte ratios usually had a high proportion of activated tumor infiltrating lymphocytes. This type of melanoma cell after treatment by allergen specific immunotherapy (ASI) is usually linked to a good clinical prognosis. The abnormal expression of PI9 in hepatocellular carcinoma may be related to the tumor immune escape mechanism, suggesting that PI9 may play an important role in the mechanism of immune escape.
The abnormal expression of PI9 is suggested to be closely related to inflammatory factors and the formation of an inflammatory environment. Liver cancer cells produce many inflammatory cytokines including interleukin (IL)-1, IL-6, IL-8, IL-1β from stromal cells, and tumor necrosis factor (TNF)-α from hepatic stellate cells. An inflammatory microenvironment of proliferation formed by inflammatory cytokines and hepatic stellate cells, macrophages, and endothelial cells of the stromal matrix environment stimulates the liver to undergo chronic damage which may lead to the development of liver cancer [14]. The cell inflammatory microenvironment is an important factor involved in accelerating the growth of HCC, and recent studies showed that TNF-α and IL-1β can induce the expression of PI9 [15,16]. We conclude that liver inflammatory cytokines increased expression of PI9 in HCC cells. PI9 inhibited the apoptosis of hepatocellular carcinoma cells and promoted the proliferation of HCC cells, which plays an important role in the occurrence and development of HCC. These findings may provide new insight into the development of HCC.
Abnormal PI9 expression has also been associated with estrogen. Studies have shown that PI9 levels are regulated by control of PI9 gene expression by estrogen [17].
The liver is the target organ of estrogen, and HCC patients often incur estrogen imbalances while tumor cells may induce the expression of ectopic hormones. Long-term use of estrogen-containing oral contraceptives increases the risk of developing breast cancer, cervical cancer, and liver cancer according to a recent report by the International Agency for Research on Cancer (WHO) [17]. Estrogen can also prevent NK cells from killing cancer cells, which may further cancer development as well as immune surveillance and immune tolerance [18]. It can be concluded that an abnormal estrogen level may lead to the expression of PI9, which may promote the occurrence and development of liver cancer.
Although many studies suggest that PI9 plays an important role in the immune escape process, liver cancer-related clinical and prognostic studies are rare. Our study showed that the expression level of PI9 was correlated with TNM stage, degree of differentiation, and tumor size in HCC. Importantly, Kaplan-Meier survival analyses showed that high PI9 expression was significantly correlated with poor overall survival. Furthermore, multivariate Cox regression analyses indicated that PI9 expression was an independent risk factor for overall survival, suggesting that the high expression of PI9 may help in the identification of HCC patients with a poor prognosis. Poor prognosis is closely related to tumor cells through PI9 to escape immune surveillance, providing an immune escape related to immune killer cells. Our study showed PI9 enhanced the apoptosis and the proliferation of HCC cells in vitro experiments, suggesting a role in promoting tumor progression. We propose that liver inflammatory cytokines promote the expression of PI9 in HCC cells, and that the development of immune escape inhibits apoptosis of hepatoma cells and plays an important role in the occurrence of HCC.
Cellular immunity has a major role in the anti-tumor immune response. Effector cells are the main activators of CTL and NK cells, and apoptosis mediated by the GrB pathway is the most important. PI9 can prevent endogenous killing through the GrB pathway [19], and MEDEMA and other recent studies found that PI9 inhibits GrB activity in non-small cell lung cancers, which indicates that it contributes to the evasion of GrB-mediated apoptosis in this cancer [20]. This mechanism may involve the residues of the P1 site of PI9, which is an acidic amino acid. The Glu340 amino acid of GrB recognizes the P1 site, which makes RCL have a similar substrate structure. Despite the inhibitory effect of PI9 on the GrB pathway [2,21], it does not interfere with the self-deletion of Fas-mediated apoptosis [7].
PI9 was shown to be highly expressed in tissues that do not express GrB, such as cancers, various types of lymphoma, and sarcoma [22]. This indicates that PI9 may be involved in the immune escape of tumors by other means. A recent study found that PI9 not only inhibits GrB-mediated apoptosis, but also inhibits apoptosis mediated by TNF, TRAIL, Fas, and caspase-1 [23]. PI9 also directly binds caspase-8 and caspase-10, and prevents further downstream caspase-3 activation to inhibit cell death [22,24]. In future research, we will continue to study the mechanism of PI9 in liver cancer signaling pathways.
In conclusion, our studies found that the expression level of PI9 is closely related to the occurrence and development of liver cancer. We therefore propose that the PI9 gene could be an important marker in the diagnosis of liver cancer, and may provide a novel target for the clinical treatment of antitumor immunity. However, further in-depth study is needed to fully understand the role of PI9 in the occurrence and development of HCC. This work provides a theoretical basis for further study and treatment mechanisms of HCC.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81070360) and Clinical medical science and technology of Jiangsu Technical Department (BL2014060); the project “medical leading talent and innovation team” (LJ201134) of Jiangsu Provincee; “CyanEngineering” of Jiangsu Province ([2010]27); Graduate innovation project (SJZZ16-0251) of Jiangsu Education Department.
Disclosure of conflict of interest
None.
References
- 1.Li W, Wang Y, Gao W, Zheng J. HCC with tumor thrombus entering the right atrium and inferior vena cava treated by percutaneous ablation. BMC Surg. 2017;17:21. doi: 10.1186/s12893-017-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun J, Ooms L, Bird CH, Sutton VR, Trapani JA, Bird PI. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9) J Biol Chem. 1997;272:15434–15441. doi: 10.1074/jbc.272.24.15434. [DOI] [PubMed] [Google Scholar]
- 3.Buzza MS, Hosking P, Bird PI. The granzyme B inhibitor, PI-9, is differentially expressed during placental development and up-regulated in hydatidiform moles. Placenta. 2006;27:62–69. doi: 10.1016/j.placenta.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.van Houdt IS, Oudejans JJ, van den Eertwegh AJ, Baars A, Vos W, Bladergroen BA, Rimoldi D, Muris JJ, Hooijberg E, Gundy CM, Meijer CJ, Kummer JA. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin Cancer Res. 2005;11:6400–6407. doi: 10.1158/1078-0432.CCR-05-0306. [DOI] [PubMed] [Google Scholar]
- 5.Oudejans JJ, Harijadi H, Kummer JA, Tan IB, Bloemena E, Middeldorp JM, Bladergroen B, Dukers DF, Vos W, Meijer CJ. High numbers of granzyme B/CD8-positive tumour-infiltrating lymphocytes in nasopharyngeal carcinoma biopsies predict rapid fatal outcome in patients treated with curative intent. J Pathol. 2002;198:468–475. doi: 10.1002/path.1236. [DOI] [PubMed] [Google Scholar]
- 6.Kaiserman D, Bird PI. Control of granzymes by serpins. Cell Death Differ. 2010;17:586–595. doi: 10.1038/cdd.2009.169. [DOI] [PubMed] [Google Scholar]
- 7.Bird CH, Sutton VR, Sun J, Hirst CE, Novak A, Kumar S, Trapani JA, Bird PI. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol Cell Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Chen P, Liao R, Li YW, Yi Y, Wang JX, Sun TW, Zhou J, Shi YH, Yang XR, Jin JJ, Cheng YF, Fan J, Qiu SJ. Overexpression of galectin-1 is associated with poor prognosis in human hepatocellular carcinoma following resection. J Gastroenterol Hepatol. 2012;27:1312–1319. doi: 10.1111/j.1440-1746.2012.07130.x. [DOI] [PubMed] [Google Scholar]
- 9.Yi Y, Wu H, Gao Q, He HW, Li YW, Cai XY, Wang JX, Zhou J, Cheng YF, Jin JJ, Fan J, Qiu SJ. Interferon regulatory factor (IRF)-1 and IRF-2 are associated with prognosis and tumor invasion in HCC. Ann Surg Oncol. 2013;20:267–276. doi: 10.1245/s10434-012-2487-z. [DOI] [PubMed] [Google Scholar]
- 10.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Stephens R, Mirza G, Kanai H, Ragoussis J, Bird PI. A serpin gene cluster on human chromosome 6p25 contains PI6, PI9 and ELANH2 which have a common structure almost identical to the 18q21 ovalbumin serpin genes. Cytogenet Cell Genet. 1998;82:273–277. doi: 10.1159/000015118. [DOI] [PubMed] [Google Scholar]
- 12.Soriano C, Mukaro V, Hodge G, Ahern J, Holmes M, Jersmann H, Moffat D, Meredith D, Jurisevic C, Reynolds PN, Hodge S. Increased proteinase inhibitor-9 (PI-9) and reduced granzyme B in lung cancer: mechanism for immune evasion? Lung Cancer. 2012;77:38–45. doi: 10.1016/j.lungcan.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Ray M, Hostetter DR, Loeb CR, Simko J, Craik CS. Inhibition of Granzyme B by PI-9 protects prostate cancer cells from apoptosis. Prostate. 2012;72:846–855. doi: 10.1002/pros.21486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carloni V, Luong TV, Rombouts K. Hepatic stellate cells and extracellular matrix in hepatocellular carcinoma: more complicated than ever. Liver Int. 2014;34:834–843. doi: 10.1111/liv.12465. [DOI] [PubMed] [Google Scholar]
- 15.Kaiserman D, Knaggs S, Scarff KL, Gillard A, Mirza G, Cadman M, McKeone R, Denny P, Cooley J, Benarafa C, Remold-O’Donnell E, Ragoussis J, Bird PI. Comparison of human chromosome 6p25 with mouse chromosome 13 reveals a greatly expanded ov-serpin gene repertoire in the mouse. Genomics. 2002;79:349–362. doi: 10.1006/geno.2002.6716. [DOI] [PubMed] [Google Scholar]
- 16.Kannan-Thulasiraman P, Shapiro DJ. Modulators of inflammation use nuclear factor-kappa B and activator protein-1 sites to induce the caspase-1 and granzyme B inhibitor, proteinase inhibitor 9. J Biol Chem. 2002;277:41230–41239. doi: 10.1074/jbc.M200379200. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Orr BA, Kranz DM, Shapiro DJ. Estrogen induction of the granzyme B inhibitor, proteinase inhibitor 9, protects cells against apoptosis mediated by cytotoxic T lymphocytes and natural killer cells. Endocrinology. 2006;147:1419–1426. doi: 10.1210/en.2005-0996. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X, Ellison SJ, Alarid ET, Shapiro DJ. Interplay between the levels of estrogen and estrogen receptor controls the level of the granzyme inhibitor, proteinase inhibitor 9 and susceptibility to immune surveillance by natural killer cells. Oncogene. 2007;26:4106–4114. doi: 10.1038/sj.onc.1210197. [DOI] [PubMed] [Google Scholar]
- 19.Lefrancois L, Obar JJ. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol Rev. 2010;235:206–218. doi: 10.1111/j.0105-2896.2010.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medema JP, de Jong J, Peltenburg LT, Verdegaal EM, Gorter A, Bres SA, Franken KL, Hahne M, Albar JP, Melief CJ, Offringa R. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc Natl Acad Sci U S A. 2001;98:11515–11520. doi: 10.1073/pnas.201398198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, Trapani JA, Bird PI. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 22.Kummer JA, Micheau O, Schneider P, Bovenschen N, Broekhuizen R, Quadir R, Strik MC, Hack CE, Tschopp J. Ectopic expression of the serine protease inhibitor PI9 modulates death receptor-mediated apoptosis. Cell Death Differ. 2007;14:1486–1496. doi: 10.1038/sj.cdd.4402152. [DOI] [PubMed] [Google Scholar]
- 23.Poe M, Blake JT, Boulton DA, Gammon M, Sigal NH, Wu JK, Zweerink HJ. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J Biol Chem. 1991;266:98–103. [PubMed] [Google Scholar]
- 24.Cunningham TD, Jiang X, Shapiro DJ. Expression of high levels of human proteinase inhibitor 9 blocks both perforin/granzyme and Fas/Fas ligand-mediated cytotoxicity. Cell Immunol. 2007;245:32–41. doi: 10.1016/j.cellimm.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]