Abstract
Early diagnosis is important to improve the prognosis of pancreatic cancer (PC). Identifying potential biomarkers is essential for the monitoring and treatment of PC. The long noncoding RNA (lncRNA) UFC1 has been identified as an oncogenic factor in many cancers. However, the expression of UFC1 and its potential role in diagnosis and prognosis of PC remain largely unknown. The present study aimed to investigate the role of serum UFC1 in diagnosis and prognosis. The results indicated that serum UFC1 expression was relatively higher in PC patients than that in healthy volunteers. ROC curve analysis revealed that the serum UFC1 levels could distinguish PC patients from healthy controls, with an AUC value of 0.810. In addition, the serum UFC1 expression level was associated with lymph nodes metastasis, distant metastasis, and clinical stage. Kaplan-Meier analysis indicated that patients with high UFC1 expression exhibited shorter progression-free survival (PFS) and overall survival (OS) than those with low UFC1 expression. Multivariate analysis demonstrated that clinical stage and UFC1 expression level were significant, independent prognostic factors in PC patients. Our data demonstrate that serum UFC1 might serve as a potential biomarker for diagnosis and prognosis of PC.
Keywords: Pancreatic cancer, long noncoding RNA, biomarker
Introduction
Pancreatic cancer (PC) is the fourth most common cause of cancer-associated mortality in the world and has a poor prognosis, with a 5-year survival rate lower than 10% [1]. Due to the hidden symptoms at early stages, fewer than 15% of patients are diagnosed with PC at a stage when they could be eligible for curative surgical resection [2]. To improve early detection and prognosis and to provide timely and effective treatment for high-risk patients, predictive biomarkers for PC are required. To date, carbohydrate antigen 199 (CA199) and CA242, which are currently used in clinical settings as serum biomarkers for PC, are inadequate for early screening and prognosis [3-5]. Therefore, novel biomarkers with better sensitivity and specificity for diagnosis and outcome prediction in PC are required.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs that are over 200 nucleotides in length and have no protein coding capacity. In recent years, lncRNAs have been shown to be involved in numerous signaling pathways. LncRNAs can regulate gene activation or inactivation by targeting different chromatin modification complexes [6]. LncRNAs can also recruit microRNAs as a scaffold or interact with mRNA-stabilizing proteins to disrupt gene regulation [7]. Accumulating evidence indicates that lncRNAs can act as biomarkers in tumor diagnosis and prognosis. For instance, the levels of the serum lncRNA LRB1 were significantly increased in patients with hepatocellular carcinoma (HCC) and positively correlated with AFP, tumor size, and tumor stage, and the serum levels of LRB1 were negatively correlated with patients’ survival [8]. Other studies demonstrated that serum HOTAIR could serve as a novel biomarker for the diagnosis of esophageal carcinoma [9]. The effect of the lncRNA UFC1 was first reported in HCC cells. The expression of UFC1 was higher in HCC tissues than in control tissues, and this lncRNA could increase the levels of β-catenin by interacting with the mRNA stabilizing protein HuR [10]. Subsequent studies validated the tumor-promoting effect of UFC1 in gastric and colorectal cancer [11,12]. However, the associations between UFC1 and PC are still unclear.
In the present study, we investigated the serum levels of UFC1 in PC patients and healthy volunteers. We further analyzed the value of UFC1 in clinical application for diagnosis and prognosis of PC.
Material and methods
Patients and serum samples
In this study, serum samples were collected from 48 patients who were diagnosed with PC at Zhejiang Cancer Hospital between January 2012 and December 2014. No patient had received any form of anti-cancer therapy before serum collection. Meanwhile, 40 age-matched healthy blood serum controls were collected from the health examination center of Zhejiang Cancer Hospital between February 2014 and December 2014. All samples were centrifuged at 10,000 rpm for 10 min at 4°C. Then, the serum supernatant was carefully transferred into a fresh tube and stored at -80°C for further analyses. This study was approved by the ethics committee of Zhejiang Cancer Hospital, and informed consent was obtained from all patients and healthy volunteers.
Quantitative RT-PCR
Total RNA was isolated from serum samples using the RNA Isolation Kit (Axygen Scientific, Inc., Union City, CA, USA) and was reverse-transcribed with a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s instructions. qRT-PCR was performed on a 7500 Fast Real-Time PCR system (Applied Biosystem, Rotkreuz, Switzerland) using FastStart Universal SYBR Green Master (Roche, Stockholm, Sweden) according to the manufacturer’s protocol. The primers were as follows: UFC1 forward, 5’-TCCAACCTGAGTGACATAGCGA-3’ and reverse, 5’-CTGACCTCCAACTCCAACGAAT-3’; U6 forward, 5’-GCTTCGGCAGCACATATACTAAAAT-3’ and reverse, 5’-CGCTTCACGAATTTGCGTGTCAT-3’. U6 was used as the endogenous control. Each experiment was performed in triplicate. The mean expression level of UFC1 was used as the cut-off value. According to the cut-off value of UFC1, all patients with PC were divided into two groups.
Statistical analysis
All statistical data were analyzed using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). Associations between UFC1 and patients’ clinicopathologic characteristics were analyzed using Pearson’s χ2 test or Fisher’s exact test. Survival curves were established using the Kaplan-Meier method, and significant differences were compared by log-rank test. The prognostic value was determined by univariate and multivariate Cox regression analysis. A p-value < 0.05 was considered significant.
Results
Expression of serum UFC1 in PC patients and its diagnostic efficacy
In this study, we first explored the expression levels of UFC1 in the sera from 48 patients who were diagnosed with PC and 40 healthy volunteers. As revealed by qRT-PCR, UFC1 expression was remarkably higher in sera from PC patients compared with that from healthy controls (P < 0.01, Figure 1A). ROC curve analysis revealed that the serum UFC1 level could distinguish PC patients from healthy controls, with an AUC value of 0.810 (95% CI: 0.719 to 0.900, P < 0.001, Figure 1B).
Figure 1.
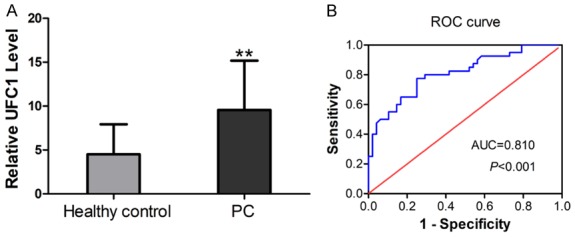
Serum UFC1 levels as a diagnostic biomarker. A. Comparison of the serum levels of UFC1 between PC patients and controls. B. ROC curve analysis for serum UFC1 in the diagnosis of PC.
Correlations of UFC1 expression with clinicopathologic features of PC patients
As shown in Table 1, the serum UFC1 expression level was associated with lymph nodes metastasis, distant metastasis and clinical stage (all P value < 0.05). However, there was no significant association with sex, age, tumor location, tumor differentiation, and local invasion. These data indicated that UFC1 may play an oncogenic role in PC progression.
Table 1.
Correlation between UFC1 expression and clinicopathologic characteristics in pancreatic cancer
| Factor | UFC1 expression | P value | |
|---|---|---|---|
|
| |||
| Low (n=29) | High (n=19) | ||
| Gender | 0.923 | ||
| Male | 21 (72.4%) | 14 (73.7%) | |
| Female | 8 (27.6%) | 5 (26.3%) | |
| Age | 0.315 | ||
| ≤ 60 | 18 (62.1%) | 9 (47.4%) | |
| > 60 | 11 (37.9%) | 10 (52.6%) | |
| Tumor location | 0.969 | ||
| Head | 19 (65.5%) | 13 (68.4%) | |
| Body | 7 (24.1%) | 4 (21.1%) | |
| Tail | 3 (10.3%) | 2 (10.5%) | |
| Differentiation | 0.945 | ||
| Well | 22 (75.9) | 15 (78.9%) | |
| Moderate | 4 (13.8%) | 2 (10.5%) | |
| Poor | 3 (10.3%) | 2 (10.5%) | |
| Local invasion | 0.057 | ||
| T1-T2 | 14 (48.3%) | 4 (21.1%) | |
| T3-T4 | 15 (51.7%) | 15 (78.9%) | |
| Lymph nodes metastasis | 0.015 | ||
| Negative | 18 (62.1%) | 5 (26.3%) | |
| Positive | 11 (37.9%) | 14 (73.7%) | |
| Distant metastasis | 0.004 | ||
| M0 | 25 (86.2%) | 9 (47.4%) | |
| M1 | 4 (13.8%) | 10 (52.6%) | |
| Clinical stage | 0.015 | ||
| I-II | 21 (72.4%) | 7 (36.8%) | |
| III-IV | 8 (27.6%) | 12 (63.2%) | |
Prognostic values of UFC1 level in PC
To investigate the correlation between UFC1 expression and PC patient prognosis, Kaplan-Meier analysis and log-rank tests were performed. As shown in Figure 2, PC patients with high UFC1 expression exhibited shorter progression-free survival (PFS) and overall survival (OS) than those with low UFC1 expression (all P value < 0.05). In addition, univariate and multivariate analyses were performed to further evaluate whether the expression level of UFC1 could be used as a prognostic factor for PC patients. As shown in Table 2, clinical stage and UFC1 expression level were significantly correlated with PFS and OS in PC patients (all P values < 0.05). Multivariate analysis demonstrated that clinical stage and UFC1 expression level were also significant, independent prognostic factors for PC patients (all P values < 0.05).
Figure 2.
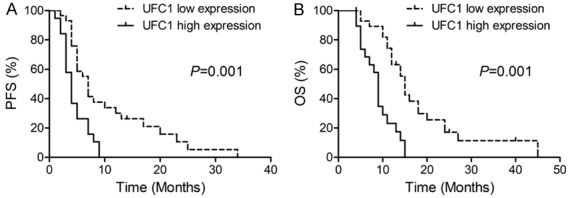
Kaplan-Meier curves of (A) PFS and (B) OS according to the level of lncRNA-UFC1 expression.
Table 2.
Univariate and multivariate Cox regression analyses of OS and PFS in pancreatic cancer patients
| Overall survival | P | Progression-free survival | P | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | HR | 95% CI | |||
| Univariate analyses | ||||||
| Gender | 0.802 | 0.397-1.619 | 0.538 | 0.818 | 0.419-1.598 | 0.557 |
| Age | 1.696 | 0.897-3.207 | 0.104 | 1.596 | 0.848-3.005 | 0.147 |
| Tumor location | 0.947 | 0.485-1.848 | 0.873 | 0.734 | 0.382-1.411 | 0.354 |
| Differentiation | 0.895 | 0.416-1.925 | 0.776 | 0.824 | 0.381-1.784 | 0.623 |
| Clinical stage | 2.315 | 1.231-4.354 | 0.009 | 2.049 | 1.099-3.819 | 0.024 |
| UFC1 expression | 3.943 | 1.943-8.004 | < 0.001 | 2.592 | 1.336-5.029 | 0.005 |
| Multivariate analyses | ||||||
| Gender | 1.013 | 0.455-2.256 | 0.974 | 0.551 | 0.255-1.188 | 0.129 |
| Age | 1.658 | 0.770-3.568 | 0.197 | 1.440 | 0.667-3.109 | 0.353 |
| Tumor location | 0.860 | 0.365-2.029 | 0.731 | 0.493 | 0.214-1.135 | 0.097 |
| Differentiation | 0.966 | 0.380-2.460 | 0.943 | 2.031 | 0.751-5.489 | 0.163 |
| Clinical stage | 2.261 | 1.134-4.506 | 0.020 | 2.091 | 1.060-4.125 | 0.033 |
| UFC1 expression | 4.410 | 2.061-9.437 | < 0.001 | 2.828 | 1.367-5.848 | 0.005 |
Discussion
To date, this is the first investigation that describes the role of UFC1 in PC. In the present study, we investigated the clinical significance of serum UFC1 in PC patients. Serum UFC1 expression was relatively higher in PC patients than that of controls. Further analysis demonstrated that UFC1 expression was correlated with lymph node metastasis and distant metastasis. Multivariate Cox regression analysis indicated that UFC1 could serve as an independent prognostic factor for PC patients. Collectively, UFC1 may serve as a biomarker for predicting the diagnosis and prognosis of PC.
PC is an aggressive tumor with poor prognosis. Early screening and prognostic analyses are important for the diagnosis and early treatment of PC. In recent decades, numerous studies have identified potential biomarkers for the prediction of diagnosis and survival in patients with cancer [13-15]. Recently, emerging evidence has demonstrated that lncRNAs can play essential roles in tumor progression and may serve as biomarkers and prognostic factors for many cancers [16-18]. For example, high levels of HOTAIR are a powerful predictor of survival and progression for many cancers, including breast cancer [19], hepatocellular carcinoma [20] and PC [21]. High levels of lncRNA MALAT1 are associated with shorter recurrence-free survival, so this lncRNA may play a role in tumor progression [22]. In this study, we demonstrated that the serum UFC1 level was relatively higher in PC patients than in control volunteers and that UFC1 levels correlate with a worse prognosis in PC patients. Importantly, UFC1 has been reported to act as an oncogene in several carcinomas, including hepatocellular [10], gastric [11], and colorectal cancer [12], which also supports the results of our study. In HCC, UFC1 is highly expressed and promotes cancer progression by increasing levels of β-catenin through interaction with the mRNA stabilizing protein HuR [10]. However, the underlying mechanisms by which UFC1 promotes PC progression remain largely unknown. The detailed mechanisms of UFC1 in PC progression will be investigated in the future.
In conclusion, the results of our study indicated that the expression of UFC1 in serum could be used as a potential diagnostic biomarker and prognosis factor for PC.
Acknowledgements
This work was supported by the grants from the Zhejiang Medical and Health Science and Technology Project (2017KY030).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakai Y, Isayama H, Sasaki T, Takahara N, Hamada T, Uchino R, Mizuno S, Miyabayashi K, Yamamoto K, Mohri D, Kogure H, Yamamoto N, Hirano K, Ijichi H, Tateishi K, Tada M, Koike K. A retrospective analysis of early CA199 change in salvage chemotherapy for refractory pancreatic cancer. Cancer Chemother Pharmacol. 2013;72:1291–1297. doi: 10.1007/s00280-013-2313-8. [DOI] [PubMed] [Google Scholar]
- 4.Parra-Robert M, Santos VM, Canis SM, Pla XF, Fradera JMA, Porto RM. Relationship between CA 19.9 and the lewis phenotype: options to improve diagnostic efficiency. Anticancer Res. 2018;38:5883–5888. doi: 10.21873/anticanres.12931. [DOI] [PubMed] [Google Scholar]
- 5.Lei XF, Jia SZ, Ye J, Qiao YL, Zhao GM, Li XH, Chang H. Application values of detection of serum CA199, CA242 and CA50 in the diagnosis of pancreatic cancer. J Biol Regul Homeost Agents. 2017;31:383–388. [PubMed] [Google Scholar]
- 6.Lin XQ, Huang ZM, Chen X, Wu F, Wu W. XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J. 2018;59:816–826. doi: 10.3349/ymj.2018.59.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malek R, Gajula RP, Williams RD, Nghiem B, Simons BW, Nugent K, Wang H, Taparra K, Lemtiri-Chlieh G, Yoon AR, True L, An SS, DeWeese TL, Ross AE, Schaeffer EM, Pienta KJ, Hurley PJ, Morrissey C, Tran PT. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77:3181–3193. doi: 10.1158/0008-5472.CAN-16-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZF, Hu R, Pang JM, Zhang GZ, Yan W, Li ZN. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett. 2018;16:1593–1601. doi: 10.3892/ol.2018.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, He X, Zheng Z, Ma X, Hu X, Wu D, Wang M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. 2017;16:75. doi: 10.1186/s12943-017-0643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–426. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Liang W, Liu J, Zang X, Gu J, Pan L, Shi H, Fu M, Huang Z, Zhang Y, Qian H, Jiang P, Xu W. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. 2018;37:134. doi: 10.1186/s13046-018-0803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu T, Shan TD, Li JY, Huang CZ, Wang SY, Ouyang H, Lu XJ, Xu JH, Zhong W, Chen QK. Knockdown of linc-UFC1 suppresses proliferation and induces apoptosis of colorectal cancer. Cell Death Dis. 2016;7:e2228. doi: 10.1038/cddis.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riely GL. What, when, and how of biomarker testing in non-small cell lung cancer. J Natl Compr Canc Netw. 2017;15:686–688. doi: 10.6004/jnccn.2017.0073. [DOI] [PubMed] [Google Scholar]
- 14.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 15.Peng HY, Chang MC, Hu CM, Yang HI, Lee WH, Chang YT. Thrombospondin-2 is a highly specific diagnostic marker and is associated with prognosis in pancreatic cancer. Ann Surg Oncol. 2019;26:807–814. doi: 10.1245/s10434-018-07109-6. [DOI] [PubMed] [Google Scholar]
- 16.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolha L, Ravnik-Glavac M, Glavac D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 19.Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, Benedetto C, Yu H. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Katsaros D, Biglia N, Shen Y, Fu Y, Loo LWM, Jia W, Obata Y, Yu H. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res Treat. 2018;171:261–271. doi: 10.1007/s10549-018-4839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


