Abstract
Background: Long non-coding RNAs (lncRNAs) have been widely confirmed to modulate many tumorigeneses, including NPC. However, the exact roles of cancer susceptibility candidate 9 (CASC9) in nasopharyngeal carcinoma (NPC) and its underlying mechanisms have not been fully established. Methods: qRT-PCR was used to determine CASC9 and miR-145 expressions. Cell apoptosis, migration, and invasion were determined by flow cytometry and transwell assays, respectively. The protein expressions of BAX, Bcl-2, MMP 9, and MMP 2 were measured by western blot. The possible binding sites between miR-145 and CASC9 were predicted by the starBase v2.0 online database and verified by a luciferase report and an RNA immunoprecipitation (RIP) assay. A xenograft tumor model was established to confirm the effects of CASC9 in NPC progression in vivo. Results: The expression level of CASC9 was upregulated in NPC tissues and cells. The knockdown of CASC9 evidently suppressed migration and invasion but promoted apoptosis in NPC cells. In addition, the inhibition of CASC9 evidently increased the BAX protein level and inhibited the expression of the Bcl-2, MMP 9, and MPP2 proteins in NPC cells. Moreover, miR-145 was directly bound to CASC9, and its inhibition reversed the inhibitory effect of CASC9 knockdown on the progression of NPC. Furthermore, the expression of miR-145 was decreased and negatively associated with CASC9 in NPC tissues and cells. Also, the knockdown of CASC9 inhibited tumor growth in vivo. Conclusion: CASC9 knockdown inhibited cell migration and invasion but increased cell apoptosis in NPC cells by regulating miR-145, providing a novel insight for the treatment of NPC.
Keywords: Nasopharyngeal carcinoma, CASC9, miR-145, migration and invasion, apoptosis
Introduction
Nasopharyngeal carcinoma (NPC) has a high malignancy and is the most prevalent head and neck malignancy in southern China and Southeast Asia [1,2]. Recently, despite advanced treatments, including radiotherapy and chemotherapy, the prognosis for advanced NPC remains quite poor [3]. Hence, elucidating the molecular mechanisms of NPC and searching for new molecular targets are of marked importance for NPC patients.
Long non-coding RNAs (lncRNAs) are a class of RNAs of more than 200 nucleotides in length that lack a protein-coding capacity [4]. Recently, lncRNAs have been reported to play important roles in tumorigenesis and progression in various cancers, including NPC [5]. For example, the lncRNA HOTAIR is a potential biomarker for the prognosis of NPC, and its dysregulation might play a vital role in NPC progression [6]. The lncRNA cancer susceptibility candidate 9 (CASC9) gene is located at 8q21.11, and it was first reported in esophageal squamous cell carcinoma (ESCC), and it is upregulated in ESCC tissues [7]. Recent evidence has indicated that CASC9 also plays key roles in other types of cancers, such as pancreatic ductal adenocarcinoma (PDAC), gastric cancer, and nasopharyngeal carcinoma [8-10]. However, the exact roles of CASC9 in the progression of NPC and its underlying mechanism remain poorly understood.
At present, increasing evidence has suggested that lncRNAs can interact with miRNAs and regulate the expression of miRNAs as competing endogenous RNA (ceRNA) [11]. MicroRNAs (miRNAs) are a group of short single-stranded non-coding RNAs, 22 nucleotides in length, that regulate gene expression by pairing to the 3’ untranslated regions (3’-UTR) of targets [12]. MiR-145 has been identified as a tumor suppressor in bladder cancer, colorectal cancer, and so on [13,14]. Moreover, a previous report showed that miR-145 acts as a tumor suppressor in NPC development and progression by targeting FSCN1 [15]. In spite of these findings, the functions and regulatory mechanisms of miR-145 in the progression of NPC need to be further investigated.
In this study, we found that the level of CASC9 was clearly increased in NPC tissues and cells, and that a higher CASC9 level was associated with the poorer overall survival of NPC patients. Moreover, we also demonstrated that the knockdown CASC9 restrained migration and invasion but promoted the apoptosis of NPC cells and tumor growth by regulating miR-145 in vivo. Collectively, our results for the first time identified the important functions of the CASC9/miR-145 axis in NPC, which may provide a new direction for the treatment of NPC.
Materials and methods
Clinical samples and cell culture
In this study, forty seven pairs of NPC tissues and adjacent normal tissues were kindly provided by the Department of Rhinology, the First Affiliated Hospital of Zhengzhou University during the period from June 2018 to November 2018. Informed consents were received from all subjects, and this study was approved by the Ethics Committee of the Department of Rhinology, the First Affiliated Hospital of Zhengzhou University. All tissues were frozen in liquid nitrogen and then stored at -80°C until the experiments were carried out.
The normal nasopharyngeal cell line NP69 and the NPC cell lines HNE1, SUNE1, CNE1, and HONE1 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in an RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Gibco, Carlsbad, CA, USA) at 37°C in an incubator supplemented with 5% CO2.
Reagent and cell transfection
Small interference RNA targeting CASC9 (si-CASC9), the siRNA negative control (sh-NC), the CASC9 overexpression vector (CASC9), the empty vector, the miR-145 mimic (miR-145), the miR-negative control (miR-NC), the miR-145 inhibitor (anti-miR-145), and the anti-miR-negative control (anti-miR-NC) were obtained from GenePharma (Shanghai, China). CNE1 and HONE1 cells were transfected with oligonucleotides or a vector using Lipofectamine 3000 (Invitrogen) referring to the manufacturer’s instructions.
qRT-PCR
Total RNA was extracted from the tissues or cells (after transfection for 48 h) according to the TRIzol kit’s instructions (Invitrogen). The extracted RNA was reverse transcribed into cDNA according to the reverse transcription kit’s instructions (Applied Biosystems, Foster City, CA, USA) or the microRNA reverse transcription Kit (Applied Biosystems). Subsequently, a qPCR analysis was performed using a SYBR Green PCR Kit (Toyobo, Tokyo, Japan) on an ABI 7500HT fast real-time PCR System (Applied Biosystems). Gene-specific RT-qPCR primers were obtained from Sangon Biotech (Shanghai, China) and were as follows: CASC9 (Forward, 5’-AGATGAAGCCGGTACCTCAGAT-3’; Reverse, 5’-TCACTTTAAAGAGGGAGAGGAG-3’); miR-145 (Forward, 5’-CAGTGCGTGTCGTGGAGT-3’; Reverse, 5’-AGGTCCAGTTTTCCCAGG-3’); β-actin (Forward, 5’-TGG ACTTCGAGCAGGAAATGG-3’; Reverse, 5’-ACGTCGCAC TTCATGATCGAG3’); U6 (Forward, 5’-ATTGGAACGATACAGAGAAGATT-3’; Reverse, 5’-GGAACGCTTCACGAATTTG3’). The results were normalized to endogenous U6 small RNA or β-actin using the 2-ΔΔCt method.
Cell migration and invasion assay
The cell migration and invasion abilities were measured using a cell migration and invasion assay. For the cell migration, CNE1 and HONE1 cells (5 × 104 cells/well) transfected with different reagents were re-suspended in 100 μl of a serum-free medium and plated in the upper chamber (Corning Costar, NY, USA) with an 8 μm porous membrane, while 500 μl of the medium supplemented with 10% FBS was placed into the lower chamber. After incubation for 24 h, the CNE1 and HONE1 cells on the upper surface of the membrane were removed, and the migrated cells on the lower surfaces were fixed with methanol formaldehyde and stained with 0.5% crystal violet (Sigma) at room temperature for 20 min. The stained cells were counted under a microscope (Olympus, Tokyo, Japan). For the migration assay, the membranes of the transwell chambers were pre-coated with Matrigel (BD, San Jose, CA, USA), and the procedure was performed in the same manner as the migration assay.
Cell apoptosis
Apoptosis in cultured cells was determined by flow cytometry using an Annexin V-FITC/PI apoptosis kit (Sigma). In brief, CNE1 and HONE1 cells were seeded in 6-well plates and transfected with different reagents. After transfection for 48 h, the CNE1 and HONE1 cells were collected and stained with Annexin V-FITC and PI for 20 min in a dark room. Finally, the cell apoptosis was quantified by flow cytometry (B.D. FACS Calibur) and analyzed using the flow cytometry software.
Western blot
The cells (after transfection for 48 h) or tissues were lysed in a RIPA lysis buffer (Thermo Fisher, Wilmington, DE, USA) and supplemented with a protease inhibitor (Sigma) on ice for 30 min, followed by centrifugation at 12,000 g for 15 min to collect the supernatant of the protein. After that, the concentration of total protein was determined using a bicinchoninic acid (BCA) Protein Assay Kit (Beyotime, Shanghai, China). Next, equal amounts of the proteins were separated by 10-12% SDS-PAGE gels and transferred onto 0.22 μm polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked in Tris-buffer saline containing 0.1% Tween 20 (TBST) supplemented with 5% non-fat milk at room temperature for 2 h, and then incubated with primary antibodies against BAX (1:1000; #14796), Bcl-2 (1:1000; #3498), MMP 9 (1:1000; #13667), MMP 2 (1:1000; #40994) and β-actin (1:1000; #4970) (Cell Signaling Technology, Beverly, MA, USA) overnight at 4°C. The membranes were subsequently incubated with HRP-conjugated secondary antibodies (1:4000, Sangon Biotech) at room temperature for 2 h. Finally, the protein signals were examined with enhanced chemiluminescence (ECL; Tanon, Shanghai, China). The relative protein expression levels were normalized to the levels of β-actin and quantified automatically using ImageJ software.
Luciferase activity assay
The putative binding sites of miR-145 and CASA9 were predicted by starBase v2.0. The sequences of wild type CASA9 (CASC9-WT) and mutant CASA9 (CASC9-MUT) with a predicted binding site to human miR-145 were amplified and cloned into the pGL3 luciferase reporter vectors (Promega, Madison, WI, USA). The miR-145 or miR-NC with CASC9-WT or CASC9-MUT reporter vectors were co-transfected into 293T cells. After incubation for 48 h, the luciferase activities were measured using a dual luciferase assay kit (Promega), following the manufacturer’s instructions.
RNA immunoprecipitation (RIP)
The relationship between miR-145 and CASA9 was detected by using a Magna RIP Kit (Millipore) according to the manufacturer’s protocol. Briefly, the CNE1 and HONE1 cells were transfected with miR-145 or miR-NC for 48 h. After that, the CNE1 and HONE1 cells were lysed using an RIP lysis buffer and incubated with magnetic beads conjugated with antibody argonaute2 (Ago2, Millipore) or against IgG (Millipore). The level of CASA9 in Ago2 or IgG immunoprecipitation complexes was measured using qRT-PCR.
Tumor xenograft model
Six-week-old female BALB/c nude mice (n = 5 per group) were purchased from the Henan Experimental Animals Center (Zhengzhou, China). The nude mice experiments were approved by the committee of Animal Research of the Department of Rhinology, the First Affiliated Hospital of Zhengzhou University. Lentivirus-mediated shRNA interference targeting (sh-CASC9) and an empty lentiviral vector (sh-NC) were constructed by GenePharma (Shanghai, China). CNE1 cells (1 × 107) transduced with sh-CASC9 or sh-NC were injected subcutaneously into BALB/c nude mice. The tumor volume was gauged every seven days with a caliper and calculated according to the formula: V = length × width2/2. All the mice were sacrificed after injection for 35 days, and the tumor specimens were isolated from all the nude mice and used for further analysis.
Statistical analysis
All of the data were repeated more than three times and as presented as the mean ± standard deviation (SD). Student’s t test was used for comparisons between two groups. P<0.05 was generally regarded as statistically significant.
Results
CASC9 expression was increased in NPC tissues and cells
To investigate the potential roles of CASC9 in NPC, its expression was determined in NPC tissues and cells. An analysis of qRT-PCR indicated that the expression level of CASC9 was clearly increased in NPC tissues (n = 47) compared with the levels in the adjacent normal tissues (Figure 1A). Similarly, the expression of CASC9 was also markedly higher in the NPC (HNE1, SUNE1, CNE1, and HONE1) cells than it was in the NP69 cells (Figure 1B). Moreover, the patients with NPC were classified into a high CASC9 expression group and a low CASC9 expression group according to the CASC9 mean value of abundance. A Kaplan-Meier analysis showed that the patients in the high CASC9 expression group had a lower survival rate than those with a low expression of CASC9 (P = 0.03) (Figure 1C). These data revealed that CASC9 might play vital roles in the development of NPC, which prompted us to investigate its biological function more deeply.
Figure 1.
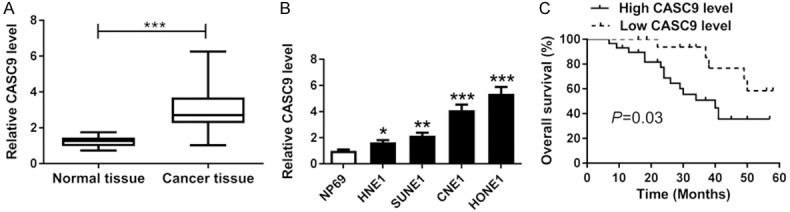
CASC9 expression was elevated in NPC tissues and cells. A. The expression level of CASC9 was measured in the NPC tissues and the adjacent normal samples by qRT-PCR. B. The abundance of CASC9 was measured in the NP69 and NPC cells (HNE1, SUNE1, CNE1 and HONE1) by qRT-PCR. C. The survival rate of NPC patients with high CASC9 expressions was significant lower than the rate of those with low CASC9 expressions. *P<0.05, **P<0.01, ***P<0.001.
The knockdown of CASC9 inhibited migration and invasion but promoted apoptosis in the NPC cells
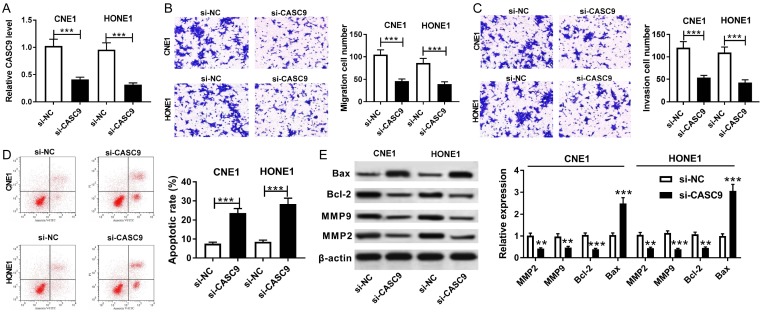
To explore the effect of CASC9 on NPC progression, si-CASC9 or si-NC was transfected into the CNE1 and HONE1 cells. The qRT-PCR assay revealed that the transfection of si-CASC9 led to a significant reduction of CASC9 expression in the CNE1 and HONE1 cells (Figure 2A). Moreover, the knockdown of CASC9 resulted in a great loss of the migration and invasion abilities in the CNE1 and HONE1 cells (Figure 2B and 2C). A flow cytometry analysis indicated that the apoptotic rate was prominently enhanced in the CNE1 and HONE1 cells transfected with si-CASC9 compared with the rate in the si-NC group (Figure 2D). Besides, the apoptosis-related and matrix metalloproteinases proteins were analyzed in the CNE1 and HONE1 cells by western blot. The results showed that the inhibition of CASC9 evidently increased the BAX protein level and prominently inhibited the expressions of the Bcl-2, MMP 9, and MPP2 proteins in CNE1 and HONE1 (Figure 2E). Our data suggest that the knockdown CASC9 restrained cell migration and invasion and elevated apoptosis in CNE1 and HONE1.
Figure 2.
The knockdown of CASC9 promoted cell apoptosis and decreased migration and invasion in NPC cells. A. The expression of CASC9 was measured in CNE1 and HONE1 transfected with si-CASC9 or si-NC by qRT-PCR. B-D. A transwell assay and flow cytometry were performed to determine the migration, invasion, and apoptosis of CNE1 and HONE1 transfected with si-CASC9 or si-NC. E. The protein expressions of BAX, Bcl-2, MMP 9, and MMP 2 were measured in CNE1 and HONE1 transfected with si-CASC9 or si-NC by western blot. **P<0.01, ***P<0.001.
miR-145 was a direct target of CASC9 in NPC cells
lncRNAs have been reported to act as a molecular sponge or as competing endogenous RNAs (ceRNAs) to regulate the accumulation of miRNA, in turn affecting its biological function. Using the web-based tool Starbase v2.0, we found that miR-145 was predicted to have complementary bases pairing with CASC9 (Figure 3A). Subsequently, the prediction was confirmed by the luciferase activity and an RIP analysis. The results showed that the transfection of miR-145 reduced the luciferase activity in 293T cells transfected with CASC9-WT, but no obvious effect was found on the relative luciferase activity in the 293T cells transfected with CASC9-MUT (Figure 3B). Moreover, an RIP assay showed that the transduction of miR-145 resulted in the substantial enrichment of CASC9 in the Ago2 group versus the IgG control group of CNE1 and HONE1 cells (Figure 3C and 3D). Taken together, these data suggest that miR-145 is a direct target of CASC9 in NPC cells.
Figure 3.
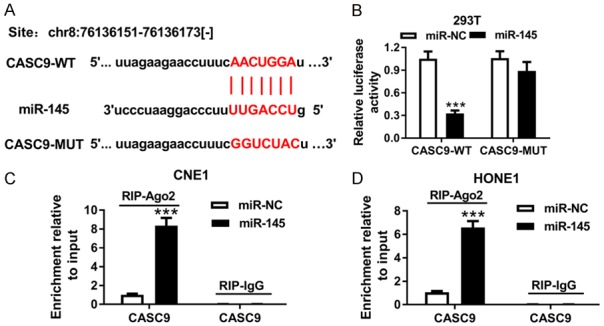
miR-145 was a direct target of CASC9. A. The potential binding sites of CASC9 and miR-145 were predicted by starBase v2.0. B. The luciferase activity in the 293T cells co-transfected with CASC9-WT or CASC9-MUT and miR-145 or miR-NC was detected using a luciferase report assay. C and D. An RIP assay was performed to detect the CASC9 enrichment level in the IgG or Ago2 immunoprecipitation complex of the CNE1 and HONE1 cells. ***P<0.001.
MiR-145 expression was downregulated and negatively associated with CASC9 in NPC tissues and cells
To probe the potential roles of miR-145 in NPC, its expression was first quantified in the NPC tissues and cells. A qRT-PCR assay revealed that the miR-145 expressions were clearly decreased in the NPC tissues and cells compared with the levels in the corresponding controls (Figure 4A and 4B). Moreover, the overexpression of CASC9 suppressed the expression of miR-145, but its knockdown showed the opposite effect in the CNE1 and HONE1 cells (Figure 4C and 4D). These data indicated miR-145 was negatively regulated by CASC9.
Figure 4.
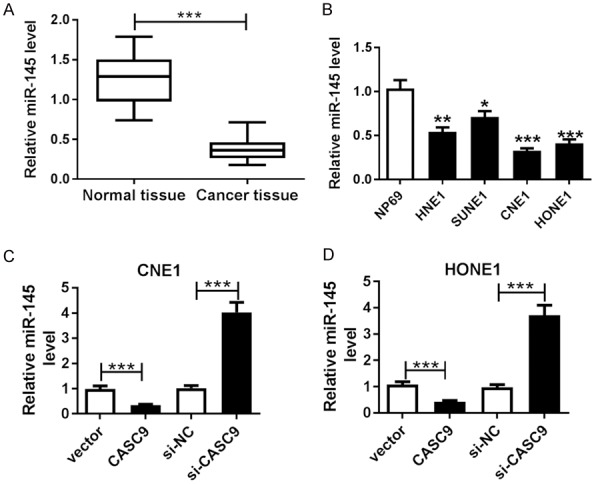
MiR-145 expression was decreased and negatively associated with CASC9 in the NPC tissues and cells. A and B. qRT-PCR was used to detect the miR-145 expression in the tissues and cells. C and D. The expression level of miR-145 was detected in the CNE1 and HONE1 cells transfected with CASCP, vector, si-CASC9 or si-NC by qRT-PCR. *P<0.05, **P<0.01, ***P<0.001.
The abrogation of miR-145 reversed the effects of the CASC9 knockdown on the migration, invasion, and apoptosis of the NPC cells
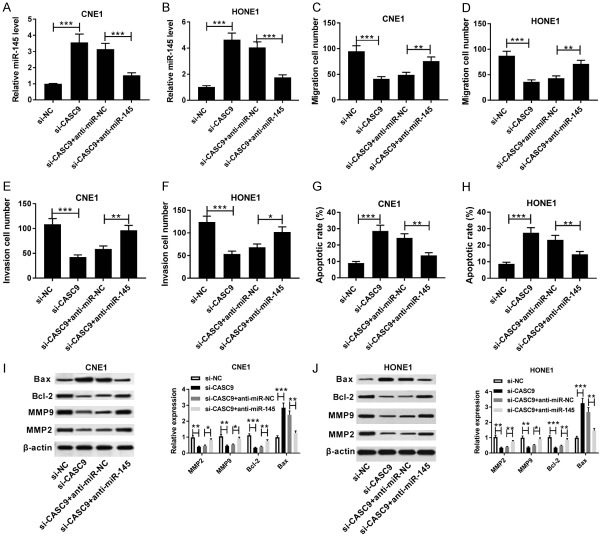
To further explore the relationship between miR-145 and CASC9 in NPC, si-CASC9, si-NC, si-CASC9+anti-miR-145 or si-CASC9+anti-miR-NC were transfected into CNE1 and HONE1 cells. The qRT-PCR assay showed that the expression of miR-145 was conspicuously elevated in the CNE1 and HONE1 cells transfected with si-CASC9, which was abated by inhibition of miR-145 (Figure 5A and 5B). Moreover, the down-regulation of miR-145 alleviated the inhibitory effect of knockdown CASC9 on the migration and invasion in the CNE1 and HONE1 cells (Figure 5C-F). Similarly, the transfection of si-CASC9 markedly limited the apoptosis rate in the CNE1 and HONE1 cells, which was reversed by the knockdown of miR-145 (Figure 5G and 5H). In addition, the abrogation of miR-145 weakened the si-CASC9-mediated promotion of BAX protein expression and the reduction of the expressions of the Bcl-2, MMP 9, and MPP 2 proteins in the CNE1 and HONE1 cells (Figure 5I and 5J). Our findings indicated that the knockdown of miR-145 reversed the CASC9 inhibition-mediated progression of NPC.
Figure 5.
The abrogation of miR-145 reversed the inhibitory effects of the CASC9 knockdown on NPC cells. A and B. qRT-PCR was used to determine the miR-145 expressions in the CNE1 and HONE1 cells transfected with si-CASCP, si-NC, si-CASCP+anti-miR-145 or si-CASCP+anti-miR-NC. C-H. A transwell assay and flow cytometry were used to determine the migration, invasion, and apoptosis of CNE1 and HONE1 transfected with si-CASCP, si-NC, si-CASCP+anti-miR-145 or si-CASCP+anti-miR-NC, respectively. I and J. The protein expressions of BAX, Bcl-2, MMP 9, and MMP 2 were measured in CNE1 and HONE1 transfected with si-CASCP, si-NC, si-CASCP+anti-miR-145 or si-CASCP+anti-miR-NC by western blot. *P<0.05, **P<0.01, ***P<0.001.
The knockdown of CASC9 decreased xenograft tumor growth in vivo
To further confirm the tumorigenicity of CASC9, CNE1 cells transfected with sh-CASC9 or sh-NC were introduced into nude mice. The tumor volumes were measured every 7 days, and the tumors’ weights and volumes were measured after 35 days of injection. The knockdown of CASC9 evidently suppressed tumor volume and weight in the CNE1 xenograft model (Figure 6A and 6B). In addition, the expression of CASC9 was clearly lower in the tumor tissues from the sh-NEAT1 group than they were in the sh-NC group (Figure 6C). Furthermore, the knockdown of CASC9 upregulated the expression level of miR-145 in tumor tissues (Figure 6D). Moreover, the deficiency of CASC9 led to a significant increase in the BAX protein abundance and the reduction of Bcl-2, MMP 9, and MMP 2 protein expressions in the CNE1 xenograft tissues (Figure 6E). Generally, these results indicated that knockdown of CASC9 suppressed NPC tumor growth in vivo.
Figure 6.
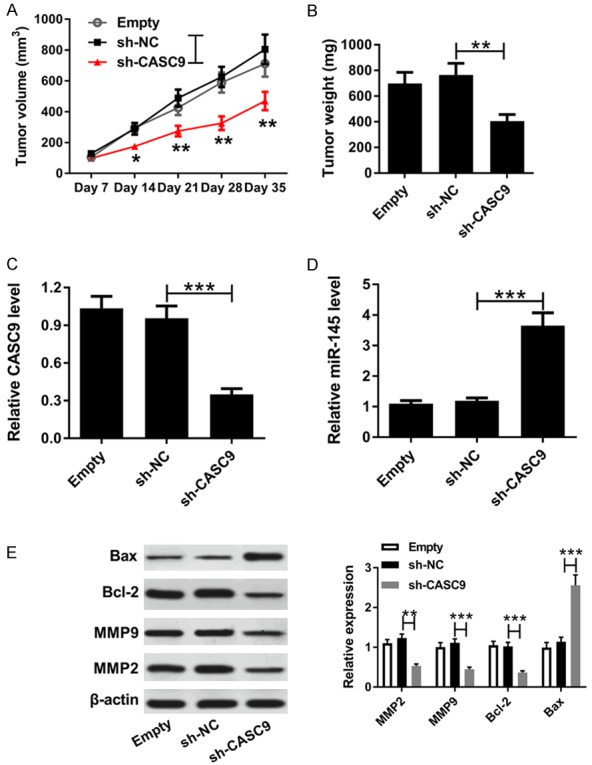
The knockdown of CASCP inhibited NPC tumor growth in vivo. A. The CNE1 cells introduced with sh-CASCP or sh-NC were injected subcutaneously into nude mice, and the tumor volumes were examined every week. B. The resected tumors were weighed after 35 days of injection. C and D. The expressions of CASCP and miR-145 were measured in the xenografted tumors using qRT-PCR analysis. E. The protein expressions of BAX, Bcl-2, MMP 9, and MMP 2 were measured in xenografted tumors by western blot. *P<0.05 **P<0.01, ***P<0.001.
Discussion
NPC has a significant ethnic and geographical distribution, but its unique and complex etiology is not completely understood [16]. Increasing lines of evidence suggest that lncRNAs are involved in the development and progression of NPC and regulate mRNA levels by competing for miRNAs [17,18]. Therefore, it is important to elucidate the molecular mechanism of lncRNAs for the treatment of NCP.
CASC9 is an lncRNA identified through a next-generation sequencing and bioinformatics analysis in ESCC, and its expression was commonly upregulated in some types of cancer. For instances, CASC9 was clearly upregulated in ESCC tissues, and the knockdown of CASC9 significantly inhibited cell migration and invasion in vitro and metastasis in nude mice in vivo [19]. In addition, CASC9 was highly expressed in glioma specimens and cells, and siRNA-mediated CASC9 silencing suppressed the proliferation and invasion in vitro and inhibited tumor growth in vivo [20]. Moreover, CASC9 was also highly expressed in the NPC tissues and as an oncogenic regulator in NPC progression by the activation of HIF1a [9]. These studies indicate that CASC9 functions as an oncogene in NPC progression, but the underlying mechanisms have not been well established.
In this report, consistent with previous observations, we found CASC9 expression was clearly increased in the NPC tissues and cells, and higher expression was associated with poor prognosis and the lower overall survival of NPC patients. Moreover, we also found that the knockdown CASC9 restrained cell migration and invasion, elevated apoptosis in CNE1 and HONE1, and evidently suppressed tumor growth in vivo.
The Bcl-2 family is composed of both proapoptotic (e.g., BAX) and anti-apoptotic (e.g., Bcl-2) family members and plays a central role in regulating apoptosis [21]. MMPs (especially MMP-2 and MMP-9) contribute to cancer cell growth, migration, and invasion, as well as metastasis and angiogenesis to promote cancer progression [22]. Our study revealed that the inhibition of CASC9 evidently increased the BAX protein level and prominently inhibited the expression of the Bcl-2, MMP 9, and MPP2 proteins in NPN cells. These results further demonstrated that the knockdown of CASC9 promoted apoptosis and inhibited migration and invasion in NPN cells.
Recent studies have shown that lncRNA can act as an endogenous miRNA sponge to bind to miRNAs and regulate their biological functions [23,24]. To explore whether CASC9 serves as an miRNA sponge in NPC, a bioinformatics analysis was carried out, and the results showed the putative binding sites between CASC9 and miR-142. Subsequently, the prediction was confirmed by the luciferase activity and an RIP analysis. These results indicated that miR-145 was a direct target of CASC9. Thus, we need to further explore their relationship in NPC cells.
A growing number of studies shows that miRNAs (as oncogenes or tumor suppressors) could be involved in many cellular processes, such as cell differentiation, cell proliferation, and apoptosis [25,26]. Earlier studies have demonstrated that the expression level of miR-145 is upregulated in several human cancers, including bladder cancer, colon cancer, and breast cancer [13,27,28]. In addition, a report found that miR-145 expression could be a novel marker for relapse in surgically treated non-small cell lung cancer (NSCLC) [29]. Moreover, it was reported that miR-145 expression was significantly decreased in NPC and inhibited invasion and metastasis by targeting Smad3 in NPC cells [30]. However, the potential molecular mechanism of miR-145 during NPC progression remains unclear.
In our study, we found that miR-145 expressions were clearly decreased in NPC tissues and cells compared with their expressions in their corresponding control, a finding consistent with prior studies. In addition, the overexpression of CASC9 inhibited the expression of miR-145, and its knockdown showed the opposite effect in NPC cells. Moreover, the abrogation of miR-145 reversed the effects of CASC9 knockdown on the migration, invasion, and apoptosis of NPC cells. These results showed that miR-145 is negatively associated with CASC9 in NPC tissues and cells.
In conclusion, the expression level of CASC9 was increased in NPC tissues and cells. The knockdown of CASC9 inhibited migration and invasion but increased apoptosis in NPC cells, which was reversed by the inhibition of miR-145. Moreover, our report presents the first evidence that miR-145 is a direct target of CASC9. In addition, miR-145 expression is downregulated and negatively associated with CASC9 in NPC tissues and cells. Also, the knockdown of CASC9 suppresses tumor growth by regulating miR-145. Therefore, these data help to better recognize the mechanism of the CASC9/miR-145 regulatory network and to explore novel, potential therapeutic strategies for NPC.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortettieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Wei KR, Zheng RS, Zhang SW, Liang ZH, Ou ZX, Chen WQ. Nasopharyngeal carcinoma incidence and mortality in China in 2010. Chin J Cancer. 2014;32:381–387. doi: 10.5732/cjc.014.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colaco RJ, Betts G, Donne A, Swindell R, Yap BK, Sykes AJ, Slevin NJ, Homer JJ, Lee LW. Nasopharyngeal carcinoma: a retrospective review of demographics, treatment and patient outcome in a single centre. Clin Oncol. 2013;25:171–177. doi: 10.1016/j.clon.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz JE, Hongjae S, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He R, Hu Z, Wang Q, Luo W, Li J, Duan L, Zhu YS, Luo DX. The role of long non-coding RNAs in nasopharyngeal carcinoma: as systemic review. Oncotarget. 2017;8:16075–16083. doi: 10.18632/oncotarget.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan N, Xiang L, Shaohua Q, Erwei S, Hua Z, Chang G. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447. doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H, Huang B. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398. doi: 10.18632/oncotarget.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 2017;36:394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Lin Y, Sui W, Zou Y, Lv Z. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6:673–680. doi: 10.1002/cam4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of mirna regulation. Genomics Proteomics Bioinformatics. 2009;7:147–54. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YQ, He QM, Ren XY, Tang XR, Xu YF, Wen X, Yang XJ, Ma J, Liu N. MiR-145 inhibits metastasis by targeting fascin actin-bundling protein 1 in nasopharyngeal carcinoma. PLoS One. 2015;10:e0122228. doi: 10.1371/journal.pone.0122228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 17.He B, Zeng J, Chao W, Chen X, Huang Y, Deng K, Huang Z, Li J, Dai M, Chen S. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8:41166–41177. doi: 10.18632/oncotarget.17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun KY, Peng T, Chen Z, Song P, Zhou XH. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging (Albany NY) 2017;9:999–1011. doi: 10.18632/aging.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, Guan X, Yang K, Bai Y. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995. doi: 10.1038/s41418-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Li C, Yang J, Sun Y, Zhang S, Yang J, Yang L, Wang Y, Jiao B. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med. 2018;22:6338–6344. doi: 10.1111/jcmm.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K. The combined functions of proapoptotic bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorina B, Paola P, Luigi M, Angela G, Sergio M, Giulia T, Raffaella G. Matrix metalloproteinases (MMP 9 and MMP 2) induce the release of vascular endothelial growth factor (VEGF) by ovarian carcinoma cells: implications for ascites formation. Cancer Res. 2003;63:5224–5229. [PubMed] [Google Scholar]
- 23.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruixi L, Ruobai S, Hicks GR, Raikhel NV. Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc Natl Acad Sci U S A. 2015;112:E89–E98. doi: 10.1073/pnas.1422656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2007;302:1–12. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 26.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2015;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Qian G, Ge S. Putative tumor suppressor mir-145 inhibits colon cancer cell growth by targeting friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Guo H, Li H, Liu Y, Liu J, Chen L, Zhang J, Zhang N. MiR-145 regulates epithelial to mesenchymal transition of breast cancer cells by targeting Oct4. PLoS One. 2012;7:e45965. doi: 10.1371/journal.pone.0045965. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Marc C, Alfons N, Nuria VO, Tania D, Rut T, Gimferrer JM, Laureano M, Cabanas ML, Jose R, Mariano M. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J. 2013;41:1172–1178. doi: 10.1183/09031936.00048712. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Sun P, Lei Z, Li M, Wang Y, Zhang HT, Liu J. miR-145 inhibits invasion and metastasis by directly targeting Smad3 in nasopharyngeal cancer. Tumour Biol. 2015;36:4123–4131. doi: 10.1007/s13277-015-3046-6. [DOI] [PubMed] [Google Scholar]