Abstract
As a new type of non-coding RNA, circular RNAs (circRNAs) have been reported to be important regulators of tumor initiation and progression. By using the high-throughput microarray, a recent study demonstrated that the expression of hsa_circ_0057481 is upregulated in human laryngeal cancer. In the present study, we aimed to elucidate the role of hsa_circ_0057481 in laryngeal cancer. We found that hsa_circ_0057481 was significantly upregulated in laryngeal cancer tissues compared with healthy normal specimens. Silencing of hsa_circ_0057481 by siRNAs suppressed cell growth, cell cycle progression, invasive and migration potential, and promoted cell apoptosis in laryngeal cancer HEP-2 and TU212 cells. Mechanistically, hsa_circ_0057481 directly sponges miR-200c in HEP-2 cells. Knockdown of hsa_circ_0057481 suppressed the expression of ZEB1, a best-known target of miR-200c. Moreover, the oncogenic effects of hsa_circ_0057481 were partly dependent on its inhibition on miR-200c. Taken together, our findings showed that hsa_circ_0057481 might act as a novel therapeutic target in laryngeal cancer.
Keywords: Laryngeal cancer, Hsa_circ_0057481, miR-200c, proliferation, apoptosis, migration
Introduction
Laryngeal carcinoma is one of the most commonly diagnosed neoplasms among head and neck tumors, which accounts for about 1.5% of all cancers and is commonly observed in old men [1,2]. Although its incidence is lower than other cancer types, laryngeal cancer is one of the major causes of tumor-associated death in China [3]. Despite surgery, radiotherapy and chemotherapy developing rapidly, the treatment outcomes of patients with laryngeal carcinoma remains very poor due to frequent metastasis and increased risk of recurrence so the five-year overall survival rate has not increased significantly [4]. Therefore, effective therapy is urgently needed. Better understanding of the underlying molecular mechanisms in laryngeal cancer initiation and development is critical to seek effective methods for early detection and treatment and to improve the survival rates.
Circular RNAs (circRNAs) are a newly recognized class of endogenous and conserved non-coding RNAs that modulate gene expression mainly at the post-transcriptional level [5,6]. These molecules have covalently closed continuous loop structure lacking 5’-cap or 3’-poly (A) tail [7]. This unique structure endows circRNAs with higher tolerance to exonucleases. Originally, circRNAs were considered to be errors or byproducts of aberrant RNA splicing without any function. Due to the newly developed technology of next generation sequencing, more and more circular RNAs have been identified in recent years. CircRNAs are now thought to be conserved, stable, and widespread in eukaryotes and participate in diverse biological processes.
CircRNAs have potential regulatory roles in a wide range of types of cancer. For instance, antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as), one of the best-known circRNAs identified recently, has been reported to act as a strong sponge for miR-7. As for the function of CDR1as in tumorigenesis, it has been reported to be involved in the carcinogenesis and metastasis by reducing the miR-7 activity and promoting cancer-associated signaling pathways [8,9]. Moreover, circRNAs have also been shown to be candidate diagnostic biomarkers for different tumors. For example, circular RNA hsa_circRNA_102958 has been demonstrated to be upregulated in gastric cancer tissues and cell lines, promotes gastric cancer progression, and may serve as a potential diagnostic marker for gastric cancer [10].
Hsa_circ_0057481, located at chr2: 190656515-190728954, has been reported to be upregulated in human laryngeal cancer [11]. This study demonstrated that hsa_circ_0057481 was significantly upregulated in laryngeal cancer specimens compared with healthy normal tissues. In addition, we found that knock-down of hsa_circ_0057481 inhibited cell proliferation, induced cell cycle arrest, inhibited cell invasion and migration, and promoted cell apoptosis in laryngeal cancer cells. Mechanistically, hsa_circ_0057481 exerts its tumor promoting effect by sponging miR-200c in HEP-2 cells. Meanwhile, knock-down of hsa_circ_0057481 suppressed the protein level of ZEB1, a well-known target gene of miR-200c. Moreover, the oncogenic effect of hsa_circ_0057481 was partly dependent on its suppression of miR-200c. Based on these observations, we suggest that hsa_circ_0057481 might act as a novel therapeutic target of laryngeal cancer.
Materials and methods
Cell culture
Both HEP-2 and TU212 laryngeal carcinoma cells were obtained from the China Infrastructure of Cell Line Resources (Beijing, China). These cells were maintained in RPMI-1640 culture medium (Gibco, Carlsbad, CA, USA) with 10% FBS (Gibco) and 100 U/ml penicillin and streptomycin (Beyotime, Shanghai, China), and incubated at 37°C in a 5% CO2 humidified atmosphere.
Clinical specimens
With the approval of the Ethics Committee of Tianjin Medical University, a total of 16 human laryngeal cancer specimens and adjacent non-cancerous tissue (1 cm distance from the primary tumor at least) were collected from the Department of Thyroid and Neck Tumor, Tianjin Medical University Cancer Institute and Hospital. All patients signed the informed consent. Tissue samples were stored at -80°C before use.
Transfection of miRNA and siRNA into cells
Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) were used for siRNAs and miRNA transfection, which followed the manufacturer’s instructions. The following two sequences (sense strand) were synthesized to target hsa_circ_0057481: (1) GAUUAUUGUAGUGACUAUTT; (2) UGUUGUUCAGGAAAUUGTT.
CCK8 assay
Cell Counting Kit-8 Assay was performed to detect the cell proliferation rate. Briefly, 2 × 10003 were seeded into the 96-well cell culture plate and were then transfected with control siRNA (100 nM) or 100 nM of hsa_circ_0057481. Cells were incubated at 37°C in 5% CO2 for 0 d, 2 d, 4 d, 6 d and 8 d. At each time point, cells were incubated in 10% CCK-8 (Beyotime) and incubated for 2 h.
RNA FISH
To detect the subcellular distribution of hsa_circ_0057481 in HEP-2 cells, RNA FISH was performed as described [12]. The sequence of the hsa_circ_0057481 probe for RNA FISH was 5’-FAM-GAAGTATTATATACAAAAAATTTTCC-3’.
Colony formation assay
A total of 1000 cells were plated onto the 6-well plate and cultured for 2 weeks and stained with 0.1% (w/v) crystal violet. The number of colonies was counted under a microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Cell cycle analysis
Flow cytometry was conducted to determine the cell cycle distribution. Briefly, Treated cells were trypsinized and fixed in 70% ethanol on ice for overnight. Subsequently, fixed cells were washed with 1 × PBS and incubated with 0.1 mg/ml of RNase A (Beyotime) and 0.05 mg/ml of propidium iodide (PI) for 30 min at 4°C for 30 min. FACSCalibur (BD Biosciences, San Jose, CA, USA) was used to determine the cell cycle distribution.
Apoptosis assay
Annexin V-FITC/PI assay were carried out to determine the apoptosis rate of indicate cells by using the Annexin V-FITC Apoptosis Detection Kit I (BD Pharmingen, San Diego, CA, USA) following the manufacturer’s instructions. Apoptotic cells were assessed by FACSCalibur in at least three independent experiments. and analyzed by the FlowJo software 7.6.1.
Transwell invasion and migration assay
Transwell invasion and migration assay were performed as described [13].
Western blot analysis
Western blot analysis was performed as previously described [14]. The antibodies are as follows: Cyclin D1 (ab134175; Abcam, Cambridge, MA, USA), p27 (ab92741; Abcam), BAX (ab182734; Abcam), BCL-2 (ab32124; Abcam), E-Cadherin (ab219332; Abcam), β-Catenin (ab32572; Abcam), ZEB1 (ab81972; Abcam), β-ACTIN (TA-09; Zhongshan Golden Bridge Biotechnology (ZSGB), Beijing China) and horseradish peroxidase (HRP)-conjugated secondary antibodies (ZDR5306 or ZDR5307; ZSGB).
qRT-PCR
qRT-PCR was performed as previously described [14]. Primers are as follows: hsa_circ_0057481, forward: 5’-GATGGGATCCAGGTATCACTTT-3’, hsa_circ_0057481, reverse: 5’-TGAAACTTCCAGGGCCAAAGA-3’; GAPDH, forward: 5’-TCGGAGTCAACGGATTTGGT-3’, GAPDH, reverse: 5’-TTGGAGGGATCTCGCTCCT-3’; Specific RT primer for miR-200c: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCATC-3’, miR-200c, forward: 5’-GTGCAGGGTCCGAGGT-3’, miR-200c, reverse: 5’-GCTAATACTGCCGGGTAATG-3’; U6, forward: 5’-CTCGCTTCGGCAGCACA-3’, U6, reverse: 5’-AACGCTTCACGAATTTGCGT-3’.
Luciferase report assay
The DNA fragment of hsa_circ_0057481 containing a miR-200c (100 bp) binding was synthesized (Sangon, Shanghai, China) and ligated into the psiCHECK-2 reporter plasmid (Promega, Fitchburg, WI, USA). For construction of hsa_circ_0057481 reporter gene plasmids with a mutant miR-200c binding site, the Site-Directed Mutagenesis System (Beyotime) was used to construct hsa_circ_0057481 reporter gene plasmids with a mutant miR-200c binding site following the manufacturer’s protocol. The primers used for mutagenesis are as follows: 5’-CTATAATTTCACTAGCAGTTACCAAATGATCATAAATATACA-3’ (hsa_circ_0057481-MUT-forward), 5’-TGTATATTTATGATCATTTGGTAACTGCTAGTGAAATTATAG-3’ (hsa_circ_0057481-MUT-reverse). The luciferase activities were measured as previously described [15].
Tumor formation assay
The xenograft assay was approved by the Animal Ethics Committee of Tianjin Medical School. HEP-2 cells were infected with pHBLV-U6-Puro-hsa_circ_0057481-sh-1 lentivirus or empty control and subcutaneously injected into two flanks of 6 weeks old nude mice (shCon: right flank; hsa_circ_0057481-sh-1: left flank). Thirty-eight days later, the following formula was used to calculate tumor volume: volume = length × width2 × 1/2. The tumor weights were also evaluated.
Statistical analysis
SPSS 17.0 (SPSS; Chicago, IL, USA) was applied for statistical analyses. The Student’s t-test or one-way ANOVA were used to analyze the Differences in the two groups. If P<0.05, differences were considered significant. All data were expressed as the mean ± standard deviation (S.D.).
Results
hsa_circ_0057481 is highly upregulated in clinical laryngeal cancer specimens and suppresses cell proliferation in HEP-2 and TU212 cells
First, to identify the role of hsa_circ-0057481 in laryngeal cancer, we examined its expression levels in laryngeal cancer tissues compared with adjacent normal tissues by qRT-PCR. In our cohort (n = 16), the expression of has_circ-0057481 was significantly increased in tumor tissues compared to the adjacent tissues (P<0.05) (Figure 1A). Next, we also detected the subcellular location of has_circ-0057481 by fluorescence in situ hybridization staining. FISH against hsa_circ-0057481 showed that hsa_circ-0057481 resided mainly within the cytosol (Figure 1B). Subsequently, to explore the exact role of hsa_circ_0057481 in laryngeal cancer, we silenced hsa_circ_0057481 in HEP-2 and TU212 cells by transfecting two specific siRNAs (Figure 1C and 1D). The specific knock-down of hsa_circ_0057481 significantly inhibited the proliferation and colony formation of HEP-2 and TU212 cells, determined by the CCK8 assay (Figure 1E and 1F) and the colony formation assay (Figure 1G and 1H), respectively. Taken together, our results implied that hsa_circ_0057481 may be a tumor promoter in laryngeal cancer cells.
Figure 1.
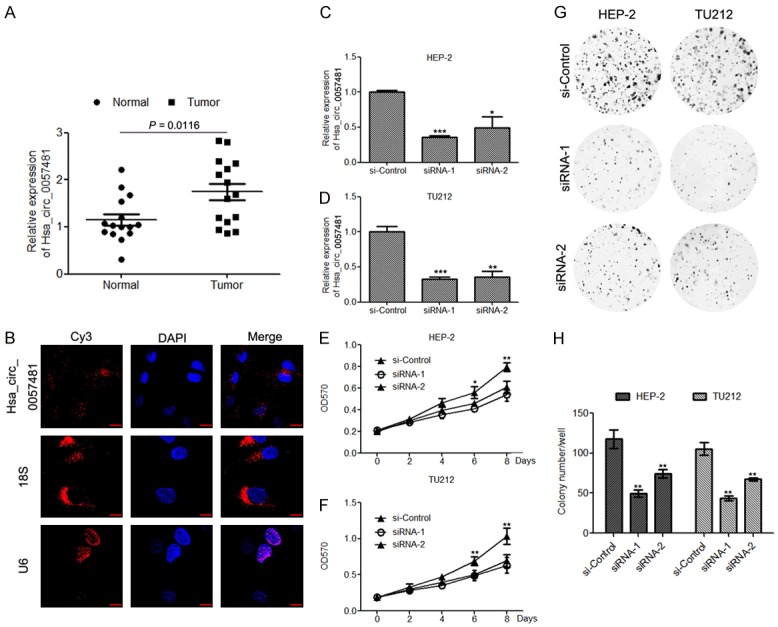
Hsa_circ_0057481 is significantly upregulated in laryngeal cancer tissues and promotes HEP-2 and TU212 cell proliferation. (A) Relative expression of hsa_circ_0057481 in 16 pairs of laryngeal cancer compared and adjacent normal tissues. (B) RNA-FISH was performed to examine the expression and localization of hsa_circ_0057481 in HEP-2 cells. DAPI was used to stain the nuclei. Scale bar, 20 μm. (C and D) qPCR analysis was performed to evaluate the Hsa_circ_0057481 expression levels in HEP-2 (C) and TU212 (D) cells after transfection with hsa_circ_0057481 specific siRNAs and control siRNA. (E and F) The cell proliferation rates of HEP-2 (E) and TU212 (F) cells transfected with hsa_circ_0057481-specific siRNAs and control siRNA were determined by the CCK8 assay. n = 6. (G, H) Colony formation assays of HEP-2 and TU212 cells transfected with hsa_circ_0057481 specific siRNAs and control siRNA. Quantification of colony numbers is shown in (H). Data = means ± S. D. *, P<0.05; **, P<0.01; ***, P<0.001.
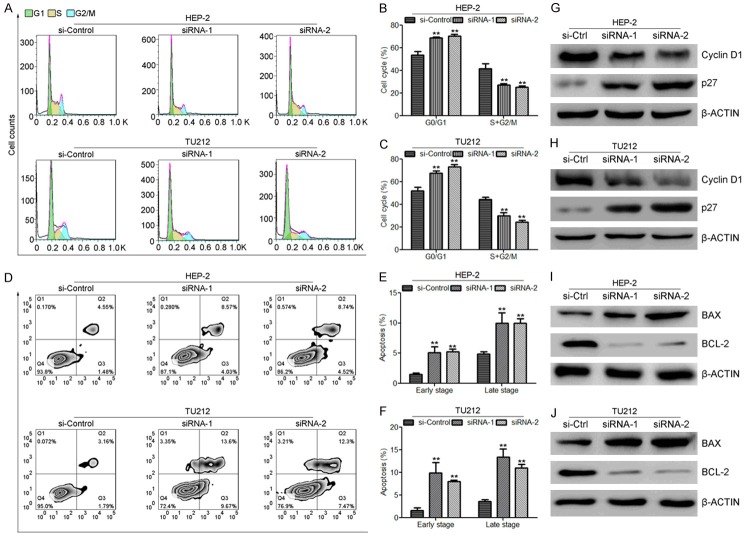
Knock-down of hsa_circ_0057481 induces cell cycle arrest and apoptosis in laryngeal cancer cells
To further figure out the role of hsa_circ_0057481 in laryngeal cancer cell cycle and cell apoptosis, flow cytometric analysis was performed. Cell cycle analysis showed that silencing of hsa_circ_0057481 induced cell cycle arrest at G0/G1 phase in both HEP-2 and TU212 cells (Figure 2A-C). As shown in Figure 2B and 2C, the cell populations in the G0/G1 phase were increased, and the cell populations in the S+G2/M phases were decreased in cells transfected with the hsa_circ_0057481 siRNAs. Additionally, as shown in Figure 2D-F, apoptosis assays showed that silencing of hsa_circ_0057481 induced laryngeal cancer cell apoptosis. To confirm that, we also detected the cell cycle-related genes Cyclin D1 and p27 and apoptosis-associated proteins BAX and BCL-2 in these groups. Western blot analysis showed that silencing of hsa_circ_0057481 suppressed the protein levels of Cyclin D1 and BCL-2, and upregulated the protein levels of p27 and BAX (Figure 2G-J), which was consistent the results of cell cycle and apoptosis assays.
Figure 2.
Knock-down of hsa_circ_0057481 suppressed cell cycle progression and induced apoptosis in HEP-2 and TU212 cells. (A-C) FACS analysis to determine cell cycle distributions of HEP-2 and TU212 cells transfected with hsa_circ_0057481 specific siRNAs and control siRNA. (D-F) Apoptotic rates of HEP-2 and TU212 cells transfected with hsa_circ_0057481 specific siRNAs compared with control siRNA, which were evaluated by Annexin V-FITC/PI double staining. (G and H) Western blot analysis to determine the protein levels of Cyclin D1 and p27 in HEP-2 (G) and TU212 (H) cells transfected with hsa_circ_0057481 specific siRNAs and control siRNA. (I and J) Western blot analysis to determine the protein levels of BAX and BCL-2 in HEP-2 (I) and TU212 (J) cells transfected with hsa_circ_0057481 specific siRNAs and control siRNA. Three independent experiments were performed for each data point. *, P<0.05. **, P<0.01. ***, P<0.001.
Silencing of hsa_circ_0057481 suppresses invasion, migration, and xenograft growth in laryngeal cancer cells
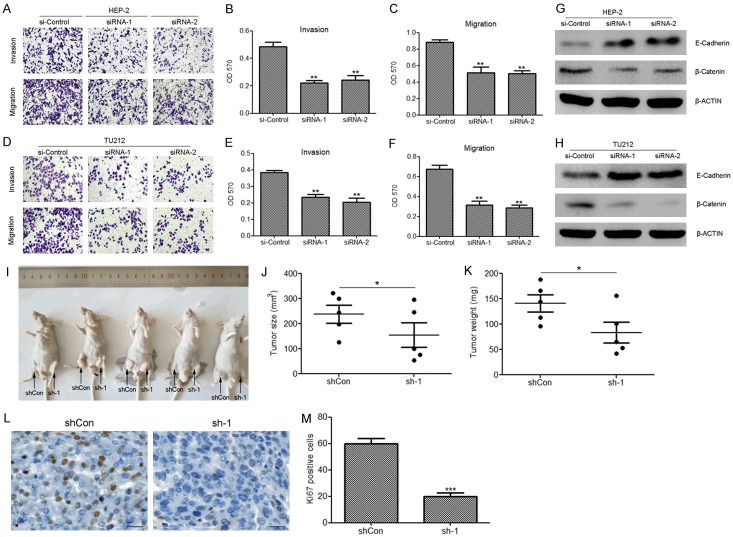
To further illustrate the role of hsa_circ_0057481 in laryngeal cancer, transwell invasion and migration assays were performed to assess the effects of hsa_circ_0057481 knock-down on the invasion and migration capacity of HEP-2 and TU212 cells. The tranwell assay revealed that hsa_circ_0057481 knockdown repressed the invasion and migration capacity of HEP-2 and TU212 cells compared with that of cells transfected with the control siRNA (Figure 3A-F). Western blot analysis also demonstrated that silencing of hsa_circ_0057481 upregulated the protein level of the epithelial marker E-cadherin, while it downregulated the protein level of the β-Catenin (Figure 3G, 3H). In a tumor formation assay in vivo, silencing of hsa_circ_0057481 significantly suppressed the ability of tumor formation in laryngeal cancer (Figure 3I-K). Moreover, silencing of hsa_circ_0057481 also suppressed the proliferation of tumor cells indexed by Ki67 staining (Figure 3L, 3M).
Figure 3.
Silencing of hsa_circ_0057481 suppresses cellular migration and invasion in HEP-2 and TU212 cells. A-F. Knockdown of hsa_circ_0057481 inhibited cell invasion and migration in HEP-2 and TU212 cells. Each data point was in triplicate and shown as means ± S. D. **, P<0.01. G and H. Determination of E-cadherin and β-catenin expression by western blot in the indicated groups. I. hsa_circ_0057481 knockdown inhibited laryngeal cancer cell HEP-2 growth in vivo. J. Tumor sizes were calculated 38 days post-injection. K. Tumor weight was analyzed 38 days post-injection. L, M. Representative xenograft tumors stained and analyzed for Ki67. Scale bars: 50 μm.
hsa_circ_0057481 acts as an miRNA sponge for miR-200c in laryngeal cancer cells
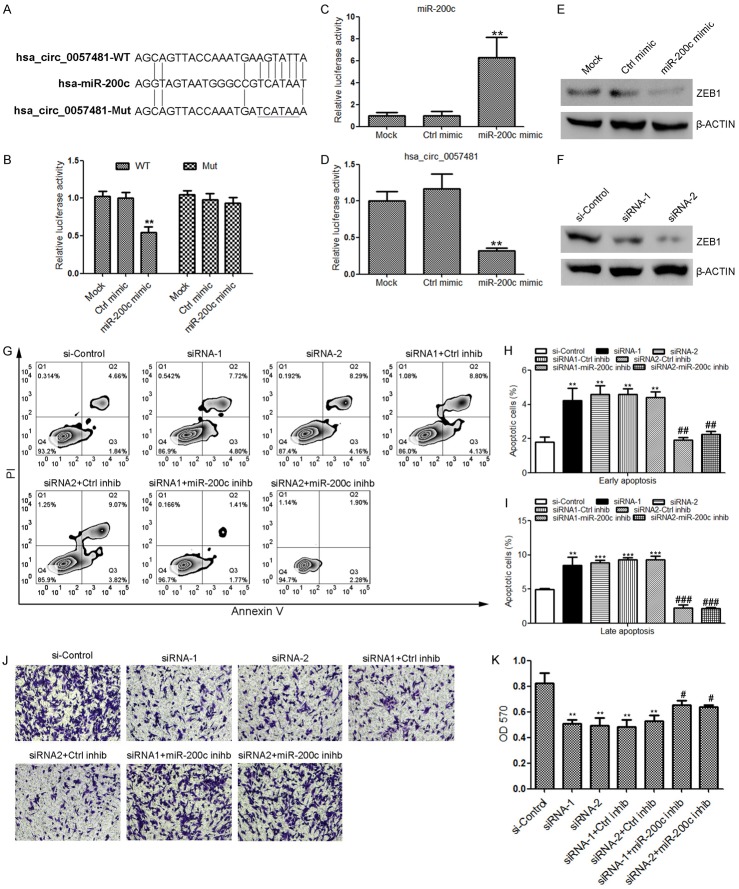
Accumulating evidence has suggested that circRNAs could function as miRNA sponges. It was hence predicted there might be miRNAs that could recognize sequences in hsa_circ_0057481 and interact with it. Based on the results of Starbase and Targetscan (http://starbase.sysu.edu.cn/; http://www.targetscan.org/vert_72/), we found that several miRNAs could potentially bind the sequence of hsa_circ_0057481, including miR-200c, which is one of the highly studied miRNAs in tumorigenesis. The predicted target sequence of miR-200c in hsa_circ_0057481 is shown in Figure 4A. To prove miR-200c was a direct target miRNA of hsa_circ_0057481, a dual-luciferase reporter assay was performed. As shown in Figure 4B, the luciferase activity was obviously lower in HEP-2 cells with the psiCHECK2-hsa_circ_0057481-WT and miR-200c co-transfection compared with controls, while that with the mutant construct was not significantly affected. These results suggest that miR-200c could directly bind hsa_circ_0057481. Moreover, overexpression of miR-200c (Figure 4C) reduced the expression of hsa_circ_0057481 in HEP-2 cells (Figure 4D). In addition, both overexpression of miR-200c and silencing of hsa_circ_0057481 suppressed the protein expression of ZEB1, a best-known target of miR-200c (Figure 4E, 4F). Importantly, inhibition of miR-200c could reverse the hsa_circ_0057481 effects of promoting apoptosis (Figure 4G-I) and inhibiting the proliferation and migration of laryngeal cancer cells (Figure 4J, 4K).
Figure 4.
Hsa_circ_0057481 sponges miR-200c in laryngeal cancer. (A) The putative wild type miR-200c binding site and its mutant in the mRNA sequence of hsa_circ_0057481. (B) Luciferase reporter assay to evaluate the effect of miR-200c on hsa_circ_0057481 expression. Data are shown as means ± S. D. (n = 6). **, P<0.01. (C and D) Overexpression of miR-200c (C) suppressed the expression of hsa_circ_0057481 (D) in HEP-2 cells, determined by quantitative RT-PCR. (E) Overexpression of miR-200c inhibited ZEB1 protein expression in HEP-2 cells, which was determined by western blot. (F) Silencing of hsa_circ_0057481 inhibited ZEB1 protein expression in HEP-2 cells, determined by western blot. (G-I) Suppression of miR-200c partially rescued the knock-down of hsa_circ_0057481 induced cell apoptosis in HEP-2 cells. Data are shown as means ± S. D. (n = 3). ** and ***, vs. si-control. P<0.01. ## and ###, vs. siRNA1+Control inhibitor. (J, K) Suppression of miR-200c partially rescued the knock-down of hsa_circ_0057481 suppressed cell migration in HEP-2 cells. Data are shown as means ± S. D. (n = 6). **, vs. si-control. P<0.01. #, vs. siRNA1+Control inhibitor.
Discussion
CircRNAs are a conserved class of covalently closed RNA with relatively low expression in cells. In the past couple of years, because of rapid progress in the next-generation sequencing and bioinformatic technologies, thousands of circRNAs are being identified and the functions of several circRNAs are being elucidated. CircRNAs can modulate parental gene expression and serve as miRNA sponges. Moreover, accumulating evidence has shown that circRNAs participate in tumorigenesis as tumor promoters or tumor suppressors. In laryngeal cancer, literature suggests that circRNAs play an important role in its tumorigenesis and progression and may serve as novel and stable biomarkers for its diagnosis. In this article, we identified one novel circRNA, hsa_circ_0057481 and found that hsa_circ_0057481 expression was higher in laryngeal cancer tissues than in the corresponding adjacent non-neoplastic tissues. The in vitro and in vivo experiments showed that silencing of hsa_circ_0057481 by specific siRNAs suppressed cell growth, cell cycle progression, invasive and migration potential, and promoted cell apoptosis in laryngeal cancer HEP-2 and TU212 cells.
Accumulating evidence has shown that circRNAs can serve as miRNA decoys or sponges, thereby regulating target mRNA expression. To elucidate the molecular mechanisms of hsa_circ_0057481 underlying laryngeal tumorigenesis, we predicted the potential miRNA targets of hsa_circ_0057481 by using online bioinformatic software. Then, miR-200c was selected, because not only it is a high-ranked target of hsa_circ_0057481 but also it plays an important role in laryngeal cancer. miR-200c was found to contain the complementary binding region of hsa_circ_0057481, which was subsequently confirmed by a dual-luciferase reporter assay. In addition, miR-200c overexpression induced hsa_circ_0057481 downregulation in laryngeal cancer cells. Moreover, rescue experiments revealed that miR-200c could reverse the inhibitory effects of hsa_circ_0057481 knock-down on HEP-2 cells. In addition, both overexpression of miR-200c and silencing of hsa_circ_0057481 suppressed the expression of ZEB1, one of best-known miR-200c target. All of these findings suggest that modulating the expression and function of miR-200c and its downstream genes may partly contribute to the effect of miR-200c on controlling the biologic behaviors of laryngeal cancer cells. However, the detailed mechanism requires further investigation.
Our study also has several limitations, one of which is we only verified the relationship between miR-200c and hsa_circ_0057481. However, the possibility that other important mechanisms may be involved could not be excluded. Potential relationships between other predicted target miRNAs and hsa_circ_0057481 remain to be further investigated.
In conclusion, our findings have demonstrated that hsa_circ_0057481 is frequently upregulated in patients with laryngeal cancer, and can be recommended as a potential oncogenic circRNA to promote laryngeal cancer development and progression. The miR-200c-ZEB1 axis might also play an important role in the regulation of malignant behavior of laryngeal cancer by hsa_circ_0057481. Therefore our study provides a novel and promising therapeutic target for laryngeal cancer treatment.
Acknowledgements
This work was partially supported by research grant from National Scientific Foundation of China (Grant No. 81702629). The first author greatly thanks her family for their great helps.
Disclosure of conflict of interest
None.
References
- 1.Re M, Magliulo G, Gioacchini FM, Bajraktari A, Bertini A, Ceka A, Rubini C, Ferrante L, Procopio AD, Olivieri F. Expression levels and clinical significance of miR-21-5p, miR-let-7a, and miR-34c-5p in laryngeal squamous cell carcinoma. Biomed Res Int. 2017;2017:3921258. doi: 10.1155/2017/3921258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghai B, Jain K, Bansal D, Bhatia N. End-tidal sevoflurane concentration for ProSeal(TM) versus Classic(TM) laryngeal mask airway insertion in unpremedicated anaesthetised adult females. Anaesth Intensive Care. 2016;44:221–226. doi: 10.1177/0310057X1604400208. [DOI] [PubMed] [Google Scholar]
- 3.Wei Q, Yu D, Liu M, Wang M, Zhao M, Liu M, Jia W, Ma H, Fang J, Xu W, Chen K, Xu Z, Wang J, Tian L, Yuan H, Chang J, Hu Z, Wei L, Huang Y, Han Y, Liu J, Han D, Shen H, Yang S, Zheng H, Ji Q, Li D, Tan W, Wu C, Lin D. Genome-wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat Genet. 2014;46:1110–1114. doi: 10.1038/ng.3090. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Hu H. Long non-coding RNA CCAT1/miR-218/ZFX axis modulates the progression of laryngeal squamous cell cancer. Tumour Biol. 2017;39:1010428317699417. doi: 10.1177/1010428317699417. [DOI] [PubMed] [Google Scholar]
- 5.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 6.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang W, Ji M, He G, Yang L, Niu Z, Jian M, Wei Y, Ren L, Xu J. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther. 2017;10:2045–2056. doi: 10.2147/OTT.S131597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as Act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Wei J, Wei W, Xu H, Wang Z, Gao W, Wang T, Zheng Q, Shu Y, De W. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2019 doi: 10.3233/CBM-182029. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang C, Qiao Y, Hu H, Liu L. RNA-Seq profiling of circular RNAs in human laryngeal squamous cell carcinomas. Mol Cancer. 2018;17:86. doi: 10.1186/s12943-018-0833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- 13.Tao F, Tian X, Ruan S, Shen M, Zhang Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J. 2018:fj201800495RR. doi: 10.1096/fj.201800495RR. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, Dong JT. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao F, Tian X, Lu M, Zhang Z. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. J Genet Genomics. 2018;45:137–145. doi: 10.1016/j.jgg.2018.03.001. [DOI] [PubMed] [Google Scholar]