Abstract
Background: The implication of miR-452-5p and its prospective machinery in hepatocellular carcinoma (HCC) remains largely unknown. For this reason, this study aimed to inspect the clinical implication of miR-452-5p expression in HCC tissues with multiple detection approaches, to analyze its potential function via in silico methods, and to validate this using a dual-luciferase reporter assay. Methods: The assessment of the expression level of miR-452-5p in HCC was conducted via four methods: 1) in-house real-time quantitative PCR (RT-qPCR), 2) miRNA-sequencing (miRNA-seq) from The Cancer Genome Atlas (TCGA), 3) miRNA microarrays from the Gene Expression Omnibus (GEO), and 4) comprehensive meta-analyses calculating the standard mean difference (SMD) and summary of receiver operator characteristic (sROC). Following the target prediction, one of the potential targets of miR-452-5p was validated through a dual-luciferase reporter assay. Results: MiR-452-5p was consistently elevated in HCC tissues via various detection methods, including in-house RT-qPCR, miRNA-seq, and miRNA microarrays. The final SMD was 0.842 for 820 cases of HCC samples. Simultaneously, the area under curve (AUC) of the sROC was 0.80 (0.76-0.83). The 1,135 predicted targets of miR-452-5p were enriched in the pathways of cytokine-cytokine receptor interaction, carbon metabolism, and complement and coagulation cascades. Among these predicted targets, CDKN1B was verified to be a real target of miR-452-5p. Conclusion: The overexpression of miR-452-5p may play a pivotal role in the carcinogenesis of HCC via targeting multiple signaling pathways and genes. The function and molecular machinery of miR-452-5p in HCC requires further in-depth exploration.
Keywords: MiR-452-5p, CDKN1B, Hepatocellular carcinoma (HCC), signaling pathways
Introduction
Liver cancer is the most prevalent cancer of the digestive system, and it can be divided into primary and secondary types. Primary liver cancer (PLC) is the main type of all liver cancers, which is composed of several histologic subtypes, such as cholangiocarcinoma (CC) and hepatocellular carcinoma (HCC), as well as the combination of the two: combined hepatocellular-cholangiocarcinoma (cHCC-CC) [1-4]. Primary liver cancer is dominated by HCC, which is the fifth most frequent malignant tumor and the third most prevalent determinant factor of cancer-related mortality globally [5]. Despite the fact rapid improvement has been achieved in the therapeutics of HCC in the last few years, HCC continues to have a high mortality rate [6-9]. Epidemiological research has indicated that 40-50% of the cases of HCC worldwide occur in China every year, which is the number two determinant of cancer death in the world. Moreover, many HCC cases are not diagnosed until they are in the advanced stage. The five-year survival rate of HCC in developing countries is extremely low, in some cases as low as 5%. Although the curative effect of comprehensive treatment of HCC, which is mainly achieved by surgery, has been greatly improved, because of the complex pathogenesis of liver cancer, the persisting survival rate and clinical cure rate of HCC are nonetheless low [10-12]. Hence, the search for new molecular mechanisms of HCC is necessary and urgent for human health protection. In fact, the molecular machinery of miRNA in liver cancer is not fully understood. Therefore, this study aimed to unravel the molecular machinery of miRNA in HCC and contribute to the discovery of new molecular mechanisms in HCC.
MiRNAs are small and non-coding RNAs, which are around 20 nucleotides long, that can repress genes expression through binding in the 3’ untranslated region (UTR) of the target mRNAs [13-17]. They adjust the expression of targets by binding to perfectly or partially matching mRNA sequences [18-21]. The evidence strongly indicates that miRNAs regulate the expression of known tumor suppressor genes and oncogenes, and they play the role of tumor suppressors [22-24]. In recent years, miRNAs have been confirmed to be abnormally expressed in HCC and have been found to be related to the occurrence and progression of HCC [25-29]. We speculate that one of the miRNAs, miR-452 may play a vital role in HCC either as a tumor suppressing gene or promoter and that it participates in and influences the pathogenesis of HCC.
MiR-452 is an important member of the miRNA family, which is divided into two subtypes: miR-452-5p and miR-452-3p. MiR-452 has been studied in a variety of cancers, including bladder cancer [30], non-small cell lung cancer (NSCLC), breast cancer, and HCC [31]. Chang et al. found that the abnormal expression of miR-452 is possibly connected with the malignant biological characteristics of cancer and is located on human chromosome Xq28 [32]. For instance, the high expression of miR-452 inhibits tumorigenesis of gliomas and metastasis of NSCLC. Liu et al. determined that miR-452, as an onco-miRNA in HCC, promotes cell metastasis [33]. Hui et al. considered miR-452-3p as a probable oncogene targeting the CPEB3/EGFR axis in HCC [34]. Zheng et al. stated that miR-452 promotes the development of stem-like cells in HCC by suppressing Sox7, which is involved in the Wnt/-catenin signaling pathway [35]. In terms of the association between miR-452-5p and HCC, the miR-452-5p expression level has been documented to be elevated in HCC. Consistent with this, Zhu et al. also mentioned this point in their research results [36]. Nevertheless, the accurate machinery of miR-452-5p in HCC remains unclear and needs to be elucidated in-depth. Zhu et al. found that LINC00052 could inhibit the cell mobility and infiltration of HCC by upregulation of EPB41L3 with miR-452-5p [37]. However, few studies have been conducted on the character of miR-452-5p in HCC, and the exact mechanism of miR-452-5p in HCC is still unclear, so it needs to be further illuminated.
In this research, we primarily evaluated the expression level of miR-452-5p in HCC with in-house real-time quantitative polymerase chain reaction (RT-qPCR), miRNA-sequencing (miRNA-seq), cancer genome sequencing using The Cancer Genome Atlas (TCGA), and miRNA microarrays from the Gene Expression Omnibus (GEO). Furthermore, a potential target of miR-452-5p was confirmed by dual-luciferase reporter gene assay. We intended to study the clinicopathological implication of miR-452-5p expression in HCC, to analyze its potential function via in silico methods, and to validate this with a dual-luciferase reporter assay. We believe that this article will contribute to the further study of the molecular mechanism of HCC and help to find more safe and effective treatment strategies for HCC.
Materials and methods
RNA extraction and RT-qPCR
An in-house RT-qPCR was conducted to evaluate the miR-452-5p level in 95 cases of HCC tissues and neighboring non-cancerous liver tissues no less than 2 cm away from the edge of the tumor. All participating patients signed an informed consent form. The RNA extraction and RT-qPCR were employed as previously reported. The sequence of miR-452-5p was aacuguuugcagaggaaacuga. Data were analyzed using the 2-ΔCq method [38-43].
MiR-452-5p expression in HCC from miRNA-seq of TCGA
In order to extend the scope of the study, we achieved the expression data of miR-452-5p in HCC using TCGA miRNA-seq, and we calculated the associations between clinical parameters and miR-452-5p. Features of HCC samples were downloaded from TCGA through the University of California, Santa Cruz (UCSC) Xena Functional Genomics Explorer (https://xena.ucsc.edu/). The downloaded data included 50 adjacent non-cancerous tissues and 373 HCC tissues, and then the extracted data were log2-transformed for subsequent analysis.
MiR-452-5p expression in HCC from other microarray and miRNA-seq data
To validate the expression of miR-452-5p in HCC, we accessed the GEO, Array Express, SRA, and Oncomine. The following key words were used in the searches for HCC: (malignan* OR neoplas* OR cancer OR carcinoma OR tumor) AND (hepatocellular OR hepatic OR liver OR HCC). All studies included had to meet to the criterion that the study was designed with a control group of human non-cancerous liver tissues and human HCC tissues as the case group. We extracted all the expression data for miR-452-5p. Only studies with proper groups and available or calculable expression data of miRNA were included. Finally, we obtained 13 eligible miRNA microarray profiles in this meta-analysis (Table 1).
Table 1.
Basic characteristics of miR-seq and microarray databases
| Dataset | Country | Sample source | Data source | Platform | Cancer | Mean1 | SD1 | Normal | Mean0 | SD0 |
|---|---|---|---|---|---|---|---|---|---|---|
| TCGA | USA | tissues | TCGA | RNA-seq | 371 | 7.419217 | 1.643848 | 49 | 5.522102 | 1.109133 |
| GSE21362 | Japan | tissues | GEO | GPL10312 | 73 | 5.723791 | 1.915917 | 73 | 4.958394 | 1.332598 |
| GSE31383 | USA | tissues | GEO | GPL10122 | 9 | 0.266314 | 0.157709 | 10 | 0.232738 | 0.075521 |
| GSE36915 | Taiwan | tissues | GEO | GPL8179 | 68 | 10.522523 | 1.404588 | 21 | 8.789549 | 1.073489 |
| GSE40744 | USA | tissues | GEO | GPL14613 | 26 | 4.825 | 1.642611 | 50 | 3.062 | 1.364373 |
| GSE41874 | Japan | tissues | GEO | GPL7722 | 6 | 1.222802 | 0.215252 | 4 | 1.036838 | 0.514752 |
| GSE5755501 | Japan | tissues | GEO | GPL16699 | 16 | 0.917185 | 0.01779 | 16 | 0.924954 | 0.023778 |
| GSE5755502 | Japan | tissues | GEO | GPL18044 | 16 | 0.966242 | 0.008091 | 16 | 0.963673 | 0.012219 |
| GSE64632 | USA | tissues | GEO | GPL18116 | 3 | 0.280778 | 0.158097 | 3 | 0.160951 | 0.063965 |
| GSE98269 | China | tissues | GEO | GPL20712 | 3 | 6.799311 | 0.9357 | 3 | 5.248964 | 0.045875 |
| GSE115016 | China | tissues | GEO | GPL21572 | 12 | 18.37839 | 29.57517 | 12 | 2.872884 | 2.150306 |
| GSE12717 | China | tissues | GEO | GPL7274 | 10 | 8.573606 | 2.382533 | 6 | 4.531271 | 0.987178 |
| GSE22058 | USA | tissues | GEO | GPL10457 | 96 | 0.750991 | 0.106841 | 96 | 0.679112 | 0.050447 |
| GSE39678 | Korea | tissues | GEO | GPL15852 | 16 | 9.021111 | 0.977739 | 8 | 7.455979 | 0.436564 |
Statistical analysis
To further evaluate the veracity of the data from these resources (TCGA, GEO, Array Express, SRA, and Oncomine), relative statistical analysis was implemented with Stata software 5 version 12.0 (StataCorp LLC, College Station, TX, USA). The continuous outcomes were assessed by the standard mean difference (SMD) with 95% confidence interval (95% CI). The heterogeneity of the analysis was assessed by the Q test (chi-squared test) and the I2 statistic value. A random-effect model was leveraged when the heterogeneity existed; if not, a fixed-effect model was preferred. Forest plots of SMDs with CIs of miR-452-5P in each group were calculated and pooled, and the publication bias was conducted using Begg’s or Egger’s funnel plots. A two-sided P-value over 0.05 indicated that no publication bias existed. Based on all available databases, the expression data were pooled together and a summary of receiver operator characteristic (sROC) was performed to elucidate the expression level and function of miR-452-5p in HCC. IBM SPSS Statistics 23.0 (IBM, Armonk, NY, USA) was used for all statistical analysis. ROC curves (sensitivity vs. 1 specificity) were generated to determine the optimal cutoff value for each protein expression. Area under the curve (AUC) of miR-452-5p was also calculated.
Functional enrichment for target genes
MiRWalk3.0 was used for the target prediction of miR-452-5p. We intersected the predicted genes from eight prediction programs among 12 in MiRWalk3.0. We received 1,135 prospective targets of miR-452-5p. In order to assess the latent function of the potential target mRNAs of miR-452-5P in HCC, we utilized the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation with R software. The GO plot package was used for mapping.
Dual-luciferase reporter assay
The miR-452-5p in HCC was selected for further verification. A dual-luciferase reporter gene assay was carried out. The human embryonic kidney HEK-293T cells were used as the tool cell line, and they were placed in a 48-well plate to grow to 70-80%. The wild-type and mutant CDKN1B 3’UTR region bound to miR-452-5p was synthesized and linked to psiCHECK-2-report luciferase reporter plasmid. The cells were transfected with miR-452-5p mimics or non-mimic controls and psiCHECK-CDKN1B 3’UTR wild type (AAACAGT) or psicheck-CDKN1B 3’UTR mutant (GTGACCA) with Lipofectamine 2000. After being cultured for another 48 h at 37°C, the activity of firefly and heparin luciferase was detected by a dual-luciferase reporter assay (Promega Corporation, Madison, WI, USA). All experiments should be carried out in triplicate.
Results
MiR-452-5p was frequently increased in HCC tissues by RT-qPCR
The miR-452-5p expression level in HCC tissues was notably increased than that in the neighboring non-cancerous liver tissues (P<0.0001, Figure 1A).
Figure 1.
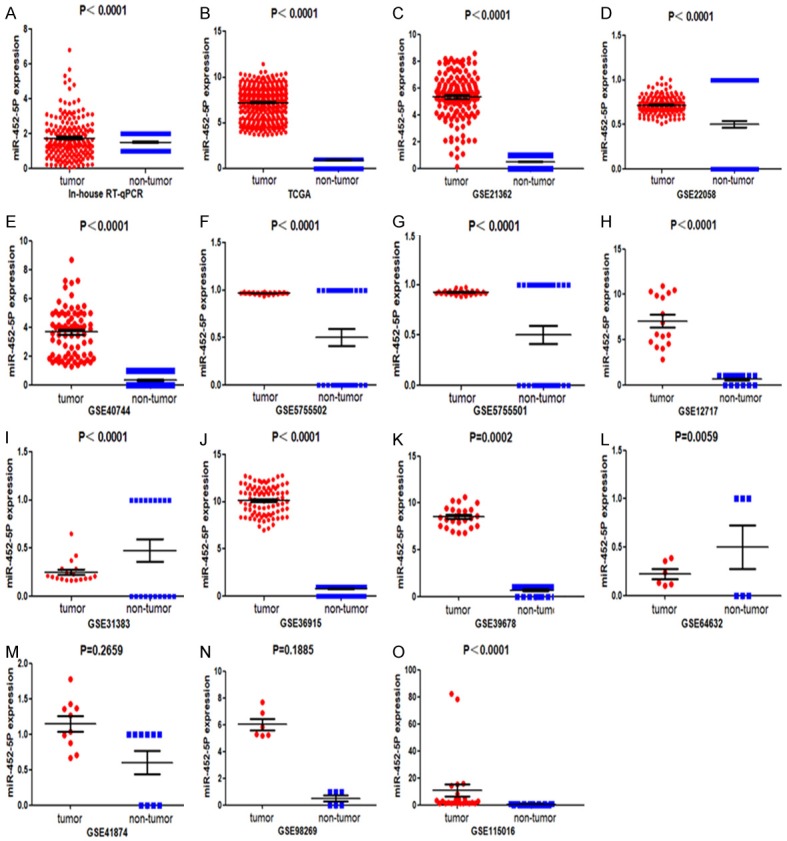
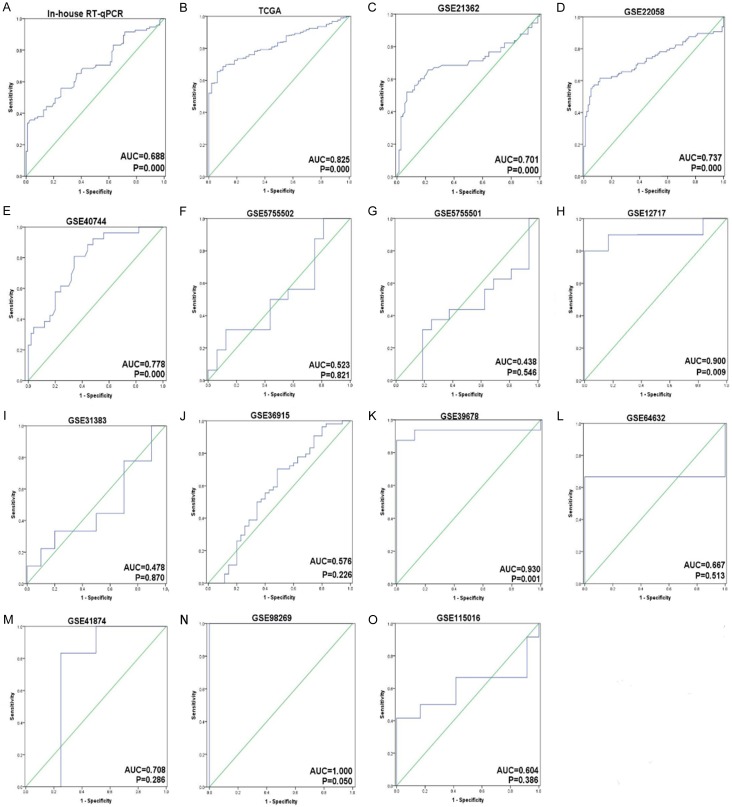
The scatterplots of individually included datasets. Based on the student’s t-test, the differences of 13 datasets were considered to be significant with P<0.05.
The increase of miR-452-5p in HCC was verified based on various databases
According to miRNA-seq data, the expression of miR-452-5p was markedly more elevated in tumors than in non-tumor tissues (P<0.0001, Figure 1B). In this study, according to the inclusion criteria, 13 GEO series (GSE) were selected from the GEO database. The expression levels of miR-452-5p in cancerous tissues were markedly up-regulated compared to the non-cancerous tissues in the 11 databases (P<0.05). However, in the rest of GSE data (GSE41874, GSE98269), no difference was noted for the miR-452-5p levels between the cancer and non-cancerous groups (Figure 1).
The meta-analysis included 15 studies from in-house RT-qPCR, miR-seq from TCGA, and microarrays from GEO mentioned above (Figure 2). The outcome of the meta-analysis notified that the expression level of miR-452-5p was observably up-regulated in 820 cases of HCC tissues compared with that in adjacent liver tissues (SMD=0.842, 95% CI [0.583,1.102], P<0.001), shown in Figure 3A. Publication bias was examined by graphic examination and quantitative assessment. No publication bias in our investigation (Pr>|z|=0.656) was detected in either Begg’s plot or Egger’s plot (Figure 3B) [44]. The root causes of heterogeneity were assessed by sensitivity analysis and no significant heterogeneity was found, as described above [45]. The ROC curves for the assessment of miR-452-5p expression in HCC were determined (Figure 4). The area under the sROC curve was 0.80 (0.76-0.83) (Figure 5A). The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) of miR-452-5p in these studies were 0.87 (95% CI [0.84, 0.89]), 0.58 (95% CI [0.54, 0.62]), 2.07 (95% CI [1.64, 2.60]), 0.31 (95% CI [0.20, 0.46]), and 7.71 (95% CI [4.77, 12.45]), respectively (Figure 5B-F) [46].
Figure 2.
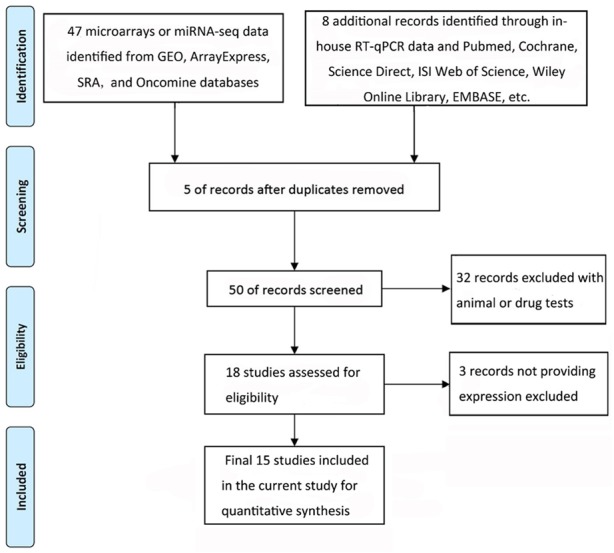
Flow chart of database search and selection.
Figure 3.
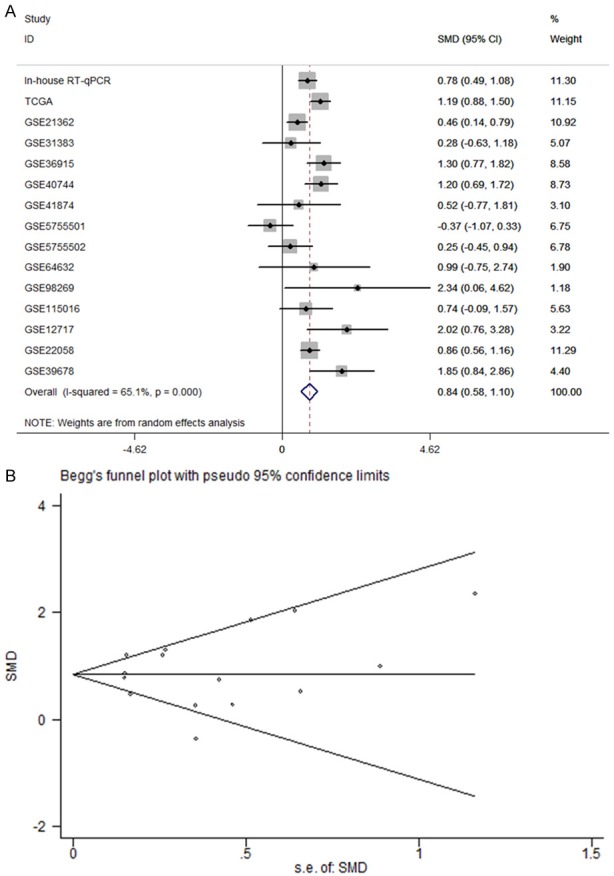
A. Forest plot of datasets evaluating miR-452-5p expression between HCC and control groups. B. The publication bias test of the meta-analysis.
Figure 4.
The ROC curves for the assessment of miR-452-5p expression in HCC.
Figure 5.
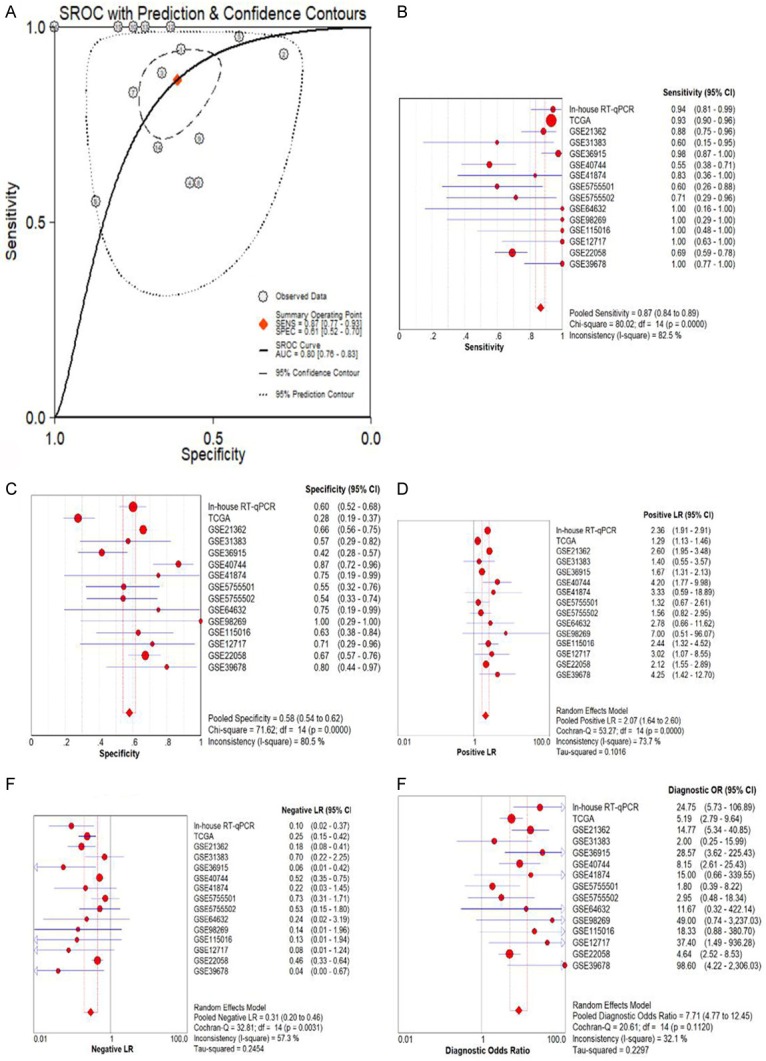
A. The SROC curves for the assessment of miR-452-5p. B-F. Forest plots showing the sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio, respectively.
Associations between clinic-pathological parameters and elevated expression of miR-452-5p in HCC tissues
The clinic-pathological characteristics of liver cancer clinical parameters downloaded from TCGA were generalized in Table 2. Expression of miR-452-5p in HCC tissues was memorably correlated with distant metastasis of liver cancer (8.302±0.198, P<0.001). No noticeable relationship was noted between miR-452-5p expression and the other clinic-pathological parameters of HCC, such as gender, event, vascular invasion, and lymph node metastasis.
Table 2.
Relationships between the expression of miR-452-5p and clinicopathological parameters in HCC analyzed by data from the TCGA database
| Clinicopathological feature | N | Relevant expression of miR-452-5P (log2x) | |||
|---|---|---|---|---|---|
|
| |||||
| Mean±SD | t | P-value | |||
| Tissue | Adjacent non-cancerous liver tissue | 49 | 5.522102±1.109133 | 10.541 | <0.001* |
| HCC | 372 | 7.419217±1.643848 | |||
| Gender | FEMALE | 119 | 7.560126±1.5700339 | 1.116 | 0.265 |
| MALE | 251 | 7.355922±1.689206 | |||
| Event | Alive | 240 | 7.307791±1.597293 | 1.771 | 0.077 |
| Dead | 129 | 7.625187±1.721841 | |||
| Stage | Stage I-II | 257 | 7.311789±1.574841 | -1.568 | 0.118 |
| Stage III-IV | 90 | 7.625093±1.785473 | |||
| Grade | G1-2 | 228 | 7.297407±1.645111 | -0.239 | 0.811 |
| G3-4 | 137 | 7.340539±1.715023 | |||
| T | T1-2 | 274 | 7.350438±1.58885 | -1.429 | 0.154 |
| T3-4 | 93 | 7.631074±1.771022 | |||
| N | NO | 253 | 7.400806±1.689829 | 0.273 | 0.785 |
| YES | 4 | 7.167555±2.213217 | |||
| M | NO | 267 | 7.395343±1.674103 | -6.362 | <0.001* |
| YES | 4 | 8.302672±0.198398 | |||
Note: Student’s unpaired t-test was used for comparison between two groups. One-way analysis of variance (ANOVA) was performed.
P<0.05 was considered statistically significant.
Functional enrichment
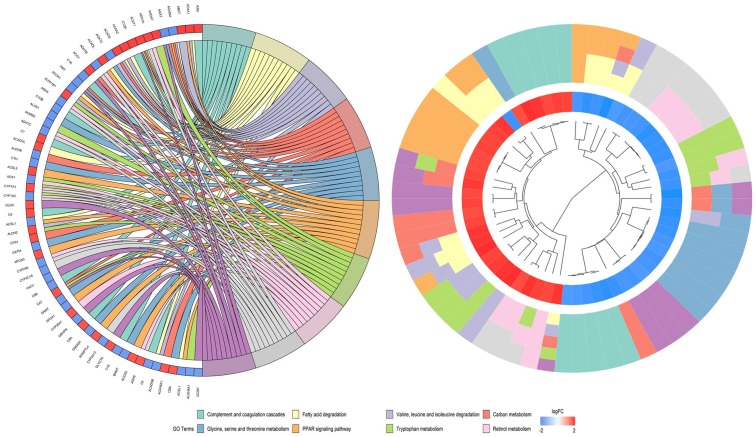
Through the verification of the literature and prediction by miRWalk3.0, we found that CDKN1B had a targeting relationship with miR-452-5p. As a result of functional enrichment, miR-452-5p plays a role in HCC tissues mainly through the small molecule catabolic process, organic acid catabolic process, and carboxylic acid catabolic process in biological processes, regarding cellular components, the most enriched pathways were mitochondrial matrix, extracellular matrix, and vesicle lumen. For molecular function, miR-452-5P was mainly aggregated in cofactor binding, coenzyme binding, and sulfur compound binding (Figure 6). Specific KEGG pathways were revealed, among which cytokine-cytokine receptor interaction, carbon metabolism, and complement and coagulation cascades deserve to be more thoroughly investigated (Figure 7).
Figure 6.
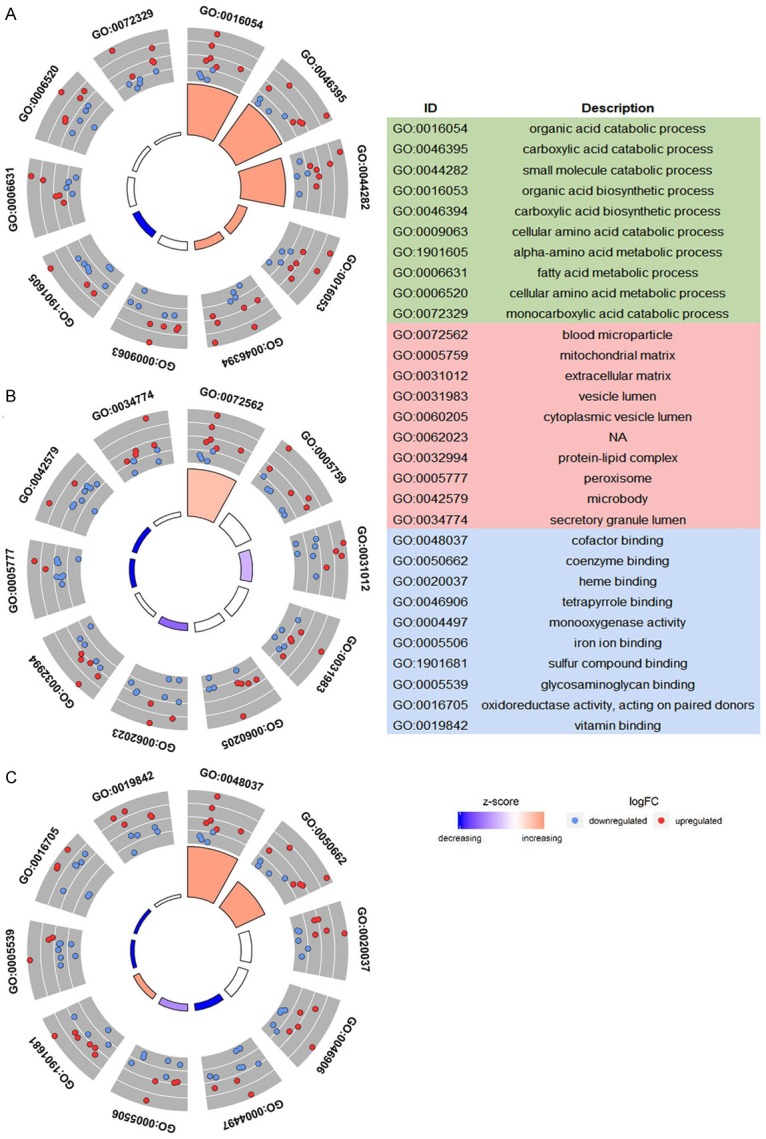
The GO of 1,135 predicted potential targets. A. Biological process. B. Cellular component. C. Molecular function.
Figure 7.
The KEGG pathways of 1,135 predicted potential targets.
CDKN1B is a direct target of miR-452-5p
We investigated whether miR-452-5p directly controls CDKN1B expression by binding to 3’UTR of CDKN1B mRNA through luciferase reporter gene detection. We found the binding site of miR-452-5p in the 3’UTR of CDKN1B mRNA. We identified that, compared with co-transfection with pmirglo-CDKN1B 3’UTR-wt and miR-452-5p mimic, the luciferase activity of pmirgl-CDKN1B 3’UTR-mut and miR-452-5p mimic was significantly reduced (P<0.05, Figure 8), indicating the specific binding of miR-452-5p and CDKN1B mRNA in 3’UTR. Thus, the result indicated that CDKN1B is a straightforward target of miR-452-5p, and miR-452-5p directly and negatively determines CDKN1B expression by binding to 3’UTR of CDKN1B mRNA.
Figure 8.
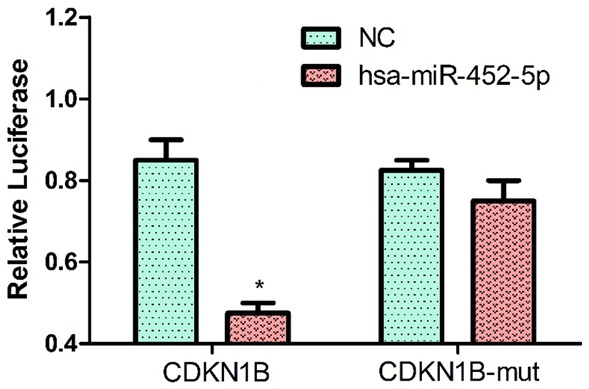
miR-452-5p suppresses expression by directly targeting the CDKN1B 3’UTR. The expression of CDKN1B transfected with miR-452-5p was significantly down-regulated compared with the control group (P<0.05). Whereas, the expression of CDKN1B-mut transfected miR-452-5p compared with the control group showed no significant difference (P>0.05).
Discussion
As far as we know, this is the first study to report the relationship of miR-452-5p with its target gene CDKN1B in HCC. The targeting regulation association between miR-452-5p and CDKN1B in HCC was investigated, and the results suggest that further exploration of the miR-452-5p underlying mechanisms in HCC may offer a theoretical basis for new treatments.
HCC is the most prevalent type of liver cancer. Due to late diagnosis and the lack of effective treatment, the mortality rate of HCC remains tremendously high [47]. It is reported that many genetic and epigenetic factors affect the incidence and progress of HCC. Qin et al. found that miRNA may play a crucial role in the pathogenesis of HCC [48]. Herein, the expression level of miR-452-5p in HCC tissues and matched adjacent normal tissues was quantified by RT-qPCR. The results indicated that the expression level of miR-452-5p in HCC tissues was significantly higher than that in adjacent normal tissues, and miR-452-5p was significantly up-regulated in HCC tissues with metastasis compared with that in HCC tissues without metastasis. This was constant with Zhu et al. [49]. The influence of miR-452-5p on the prognosis of HCC was also verified. These results suggest that miR-452-5p might play an important role in HCC as an oncogene and a biomarker with poor prognosis. Thus, miR-452-5p could be a new marker for the diagnosis and treatment of HCC.
Recently, an increasing amount of evidence has supported the impression that the miRNAs play a prime role in the molecular mechanism of HCC through changing their biological behavior in HCC. Zheng and others have shown that the expression of miR-452 is pronouncedly augmented in HCC tissues, miR-452 greatly accelerates the overexpression of proliferation, it significantly improves the migration and invasion of qgy-7703 and HepG2 cells in vitro, and it directly targets CDKN1B in the 3’UTR. The ectopic expression of miR-452 inhibits the expression level of CDKN1B mRNA and protein. Studies have shown that increased expression of miR-452-5p was increased in HCC tissues and that the high expression of miR-452-5p significantly improved cell growth, migration, and invasion in vitro. Furthermore, our results confirmed that CDKN1B is determined as a direct target gene of miR-452-5p in HCC by luciferase reporter assay. From this, we hold the opinion that miR-452-5p acts as a cancer promotor in HCC and its upregulation enhances metastasis of HCC.
Although miR-452-5p can target a great amount of predictive genes, our findings indicate that miR-452-5p can also target CDKN1B. In our research, we explored the potential molecular characterization of miR-452-5p mediated by CDKN1B in HCC.
CDKN1B, a member of the Cip/Kip family of cyclin/cyclin-dependent kinase inhibitors (CKIs), promotes the migration of metastatic HCC cells by regulating RhoA activity. Research has shown that the lack of CDKN1B in tumor cells infiltrating inflammatory cells and inflammatory cytokines in rising and STAT3 signaling pathways promotes the occurrence of carcinogen-induced liver cancer. Thus, the continued lack of cell cycle protein kinase inhibitor CDKN1B, traditionally considered as a result of DNA damage, by enhancing inflammation, in turn, promotes the progress of HCC, which can prevent the occurrence of cancer of the liver and the progress of promising therapeutic targets. Since the expression level of CDKN1B decreased in the occurrence and progress of HCC, the expression of CDKN1B in HCC has been considered as an indicator of the diagnosis and prognosis of HCC. Studies have shown that the expression of CDKN1B has something to do with tumor grade reduction and liver cancer recurrence. CDKN1B can decrease the proliferation and death of HCC cells and the accumulation of DNA damage [50]. In our research, we primarily concentrated on investigating the expressions of miR-452-5p and evaluated its function in HCC tissues. At the same time, we also concentrated on probing the targeted regulatory relationship between miR-452-5p and CDKN1B. Despite a large number of articles in the literature that have shown that CDKN1B is poorly expressed in liver cancer and inhibits the proliferation, migration, and invasion of tumor cells, the up-regulation of miR-452-5p could reverse these effects to a large extent. Luciferase analysis has provided evidence that CDKN1B is directly targeted by miR-452-5p. Thus, we speculated that high expression of miR-452-5p can play a role in promoting HCC by down-regulating CDKN1B.
Furthermore, the prospective mechanism of miR-452-5p in HCC was elucidated in respect to possible signaling pathways. A total of 1,135 predicted potential targets were annotated to investigate how miR-452-5p works on HCC. The GO analysis results were mainly related to metabolism, including the small molecule catabolic process, organic acid catabolic process, and carboxylic acid catabolic process. Studies have shown that miR-452 is elevated in HCC and predicts survival and advanced TNM staging in HCC patients, and miR-452 has been found to accelerate tumor stem cells via the activation of the Wnt signaling pathway by inhibiting Sox7. However, there is no direct evidence that miR-452-5p is related to the catabolic process of small molecules, the catabolic process of organic acids, or the catabolic process of carboxylic acids. In terms of biological engineering, this study hypothesized that miR-452-5p may play an anti-tumor role by activating the small molecule catabolic process, organic acid catabolic process, and carboxylic acid catabolic process. However, more research is needed to verify this.
In order to explain the potential machinery of miR-452-5p in HCC, we conducted KEGG pathway enrichment for 1,135 predicted potential targets, and the results showed that the cytokine-cytokine receptor interaction was the main pathway in which miR-452-5p acted in HCC. Cytokines are low molecular weight (15~30 KD) proteins or glycoproteins secreted by a variety of cells. They regulate cell growth and differentiation by binding to the corresponding receptors and participate in immune inflammatory reactions and wound healing. Cytokine-cytokine receptor interaction is the key to the study of cytokine function. Some studies indicated that there is a cytokine-cytokine interface in the assembly of higher-order structure and stimulation of the interleukin-3 receptor complex [51]. Ye et al. argue that comparative transcriptomic analysis of porcine peripheral blood discloses aberrantly expressed genes from the cytokine-cytokine receptor interaction pathway correlated with health status [52]. In this study, we suggest that miR-452-5p may exert its effect on HCC by affecting the cytokine-cytokine receptor interaction pathway.
The second abundant KEGG pathway is “carbon metabolism”, whose mechanism is quite complicated in HCC. Previous studies have reported that the polymorphism of OCM-related donor genes plays an important role in HCC recurrence after transplantation [53]. Genetic variations in the one-carbon metabolism gene may contribute to HCC susceptibility [54]. Also, research has indicated that one-carbon metabolism and nucleotide biosynthesis are attractive targets for anticancer therapy [55]. These results suggest that miR-452-5p may play a substantial role in the carcinogenesis of liver cancer through carbon metabolism.
At present, the study of miR-452-5p in HCC is superficial, and the discussion is not deep enough. However, we hope that the results of this study can supplement the pathogenesis of HCC. Luciferase reporter assay results showed that CDKN1B is determined as a direct target gene of miR-452-5p in HCC. This is powerful evidence proving that miR-452-5p plays an essential role in the development of HCC. Based on our findings, more research must be conducted to address the shortcomings of our study. The knockdown and overexpression may need to validate the role of miR-452-5p by in vitro experimentation. Moreover, a larger cohort is desirable to confirm the clinical role and prognostic value of CDKN1B in HCC patients. These findings can help us to explore more new prognostic markers and potential targets supporting the treatment strategy of liver cancer in the near future.
In summary, we determined that miR-452-5p is overexpressed in HCC tissues. According to our research, we proved that miR-452-5p may be a new marker for HCC. The overexpression of miR-452-5p may play a pivotal role in the carcinogenesis of HCC via targeting multiple signaling pathways and genes.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (NSFC 81860596), Guangxi Natural Scientific Research (No. 2016GXNSFAA380022) and Guangxi Zhuang Autonomous Region University Student Innovative Plan (2018229).
Disclosure of conflict of interest
None.
References
- 1.Massarweh NN, El-Serag HB. Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Control. 2017;24:1073274817729245. doi: 10.1177/1073274817729245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, Forgues M, Pomyen Y, Bhudhisawasdi V, Lertprasertsuke N, Chotirosniramit A, Pairojkul C, Auewarakul CU, Sricharunrat T, Phornphutkul K, Sangrajrang S, Cam M, He P, Hewitt SM, Ylaya K, Wu X, Andersen JB, Thorgeirsson SS, Waterfall JJ, Zhu YJ, Walling J, Stevenson HS, Edelman D, Meltzer PS, Loffredo CA, Hama N, Shibata T, Wiltrout RH, Harris CC, Mahidol C, Ruchirawat M, Wang XW. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell. 2017;32:57–70. e53. doi: 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo EC, N Rucker A, Federle MP. Hepatocellular carcinoma and intrahepatic cholangiocarcinoma: imaging for diagnosis, tumor response to treatment and liver response to radiation. Semin Radiat Oncol. 2016;28:267–276. doi: 10.1016/j.semradonc.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Labib PL, Goodchild G, Pereira SP. Molecular pathogenesis of cholangiocarcinoma. BMC Cancer. 2019;19:185. doi: 10.1186/s12885-019-5391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Li ZY, Hou XX, Wang X, Luo YH, Ying YP, Chen G. Clinical significance and effect of AEG-1 on the proliferation, invasion, and migration of NSCLC: a study based on immunohistochemistry, TCGA, bioinformatics, in vitro and in vivo verification. Oncotarget. 2017;8:16531–16552. doi: 10.18632/oncotarget.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi B, Luo Y, Jian Z, Zhou X. Prognostic value of plasma fibrinogen in hepatocellular carcinoma: a meta-analysis. Cancer Manag Res. 2018;10:5027–5041. doi: 10.2147/CMAR.S175780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosseinzadeh F, Verdi J, Ai J, Hajighasemlou S, Seyhoun I, Parvizpour F, Hosseinzadeh F, Iranikhah A, Shirian S. Combinational immune-cell therapy of natural killer cells and sorafenib for advanced hepatocellular carcinoma: a review. Cancer Cell Int. 2018;18:133. doi: 10.1186/s12935-018-0624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Ruan Z, Yu S, Tian T, Liang X, Jing L, Li W, Wang X, Xiang L, Claret FX, Nan K, Guo H. A four-methylated mRNA signature-based risk score system predicts survival in patients with hepatocellular carcinoma. Aging (Albany NY) 2019;11:160–173. doi: 10.18632/aging.101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mocan T, Sparchez Z, Craciun R, Bora CN, Leucuta DC. Programmed cell death protein-1 (PD-1)/programmed death-ligand-1 (PD-L1) axis in hepatocellular carcinoma: prognostic and therapeutic perspectives. Clin Transl Oncol. 2019;21:702–712. doi: 10.1007/s12094-018-1975-4. [DOI] [PubMed] [Google Scholar]
- 11.Schaub SK, Hartvigson PE, Lock MI, Hoyer M, Brunner TB, Cardenes HR, Dawson LA, Kim EY, Mayr NA, Lo SS, Apisarnthanarax S. Stereotactic body radiation therapy for hepatocellular carcinoma: current trends and controversies. Technol Cancer Res Treat. 2018;17:1533033818790217. doi: 10.1177/1533033818790217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W, Tan HY, Wang N, Wang X, Feng Y. Deciphering hepatocellular carcinoma through metabolomics: from biomarker discovery to therapy evaluation. Cancer Manag Res. 2018;10:715–734. doi: 10.2147/CMAR.S156837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 14.Pan D, Lin P, Wen D, Wei Y, Mo Q, Liang L, Chen G, He Y, Chen J, Yang H. Identification of down-regulated microRNAs in thyroid cancer and their potential functions. Am J Transl Res. 2018;10:2264–2276. [PMC free article] [PubMed] [Google Scholar]
- 15.Ye D, Shen Z, Zhou S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag Res. 2019;11:969–979. doi: 10.2147/CMAR.S191696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang CZ, Deng F, Li H, Wang DD, Zhang W, Ding L, Tang JH. MiR-101: a potential therapeutic target of cancers. Am J Transl Res. 2018;10:3310–3321. [PMC free article] [PubMed] [Google Scholar]
- 17.Hua KT, Liu YF, Hsu CL, Cheng TY, Yang CY, Chang JS, Lee WJ, Hsiao M, Juan HF, Chien MH, Yang SF. 3’UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci Rep. 2017;7:4466. doi: 10.1038/s41598-017-04732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YJ, Guo YN, Shi K, Huang HM, Huang SP, Xu WQ, Li ZY, Wei KL, Gan TQ, Chen G. Down-regulation of microRNA-144-3p and its clinical value in non-small cell lung cancer: a comprehensive analysis based on microarray, miRNA-sequencing, and quantitative real-time PCR data. Respir Res. 2019;20:48. doi: 10.1186/s12931-019-0994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayya VK, Duchaine TF. Ciphers and executioners: how 3’-untranslated regions determine the fate of messenger RNAs. Front Genet. 2019;10:6. doi: 10.3389/fgene.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Liang X, Li X, Zhang Y, Sun Z, Liu Y, Wang J. MicroRNA-195: a review of its role in cancers. Onco Targets Ther. 2018;11:7109–7123. doi: 10.2147/OTT.S183600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Zheng H, Chan MT, Wu WKK. MicroRNAs: new players in cataract. Am J Transl Res. 2017;9:3896–3903. [PMC free article] [PubMed] [Google Scholar]
- 22.Song C, Chen H, Song C. Research status and progress of the RNA or protein biomarkers for prostate cancer. Onco Targets Ther. 2019;12:2123–2136. doi: 10.2147/OTT.S194138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Zhou S, Fan K, Jiang C. MicroRNA-21 and its impact on signaling pathways in cervical cancer. Oncol Lett. 2019;17:3066–3070. doi: 10.3892/ol.2019.10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei DM, Jiang MT, Lin P, Yang H, Dang YW, Yu Q, Liao DY, Luo DZ, Chen G. Potential ceRNA networks involved in autophagy suppression of pancreatic cancer caused by chloroquine diphosphate: a study based on differentially expressed circRNAs, lncRNAs, miRNAs, and mRNAs. Int J Oncol. 2019;54:600–626. doi: 10.3892/ijo.2018.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mace TA, Collins AL, Wojcik SE, Croce CM, Lesinski GB, Bloomston M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J Surg Res. 2013;184:855–60. doi: 10.1016/j.jss.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Yang Y, Xia L, Yang Y, Wang F, Song M, Chen X, Liu J, Song Y, Zhao Y, Yang C. MiR-221 promotes Capan-2 pancreatic ductal adenocarcinoma cells proliferation by targeting PTEN-Akt. Cell Physiol Biochem. 2016;38:2366–2374. doi: 10.1159/000445589. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The role of MicroRNAs in hepatocellular carcinoma. J Cancer. 2018;9:3557–3569. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puerta-Gil P, García-Baquero R, Jia AY, Ocaña S, Alvarez-Múgica M, Alvarez-Ossorio JL, Cordon-Cardo C, Cava F, Sánchez-Carbayo M. MiR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–1815. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Q, Sheng Q, Jiang C, Shu J, Chen J, Nie Z, Lv Z, Zhang Y. MicroRNA-452 promotes tumorigenesis in hepatocellular carcinoma by targeting cyclin-dependent kinase inhibitor 1B. Mol Cell Biochem. 2014;389:187–195. doi: 10.1007/s11010-013-1940-z. [DOI] [PubMed] [Google Scholar]
- 32.Chang IF. Proteomic characterization of evolutionarily conserved and variable proteins of arabidopsis cytosolic ribosomes. Plant Physiol. 2005;137:848–862. doi: 10.1104/pp.104.053637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F, Guo L, Xin G, Wang Z. MiR-452 promotes cell metastasis and the epithelial to mesenchymal by targeting SOX7 in clear-cell renal-cell carcinoma. J Cell Biochem. 2018 doi: 10.1002/jcb.28125. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Tang H, Zhang J, Yu Z, Ye L, Li K, Ding F, Feng X, Meng W. Mir-452-3p: a potential tumor promoter that targets the CPEB3/EGFR Axis in human hepatocellular carcinoma. Technol Cancer Res Treat. 2017;16:1136–1149. doi: 10.1177/1533034617735931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, Lin B, Chen T, Xing C, Liu Z, Song P, Yin S, Zheng S, Zhou L. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:28000–28012. doi: 10.18632/oncotarget.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Wu Y, Li P. MicroRNA-452 suppresses pancreatic cancer migration and invasion by directly targeting B-cell-specific Moloney murine leukemia virus insertion site 1. Oncol Lett. 2017;14:3235–3242. doi: 10.3892/ol.2017.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Qiao N, Wang Y, Jiang M, Wang S, Wang C, Hu L. Association between the telomerase reverse transcriptase (TERT) rs2736098 polymorphism and cancer risk: evidence from a case-control study of non-small-cell lung cancer and a meta-analysis. PLoS One. 2013;8:e76372. doi: 10.1371/journal.pone.0076372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He RQ, Yang X, Liang L, Chen G, Ma J. MicroRNA-124-3p expression and its prospective functional pathways in hepatocellular carcinoma: a quantitative polymerase chain reaction, gene expression omnibus and bioinformatics study. Oncol Lett. 2018;15:5517–5532. doi: 10.3892/ol.2018.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He RQ, Wu PR, Xiang XL, Yang X, Liang HW, Qiu XH, Yang LH, Peng ZG, Chen G. Downregulated miR-23b-3p expression acts as a predictor of hepatocellular carcinoma progression: a study based on public data and RT-qPCR verification. Int J Mol Med. 2018;41:2813–2831. doi: 10.3892/ijmm.2018.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang L, Zeng JH, Wang JY, He RQ, Ma J, Chen G, Cai XY, Hu XH. Down-regulation of miR-26a-5p in hepatocellular carcinoma: a qRT-PCR and bioinformatics study. Pathol Res Pract. 2017;213:1494–1509. doi: 10.1016/j.prp.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Liang HW, Ye ZH, Yin SY, Mo WJ, Wang HL, Zhao JC, Liang GM, Feng ZB, Chen G, Luo DZ. A comprehensive insight into the clinicopathologic significance of miR-144-3p in hepatocellular carcinoma. Onco Targets Ther. 2017;10:3405–3419. doi: 10.2147/OTT.S138143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding H, Ye ZH, Wen DY, Huang XL, Zeng CM, Mo J, Jiang YQ, Li JJ, Cai XY, Yang H, Chen G. Downregulation of miR1365p in hepatocellular carcinoma and its clinicopathological significance. Mol Med Rep. 2017;16:5393–5405. doi: 10.3892/mmr.2017.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu L, Yang N, Chen J, Zeng T, Yan S, Liu Y, Yu G, Chen Q, Du G, Pan W, Li X, Zhou H, Huang A, Tang H. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget. 2017;8:63724–63737. doi: 10.18632/oncotarget.18892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei L, Tan G, Wang L, Guo W, Xiao B, Gao X, Wang L, Li H, Xu Z, Zhang X, Zhao J, Yi J, Huang Y. Comparison of combined general-epidural anesthesia with general anesthesia effects on survival and cancer recurrence: a meta-analysis of retrospective and prospective studies. PLoS One. 2014;9:e114667. doi: 10.1371/journal.pone.0114667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen DY, Lin P, Liang HW, Yang X, Li HY, He Y, Yang H, Chen G. Up-regulation of CTD-2547G23. 4 in hepatocellular carcinoma tissues and its prospective molecular regulatory mechanism: a novel qRT-PCR and bioinformatics analysis study. Cancer Cell Int. 2018;18:74. doi: 10.1186/s12935-018-0566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 48.Qin Y, Dang X, Li W, Ma Q. miR-133a functions as a tumor suppressor and directly targets FSCN1 in pancreatic cancer. Oncol Res. 2013;21:353–363. doi: 10.3727/096504014X14024160459122. [DOI] [PubMed] [Google Scholar]
- 49.Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, Lin B, Chen T, Xing C, Liu Z, Song P, Yin S, Zheng S, Zhou L. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:28000–28012. doi: 10.18632/oncotarget.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranchal I, González R, Bello RI, Ferrín G, Hidalgo AB, Linares CI, Aguilar-Melero P, González-Rubio S, Barrera P, Marchal T, Nakayama KI, de la Mata M, Muntané J. The reduction of cell death and proliferation by p27 (Kip 1) minimizes DNA damage in an experimental model of genotoxicity. Int J Cancer. 2009;125:2270–80. doi: 10.1002/ijc.24621. [DOI] [PubMed] [Google Scholar]
- 51.Dey R, Ji K, Liu Z, Chen L. A cytokine-cytokine interaction in the assembly of higher-order structure and activation of the interleukine-3: receptor complex. PLoS One. 2009;4:e5188. doi: 10.1371/journal.pone.0005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye MH, Bao H, Meng Y, Guan LL, Stothard P, Plastow G. Comparative transcriptomic analysis of porcine peripheral blood reveals differentially expressed genes from the cytokine-cytokine receptor interaction pathway related to health status. Genome. 2017;60:1021–1028. doi: 10.1139/gen-2017-0074. [DOI] [PubMed] [Google Scholar]
- 53.Zheng GY, Zhao R, Feng XH. [Dot-immunobinding assay with sheep hydatid fluid antigen for the diagnosis of hydatidosis] . Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing ZaZhi. 1988;6:87–9. [PubMed] [Google Scholar]
- 54.Zhang H, Liu C, Han YC, Ma Z, Zhang H, Ma Y, Liu X. Genetic variations in the one-carbon metabolism pathway genes and susceptibility to hepatocellular carcinoma risk: a case-control study. Tumour Biol. 2015;36:997–1002. doi: 10.1007/s13277-014-2725-z. [DOI] [PubMed] [Google Scholar]
- 55.Shuvalov O, Petukhov A, Daks A, Fedorova O, Vasileva E, Barlev NA. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget. 2017;8:23955–23977. doi: 10.18632/oncotarget.15053. [DOI] [PMC free article] [PubMed] [Google Scholar]