Abstract
Gastric cancer (GC) is one of the most common malignant tumors of the digestive system. A deeper understanding of the mechanism of proliferation and metastasis is needed to improve patient survival. T cell lymphoma invasion and metastasis 1 (TIAM1) has been proven to play an essential role in the proliferation and metastasis of GC. The aim of this study was to explore the relevant upstream regulatory mechanism of TIAM1. Bioinformatic analysis, RT-qPCR, and dual luciferase reporter assays were used to predict and validate microRNAs that target the TIAM1 gene. Among eleven predicted microRNAs, eight (miR-10b-5p, miR-589-3p, miR-651-3p, miR-335-3p, miR-653-5p, miR-373-3p, miR-372-3p, and miR-205-3p) affected TIAM1 expression; and only miR-10b-5p regulated TIAM1 expression by directly binding to the 3’-UTR of TIAM1 mRNA. miR-10b-5p levels were determined in both normal and cancerous tissues retrieved from GC patients. We observed that by targeting TIAM1 expression, miR-10b-5p inhibited the proliferation, migration, and invasion of GC cells. To verify our observations, we evaluated the participation of runt-related transcription factor 3 (RUNX3), a known regulator of microRNA expression and tumor suppressor. Tumor-suppressor RUNX3 combined with core-binding factor subunit beta (CBFβ) upregulated miR-10b-5p and suppressed GC. In conclusion, we identified a CBFβ/RUNX3-miR10b-TIAM1 molecular axis that inhibits GC progression and metastasis and may provide suitable treatment targets for GC.
Keywords: Gastric cancer, T cell lymphoma invasion and metastasis 1, microRNA-10b-5p, runt-related transcription factor 3, core-binding factor subunit beta
Introduction
Gastric cancer (GC) has an extremely high rate of mortality and is one of the leading causes of cancer-related death worldwide [1,2]. Although the 5-year overall survival rate of patients with GC has been prolonged to some degree by endoscopic screening and treatment [3], it remains difficult to inhibit tumor proliferation and prevent metastasis [4]. A detailed understanding of the molecular mechanisms of GC will contribute to improving patient survival [4-6]. Among the various signaling pathways and mechanisms regulating GC, T cell lymphoma invasion and metastasis 1 (TIAM1) encodes a RAC1-specific guanine nucleotide exchange factor (GEF), and, by regulating RAC1 signaling pathways, this gene plays an important role in carcinogenesis, progression, invasion, and metastasis including in GC [7-10]. Knowledge of the relevant regulatory mechanisms of TIAM1 may facilitate further research on the mechanisms underlying metastasis and on treatment targets for GC.
As the important roles of microRNAs are increasingly recognized [11] and microRNA-based therapeutics in various tumors have been reported [12], we focused on the upstream regulatory mechanism of TIAM1-related microRNAs. Although numerous microRNAs targeting TIAM1 are predicted according to online databases, only a few of them have been experimentally validated [13], not to mention the genes that can regulate those microRNAs.
Although no genes have been reported to regulate TIAM1-related microRNAs in GC, previous studies have shown that runt-related transcription factor (RUNX) is related to the regulation of other tumor-related microRNAs [14-16]. RUNX, also known as CBFα, typically combines with core-binding factor subunit beta (CBFβ) to form a heterodimeric core-binding transcription factor that plays important roles in various diseases including cancers [17-19]. The three members of the RUNX family, RUNX1, RUNX2, and RUNX3, contain the same runt domain and show distinct tissue-specific expression patterns [20,21]. RUNX3, rather than RUNX2 or RUNX1, is most closely associated with GC, and its inactivation has been established as a causative factor in tumorigenesis [20-23]. Thus, a relationship may exist between RUNX3, TIAM1-related microRNAs, and the malignancy of GC.
In this study, we investigated the microRNAs that target TIAM1 using bioinformatic analysis based on seven online databases, RT-qPCR, and dual luciferase reporter assays. In addition, the relevant effects on proliferation, invasion, and migration of GC were observed using CCK and transwell assays in BGC823 cells. The involvement of the CBFβ-RUNX3 interaction in the regulatory mechanism of GC was tested in CBFB-knockdown BGC823 cells.
Materials and methods
Human tissues
A total of 38 tissues including 19 GC tissues and 19 corresponding adjacent normal gastric epithelial tissues were obtained from patients with primary GC. The fresh tissues were stored in liquid nitrogen within 30 min after being excised during surgery until RNA extraction. The clinicopathologic characteristics of the patients with GC involved in this study are shown in Table 1. The research protocol was reviewed and approved by the approval of the Ethics Review Committees of the First Hospital of Jilin University, and written informed consent was obtained from each patient.
Table 1.
Characteristics of patients with GC
| Finding | No. of cases | % |
|---|---|---|
| Gender | ||
| Male | 10 | 53 |
| Female | 9 | 47 |
| Age | ||
| ≥ 60 years | 14 | 74 |
| < 60 years | 5 | 26 |
| Pathological stage | ||
| I-II | 2 | 11 |
| III-IV | 17 | 89 |
Cell lines
All cell lines in this study, including BGC823, MGC803, SGC7901, and GES-1 cells, were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were routinely cultured in RPMI 1640 (HyClone, Logan, UT, USA) (BGC823, MGC803, and SGC7901 cells) or DMEM (HyClone) (GES-1) containing 10% fetal bovine serum (Biological Industries, Beit-Haemek, Israel) and 1% penicillin-streptomycin antibiotics (Biological Industries) at 37°C in a 5% CO2 constant-temperature incubator.
microRNA prediction
Seven online databases were searched for microRNAs: Targetscan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), MicroT-CDS (http://www.microrna.gr/mic-roT-CDS/), miRNet (http://www.mirnet.ca/), miRSystem (http://mirsystem.cgm.ntu.edu.tw/), and miRNAMap (http://mirnamap.mbc.nctu.edu.tw/). MicroRNAs predicted by at least three online databases were selected for further research.
Plasmid construction
The entire 3’-UTR (1950 bp in length) of TIAM1 was cloned into the psiCHECKTM-2 vector (Promega) that contained firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferase reporter genes. The 3’-UTR of TIAM1 was inserted into the 3’-end of the Renilla luciferase gene. The firefly luciferase gene served as an internal control to normalize Renilla luciferase expression. The vector map (psi-TIAM1-3’-UTR) (created by SnapGene®) is shown in Figure S1. The overexpression vector plasmids expressing myc-tagged RUNX3 were a gift from Dr. A. Friedman. The construction of pc-CBFB-myc expressing myc-tagged CBFβ has been described previously [24].
Transfection and lentivirus infection
The mimic/inhibitor of microRNAs (Mimics and Inhibitors) and corresponding negative controls (Ribobio, Shanghai, China) were transfected using LipofectamineTM RNAiMax (Invitrogen, Carlsbad, CA, USA) based on the manufacturer’s protocol. The plasmids were transfected using Lipofectamine3000 (Invitrogen). Lentiviral vectors (pLK0.1-puro/shCBFB-puro, RRE, REV, Vsvg) were co-transfected into 293T cells using Lipofectamine3000. Twenty-four hours after transfection, the culture medium was changed. On the next day, the supernatants were collected and filtered through a 0.22-μm membrane. GC cells were infected with virus in the presence of 0.1% DEAE-Dextran (Sigma, St. Louis, MO, USA) for 48 h. Next, culture medium containing 3 µg/mL of puromycin (Sigma) was used to select cells for one week to obtain stably infected cells.
Dual-luciferase reporter gene assay
Vectors psi-TIAM1-3’-UTR or psi-TIAM1-3’-UTR-10b-MU were transfected into GC cells. After 48 h, the Renilla luciferase activity (psi-TIAM1-3’-UTR and psi-TIAM1-3’-UTR-10b-MU, normalized to firefly luciferase activity) was measured using a luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
RNA extraction and real-time PCR
RNAs were extracted from tissues or cells using the EasyPure ® miRNA kit (Transgene, Beijing, China) or EasyPure ® RNA kit (Transgene) according to the manufacturer’s instructions. Next, cDNA was synthesized using the EasyScript ® First-Step cRNA Synthesis SuperMix kit (Transgene) and RT-qPCR was performed using the TransStart® Tip-qMix kit (Transgene). miR-10b-5p and U6 were quantified with the Bulge-loopTM miRNA RT-qPCR Premier Set (Ribobio). The other primer sequences were as follows: 5’-AGATGGCAAGAGGGAGAAAGAAG-3’ (forward) and 5’-GACAGAACCAGGATGGAGTGAAATAC-3’ (reverse) for TIAM1; 5’-GCAGGGAGAACAGCGACAAA-3’ (forward) and 5’-CAAACTCCAGACAGCCCATACC-3’ (reverse) for CBFB; 5’-ACCGAGCGCGGCTACAG-3’ (forward) and 5’-CTTAATGTCACGCACGATTTCC-3’ (reverse) for β-actin. The relative expression levels of TIAM1, CBFB and miR-10b-5p were calculated using the 2-ΔΔCt method and normalized to the levels of β-actin and U6.
CCK-8 assay for cell proliferation
Cells were transferred into 96-well plates 24 h after transfection at a density of 5 × 103 per well. The cells were cultured for 0, 24, 48, and 72 h in medium. At each time point, 10 µL CCK solution was added to each well. Next, the 96-well plates were incubated for 2 h before elucidating the optical density (OD) values at 450 nm, which were then used to determine the efficiency of cell proliferation.
Transwell assay for cell migration and invasion
Transwell chambers (8.0-µm, Corning, Inc., Corning, NY, USA) without Matrigel coating (Corning) were used for cell migration analysis and coated chambers were used for cell invasion analysis. The cells were resuspended in 200 µL serum-free medium and transferred into the upper compartment 24 h after transfection. Culture medium containing 10% fetal bovine serum was added to the lower compartments. Each chamber was seeded with the same number of cells. Before staining and counting, migrated cells were incubated for 24 h and invading cells were incubated for 48 h. Cells with stronger migration or invasion capabilities showed a greater ability to pass through the uncoated or coated Transwell chamber membrane. The average numbers of migrating or invading cells in each group were calculated in at least five fields.
Western blot antibodies
Cells and tissues were lysed by RIPA lysis buffer (Sigma, USA). Protein extract was conducted by bicinchoninic acid (BCA) method. The following antibodies were used in this study: anti-myc (Covance, Princeton, NJ, USA), anti-β-actin (Covance, Princeton, NJ, USA), anti-histone (Abcam, Cambridge, UK), anti-CBFβ (Abcam, Cambridge, UK), anti-TIAM1 (Abcam, Cambridge, UK) and anti-RUNX3-endo (Abcam, Cambridge, UK).
Statistical analysis
All data were analyzed using SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA). Significant differences between groups were evaluated by Student’s t-test. The results are displayed as the mean ± standard deviation, with p values less than 0.05 considered significant.
Results
TIAM1 expression is affected by eight microRNAs and miR-10b-5p directly targets TIAM1 mRNA-3’-UTR
To verify the reported association between GC and TIAM1, we determined the relative expression of TIAM1 in vitro using RT-qPCR and western blotting. The results show increased expression of TIAM1 in GC cells (BGC823, MGC823, and SGC7901) compared to the normal gastric mucosal epithelial cell line (GES-1) both in mRNA and protein levels (Figure 1A and 1B).
Figure 1.
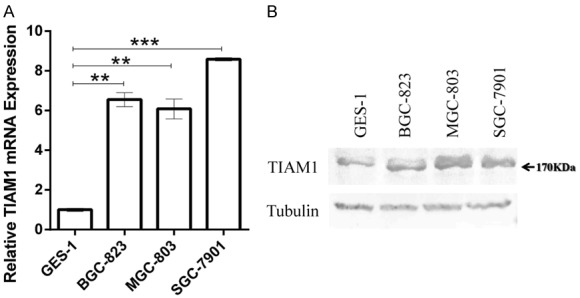
TIAM1 expression levels increased in gastric cancer (GC) cells (BGC823, MGC803, and SGC7901) compared to normal gastric epithelial cells (GES-1) both at mRNA (A) and protein (B) levels. (A) Bar charts showing the relative expression levels of TIAM1 mRNA. (B) Western blotting results of TIAM1 protein. **P < 0.01, ***P < 0.001.
To identify TIAM1-related microRNAs, we searched more than 20 online databases (Table S1) and selected seven databases based on the study design, update time, inclusion range, and algorithms as diverse as reasonably possible. Eleven microRNAs predicted by at least three online databases and most likely targeting the 3’-UTR of TIAM1 mRNA were included in this study (Table 2). BGC823 cells were used for TIAM1-related microRNAs experimental verification and screening because of the high transfection efficiency. Mimics, inhibitors, and the corresponding negative controls of eleven predicted microRNAs were transfected into BGC823 cells. After 48 h, TIAM1 mRNA expression levels were evaluated using RT-qPCR. The results show that the expression levels of TIAM1 mRNA were affected by miR-10b-5p, miR-589-3p, miR-651-3p, miR-335-3p, miR-653-5p, miR-373-3p, miR-372-3p, and miR-205-3p, but not by miR-4770, miR-592, or miR-7844-5p (Figure 2).
Table 2.
MicroRNAs predicted to target TIAM1 mRNA 3’UTR
| No. | Name | Targetscan | miRDB | miRWalk | MicroT-CDS | miRNAMap | miRNet | miR-System |
|---|---|---|---|---|---|---|---|---|
| No.1 | miR-10b-5p | Yes | Yes | No | Yes | No | No | Yes |
| No.2 | miR-589-3p | Yes | No | Yes | Yes | Yes | Yes | No |
| No.3 | miR-651-3p | Yes | Yes | Yes | No | No | Yes | No |
| No.4 | miR-335-3p | Yes | Yes | Yes | Yes | Yes | Yes | No |
| No.5 | miR-653-5p | Yes | Yes | Yes | Yes | No | Yes | Yes |
| No.6 | miR-373-3p | Yes | Yes | Yes | Yes | No | Yes | Yes |
| No.7 | miR-372-3p | Yes | Yes | Yes | Yes | No | Yes | Yes |
| No.8 | miR-205-3p | Yes | No | Yes | Yes | No | Yes | No |
| No.9 | miR-4770 | Yes | Yes | No | No | No | Yes | No |
| No.10 | miR-592 | Yes | No | No | Yes | No | Yes | Yes |
| No.11 | miR-7844-5p | Yes | No | Yes | Yes | No | Yes | No |
Figure 2.
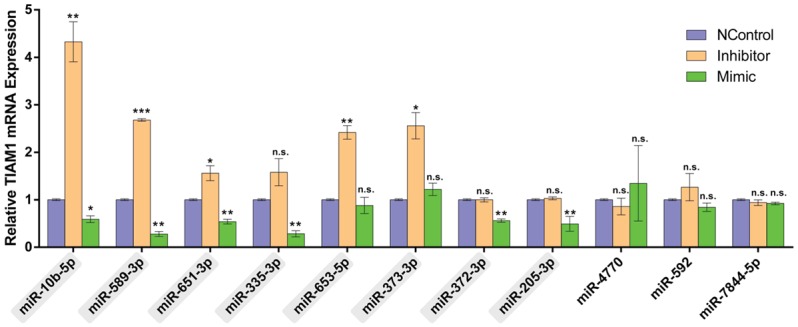
TIAM1 expression levels were affected by several microRNAs. Bar charts indicating the relative expression levels of TIAM1 mRNA after transfection with mimics or inhibitors of different microRNAs. The results show that miR-10b-5p, miR-589-3p, miR-651-3p, miR-335-3p, miR-653-5p, miR-373-3p, miR-372-3p, and miR-205-3p affected TIAM1 expression, but not miR-4770, miR-592, or miR-7844-5p. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. P > 0.05.
Next, we used dual-luciferase reporter gene assays to confirm a direct relationship between the above eight microRNAs and TIAM1 mRNA-3’-UTR. As shown in Figure 3A, the inhibition or overexpression of miR-10b-5p significantly increased or decreased, respectively, the relative activity of luciferase in BGC823 cells. For the other seven TIAM1-related microRNAs, dual-luciferase assay results did not support a direct relationship (Figure S2). We then mutated the miR-10b-5p binding site in the TIAM1-3’-UTR in the psi-TIAM1-3’-UTR plasmid (Figure 3B). After co-transfection of miR-10b-mimic or miR-10b-inhibitor and psi-TIAM1-3’-UTR-mu, no obvious changes in the relative activity of luciferase activity were observed (Figure 3A). These findings suggest that miR-10b-5p negatively regulates TIAM1 through direct interaction.
Figure 3.
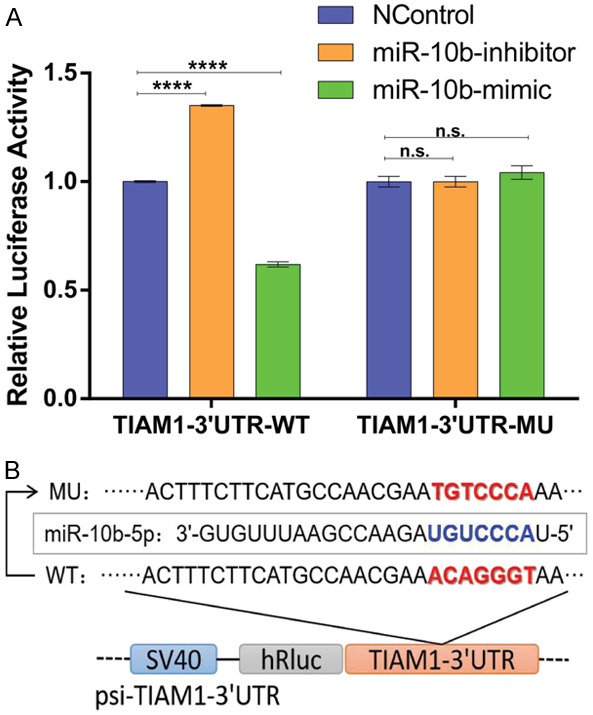
miR-10b-3p directly targeted TIAM1 mRNA 3’-UTR. A. Bar charts showing the relative luciferase activity after co-transfection with psi-TIAM1-3’-UTR/psi-TIAM1-3’-UTR-10b-MU and the mimcs/inhibitors of miR-10b-5p. B. Schematic diagram of mutant miR-10b-5p binding site. ****P < 0.0001, n.s. P > 0.05.
miR-10b-5p inhibits the proliferation, migration, and invasion of GC cells and RUNX3 upregulates miR-10b-5p expression
To determine the roles of miR-10b-5p in GC, we first investigated its expression levels both in GC tissues and cell lines. The results showed that miR-10b-5p was down-regulated in GC tissues compared to in adjacent normal gastric mucosal epithelium (P = 0.0041) (Figure 4A). Similarly, reduced miR-10b-5p expression was observed in all GC cell lines (BGC823, MGC803, SGC7901) compared to the normal gastric epithelial cell line (GES-1) (Figure 4B). The GC cell line BGC823 was further evaluated because of its high transfection efficiency. The function of miR-10b-5p in GC cell proliferation was determined by CCK-8 assay. The relative OD450 values of GC cells transfected with the miR-10b-mimic were significantly lower than those of GC cells transfected the NC-mimic in CCK-24 h, CCK-48 h, and CCK-72 h, whereas inhibition of miR-10b-5p showed the opposite results (Figure 4C). These results suggest that the miR-10b-mimic suppressed the proliferation of BGC823 cells. The migration and invasion abilities of miR-10b-5p in GC cells were investigated by Transwell assay. The results revealed that the numbers of migrated and invaded cells in the miR-10b-mimic group were significantly lower than in the NC-mimic groups, whereas inhibition of miR-10b-5p showed the opposite results (Figure 4D-F). This indicates that miR-10b-mimic can suppress the migration and invasion of BGC823 cells. Hence, it suggests that miR-10b-5p inhibits the proliferation, migration, and invasion of GC cells.
Figure 4.
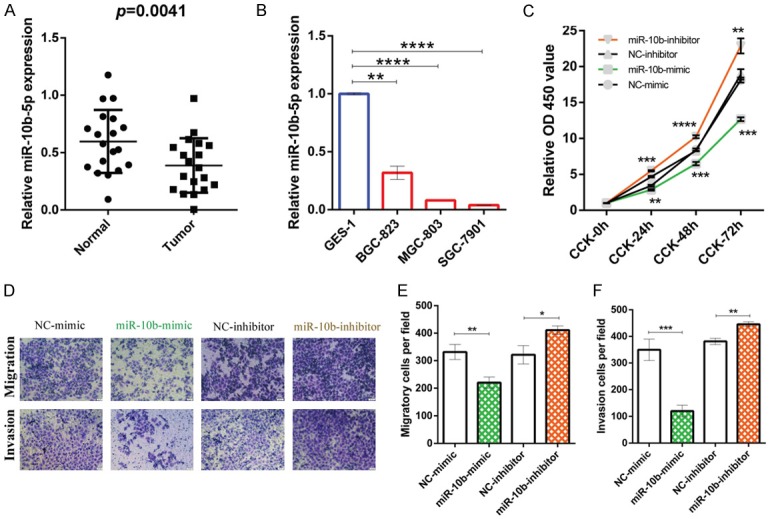
miR-10b-5p is decreased in GC and miR-10b-5p suppresses the proliferation, migration, and invasion of GC cells. (A) Columnar scatterplot illustrates the reduced expression of miR-10b-5p in gastric cancer tissues compared to paired adjacent normal gastric mucosal tissues. The relative expression levels of miR-10b-5p of 19 paired gastric cancer tissues and corresponding normal gastric mucosal tissues were measured by RT-qPCR and normalized to U6. (B) Bar charts indicating the reduced expression of miR-10b-5p detected by RT-qPCR in gastric cancer cells (BGC823, MGC803, and SGC7901) compared to normal gastric epithelial cells (GES-1). (C-F) Results of CCK-8 assay and Transwell assay indicate the inhibitory effects of miR-10b-5p on the proliferation (C), migration (D, E), and invasion (D, F) of gastric cancer cells. BGC823 cells were transfected with miR-10b-mimic, miR-10b-inhibitor, or corresponding negative control. Refer to Methods for more details. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To investigate the upstream transcriptional regulation of miR-10b-5p, we investigated RUNX3, a tumor suppressor gene. Our RT-qPCR and western blotting results show a decrease in RUNX3 expression in GC tissues (Figure 5A and 5B). After transfection, RUNX3 increased miR-10b-5p expression (Figure 5C) and inhibited proliferation, migration, and invasion of GC cells (Figure 5D-G). Based on the above results, we conclude that RUNX3 upregulates miR-10b-5p to suppress the proliferation, migration, and invasion of GC.
Figure 5.
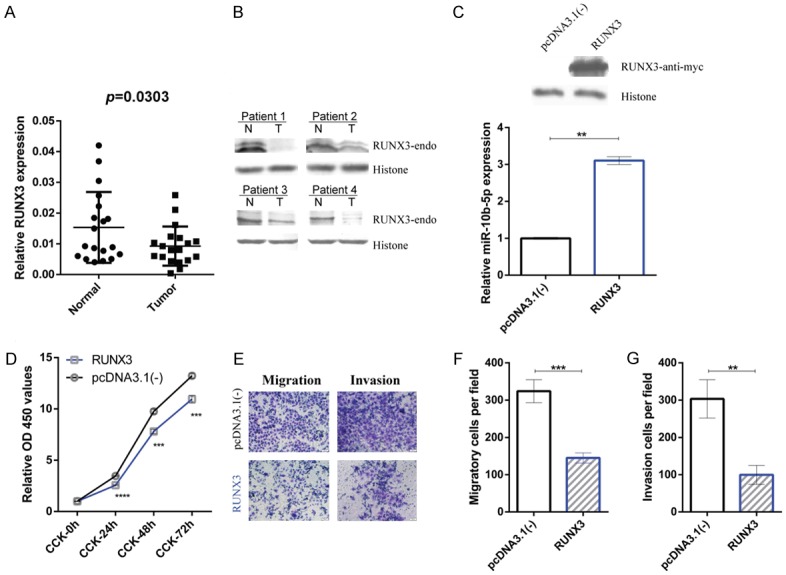
RUNX3 is decreased in GC and RUNX3 upregulates miR-10b-5p as well as suppresses the proliferation, migration, and invasion of GC cells. (A) Columnar scatterplots show the reduced expression of RUNX3 in gastric cancer tissues compared to paired adjacent normal gastric mucosal tissues. The relative expression levels of RUNX3 of 19 paired gastric cancer tissues and corresponding normal gastric mucosal tissues were measured using RT-qPCR and normalized to β-actin. (B) Western blotting shows the expression of RUNX3 in gastric cancer patients. The RUNX3 mRNA levels of the four patients are included in (A). (C) BGC823 cells were transfected with RUNX3 or control [pcDNA3.1(-)]. Relative miR-10b-5p expression (lower, measured by RT-qPCR, and shown by bar charts) suggests the upregulation effects of RUNX3 on miR-10b-5p and corresponding western blot results (C upper). (D-G) CCK-8 assay and Transwell assay results reveal the inhibitory effects of RUNX3 on the proliferation (D), migration (E, F), and invasion (E, G) of gastric cancer cells. Refer to Methods for more details. **P < 0.01, ***P < 0.001, ****P < 0.0001.
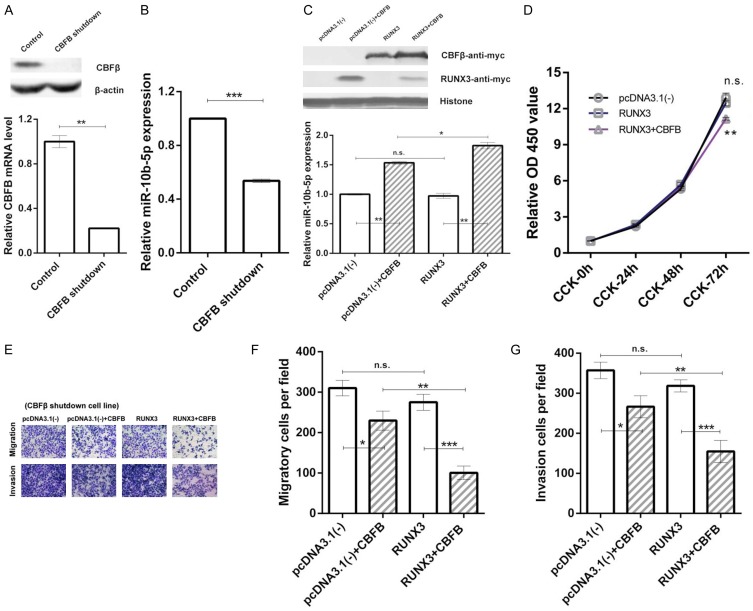
CBFβ/RUNX3 regulates miR-10b-5p expression and inhibits the proliferation, migration, and invasion of GC cells
To determine whether CBFβ is involved in RUNX3-mediated miR-10b regulation, endogenous expression of CBFβ in BGC823 cells was knocked down by transduction with a CBFB-specific short hairpin RNA. A stable CBFB-knockdown cell line was obtained (Figure 6A). In this cell line, the relative expression level of miR-10b-5p was decreased (Figure 6B). When we transfected the CBFβ expression vector (pc-CBFB-myc, CBFB) into these cells, the relative expression level of miR-10b-5p increased compared to in the control group [pcDNA3.1(-)] (Figure 6C). Moreover, in stable CBFB-knock-down cells, RUNX3 no longer upregulated miR-10b-5p expression. When we co-transfected RUNX3 and CBFB, miR-10b-5p expression was upregulated again (Figure 6C). Our results suggest that CBFβ plays an indispensable role in RUNX3 regulation of miR-10b-5p and the CBFβ-RUNX3 interaction significantly increases miR-10b-5p expression.
Figure 6.
CBFβ/RUNX3 upregulates miR-10b-5p expression and enhances the inhibitory effects on the proliferation, migration, and invasion of gastric cancer cells. (A) Reduced CBFB mRNA (lower, RT-qPCR results shown by bar charts) and CBFβ protein (upper, western blot results) in stable CBFB-knockdown BGC823 cell lines. (B) Bar charts showing decreased miR-10b-5p expression in stable CBFB-knockdown BGC823 cells. (C) Relative miR-10b-5p expression (lower, detected by RT-qPCR, and shown by bar charts) and corresponding western blot results (upper) of stable CBFB-knockdown BGC823 cells transfected with pcDNA3.1(-) (control), pcDNA3.1 (-)+CBFB, RUNX3, RUNX3+CBFB. (D) Results of CCK-8 assay of stable CBFB-knockdown BGC823 cells transfected with pcDNA3.1(-) (control), RUNX3, or RUNX3+CBFB. (E-G) Results of Transwell assay of stable CBFB-knockdown BGC823 cells transfected with pcDNA3.1(-) (control), pcDNA3.1(-)+CBFB, RUNX3, RUNX3+CBFB. Refer to Methods and Results for more details of (C-G). *P < 0.05, **P < 0.01, ***P < 0.001, n.s. P > 0.05.
To investigate the effects of CBFβ/RUNX3 on the proliferation of GC cells, we conducted a CCK-8 assay in the stable CBFB-knockdown cell line. The relative OD450 values of GC cells transfected RUNX3 were the same as those in the control group. After co-transfecting RUNX3 and CBFB, the relative OD450 values were significantly lower than in the control group in CCK-72 h (Figure 6D). The results indicate that the CBFβ-RUNX3 interaction is required for RUNX3 suppression of GC cell proliferation.
To test the effects of CBFβ/RUNX3 on the migration and invasion abilities of GC cells, we conducted transwell assays using the stable CBFB-knockdown cell line. The numbers of migrated and invaded cells did not differ in the RUNX3 group compared to the control group [pcDNA3.1(-)]. After co-transfecting RUNX3 and CBFB into these cells, the numbers of migrating and invaded cells were significantly lower than in the control group [pcDNA3.1(-)]. The results are shown in Figure 6E-G, which reveals that the CBFβ-RUNX3 interaction plays an essential role in inhibiting the migration and invasion of GC cells.
RUNX3 suppresses TIAM1 protein expression by upregulating miR-10b-5p
To verify the relationship between RUNX3 and TIAM1, we used RT-qPCR and western blotting to determine the TIAM1 mRNA and protein levels after co-transfecting RUNX3 and miR-10b-inhibitor in BGC823 cells. The results showed that RUNX3 decreased both TIAM1 mRNA (Figure 7A) and protein (Figure 7B) levels. After inhibiting miR-10b-5p, RUNX3 no longer reduced TIAM1 expression (Figure 7B). Our data reveal that RUNX3 inhibits TIAM1 expression by upregulating miR-10b-5p. We conclude that a CBFβ/RUNX3-miR10b-TIAM1 pathway plays an important role in GC progression and metastasis. A graphical presentation of this pathway is shown in Figure 8.
Figure 7.
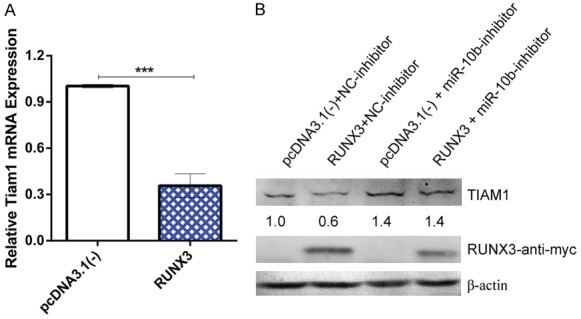
RUNX3 suppresses TIAM1 expression by upregulating miR-10b-5p. A. Bar chart showing the relative TIAM1 mRNA expression after transfection with RUNX3 and control. B. Western blot showing the TIAM1 protein levels after co-transfection with RUNX3 and miR-10b-inhibitor. RUNX3 inhibits TIAM1 expression by upregulating miR-10b-5p. ***P < 0.001.
Figure 8.
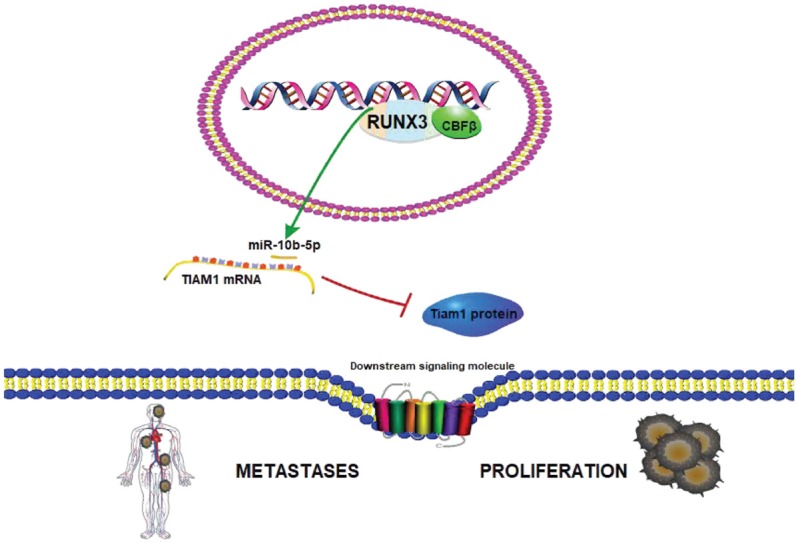
Schematic presentation of the CBFβ/RUNX3-miR10b-TIAM1 pathway.
Discussion
Studies have shown that TIAM1 is overexpressed in tumor tissues and the inhibition of TIAM1 can prevent tumor metastasis, including GC [9,10,25]. In this study, we investigated the upstream regulatory mechanisms of TIAM1. Our study demonstrated a CBFβ/RUNX3-miR10b-TIAM1 molecular axis that inhibits the proliferation, migration, and invasion of GC cells.
In this study, bioinformatic prediction and RT-qPCR validation indicated that miR-653-5p, miR-589-3p, miR-651-3p, miR-335-3p, miR-373-3p, miR-372-3p, miR-205-3p, and miR-10b-5p all can affect TIAM1 expression. As previously reported, some of them are related to invasion, migration, and epithelial-mesenchymal transition in GC [26-28]. However, according to the dual luciferase reporter assay results, none of them directly targets the TIAM1 mRNA 3’-UTR with the exception of miR-10b-5p, although these molecules may still be considered in future studies on metastasis of GC.
Although miR-10b-5p is a promising biomarker and therapy target for various metastatic tumors [29,30], the relationship between miR-10b-5p and GC remains controversial [31-34]. In this study, we evaluated a known tumor suppressor, RUNX3 [14,35,36], to evaluate the role of miR-10b in GC. Our data confirmed the suppressive effects of both miR-10b and RUNX3 on the proliferation, migration, and invasion of GC cells. Further analysis indicated that RUNX3 regulates GC at least partially through by increasing the endogenous levels of miR-10b-5p. Therefore, miR-10b-5p plays an inhibitory role in GC.
Additionally, because the CBFβ-RUNX3 interaction typically enhances the function of RUNX3 [37,38], we also explored how RUNX3 upregulates miR-10b-5p expression. Our study showed that CBFβ-RUNX3 plays critical roles in regulating miR-10b-5p to inhibit the proliferation, migration, and invasion of GC cells. CBFβ-RUNX3-related regulation may serve as a protective regulatory mechanism in normal gastric epithelial cells and its dysfunction may reduce miR-10b-5p levels, leading to tumorigenesis, progression, and metastasis. Additionally, as reported in preclinical research, RUNX3 and miR-10b-5p may be potential targets for tumor treatment [36,39]. The discovery of CBFβ’s involvement in RUNX3-mediated tumor suppression and miR-10b-5p upregulation provides insight for individual targeted treatment.
The present study identified a CBFβ/RUNX3-miR10b-TIAM1 molecular axis that provides two major directions for future clinically relevant work. First, miR-10b-based therapeutics have been fully investigated in other cancers in vitro and in animal cancer models [39], and the confirmed inhibitory function of miR-10b-5p on GC suggests further research and therapeutic target evaluation. Second, not only the target genes but also the upstream regulatory genes of microRNAs influence the treatment efficacy of potential miR-targeting drugs. The CBFβ-RUNX3 interaction with relevant regulatory effects found in the present study may be useful in future research.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 81672106 and 81601363].
Disclosure of conflict of interest
None.
Abbreviations
- GC
gastric cancer
- TIAM1
T cell lymphoma invasion and metastasis 1
- RUNX3
runt-related transcription factor 3
- CBFβ
core-binding factor subunit beta
- GEF
guanine nucleotide exchange factor
Supporting Information
References
- 1.Bray F, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018 GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, Ogunbiyi OJ, Azevedo E Silva G, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP CONCORD Working Group. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM, Niyazov J, Zucker A, Karchin R, Kinzler KW, Iacobuzio-Donahue CA, Vogelstein B, Nowak MA. Minimal functional driver gene heterogeneity among untreated metastases. Science. 2018;361:1033–1037. doi: 10.1126/science.aat7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poudel KR, Roh-Johnson M, Su A, Ho T, Mathsyaraja H, Anderson S, Grady WM, Moens CB, Conacci-Sorrell M, Eisenman RN, Bai J. Competition between TIAM1 and membranes balances endophilin A3 activity in cancer metastasis. Dev Cell. 2018;45:738–752. e6. doi: 10.1016/j.devcel.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J, Feng F. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;19:17559–17569. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi YL, Miao RZ, Cheng L, Guo XB, Yang B, Jing CQ, Zhang L, Jin X, Li LP. Up-regulation of T-lymphoma and metastasis gene 1 in gastric cancer and its involvement in cell invasion and migration. Chin Med J (Engl) 2013;126:640–645. [PubMed] [Google Scholar]
- 11.Chen Z, Li Z, Soutto M, Wang W, Piazuelo MB, Zhu S, Guo Y, Maturana MJ, Corvalan AH, Chen X, Xu Z, El-Rifai WM. Integrated analysis of mouse and human gastric neoplasms identifies conserved microRNA networks in gastric carcinogenesis. Gastroenterology. 2019;156:1127–1139. e8. doi: 10.1053/j.gastro.2018.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miroshnichenko S, Patutina O. Enhanced inhibition of tumorigenesis using combinations of miRNA-targeted therapeutics. Front Pharmacol. 2019;10:488. doi: 10.3389/fphar.2019.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Tian X, Chang J, Liu P, Zhang R. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomed Pharmacother. 2017;96:782–791. doi: 10.1016/j.biopha.2017.08.144. [DOI] [PubMed] [Google Scholar]
- 15.Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J, Liu Z. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 2016;381:138–148. doi: 10.1016/j.canlet.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Chen L, Zhang X, Xu X, Xing H, Zhang Y, Li W, Yu H, Zeng J, Jia J. RUNX3 regulates vimentin expression via miR-30a during epithelial-mesenchymal transition in gastric cancer cells. J Cell Mol Med. 2014;18:610–623. doi: 10.1111/jcmm.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegg HJ, Harrison H, Rogerson C, Shore P. The RUNX transcriptional coregulator, CBF beta, suppresses migration of ER(+) breast cancer cells by repressing ERalpha-mediated expression of the migratory factor TFF1. Mol Cancer Res. 2019;17:1015–1023. doi: 10.1158/1541-7786.MCR-18-1039. [DOI] [PubMed] [Google Scholar]
- 18.Mendoza-Villanueva D, Deng W, Lopez-Camacho C, Shore P. The Runx transcriptional co-activator, CBFbeta, is essential for invasion of breast cancer cells. Mol Cancer. 2010;9:171. doi: 10.1186/1476-4598-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, Rückert B, Meiler F, Akdis M, Littman DR, Akdis CA. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 21.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 22.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 23.Ito K, Chuang LS, Ito T, Chang TL, Fukamachi H, Salto-Tellez M, Ito Y. Loss of Runx3 is a key event in inducing precancerous state of the stomach. Gastroenterology. 2011;140:1536–1546. doi: 10.1053/j.gastro.2011.01.043. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2011;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed M, Sottnik JL, Dancik GM, Sahu D, Hansel DE, Theodorescu D, Schwartz MA. An osteopontin/CD44 axis in RhoGDI2-mediated metastasis suppression. Cancer Cell. 2016;30:432–443. doi: 10.1016/j.ccell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Li K, Pan M, Li W, Wu J, Li M, Zhao L, Wang H. miR-589 promotes gastric cancer aggressiveness by a LIFR-PI3K AKT-c-Jun regulatory feedback loop. J Exp Clin Cancer Res. 2018;37:153. doi: 10.1186/s13046-018-0821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing JJ, Wang ZY, Li H, Sun LP, Yuan Y. Key elements involved in Epstein-Barr virus-associated gastric cancer and their network regulation. Cancer Cell Int. 2018;18:146. doi: 10.1186/s12935-018-0637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu C, Li M, Zhang L, Bi Y, Wang P, Li J, Jiang X. MicroRNA-205 suppresses the invasion and epithelial-mesenchymal transition of human gastric cancer cells. Mol Med Rep. 2016;13:4767–4773. doi: 10.3892/mmr.2016.5118. [DOI] [PubMed] [Google Scholar]
- 29.Sheedy P, Medarova Z. The fundamental role of miR-10b in metastatic cancer. Am J Cancer Res. 2018;8:1674–1688. [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo B, Greninger P, Stein GT, Egan RK, McClanaghan J, Moore A, Benes CH, Medarova Z. Potent and selective effect of the mir-10b inhibitor MN-anti-mir10b in human cancer cells of diverse primary disease origin. PLoS One. 2018;13:e0201046. doi: 10.1371/journal.pone.0201046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 32.Kim K, Lee HC, Park JL, Kim M, Kim SY, Noh SM, Song KS, Kim JC, Kim YS. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6:740–51. doi: 10.4161/epi.6.6.15874. [DOI] [PubMed] [Google Scholar]
- 33.Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W, Chen Y, Fan Y, Qi L, Yin Y, Zhu W, Shu Y, Liu P. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188–196. doi: 10.1158/1055-9965.EPI-16-0607. [DOI] [PubMed] [Google Scholar]
- 34.Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX, Tao HQ, Wang HJ, He XJ. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum Pathol. 2013;44:1278–1285. doi: 10.1016/j.humpath.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, Ma G, Jiang P, Zhang W. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46:2445–2459. doi: 10.1159/000489651. [DOI] [PubMed] [Google Scholar]
- 36.Lim J, Duong T, Do N, Do P, Kim J, Kim H, El-Rifai W, Ruley HE, Jo D. Antitumor activity of cell-permeable RUNX3 protein in gastric cancer cells. Clin Cancer Res. 2013;19:680–690. doi: 10.1158/1078-0432.CCR-12-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschos K, Bazot Q, Ho G, Parker GA, Lees J, Barton G, Allday MJ. Core binding factor (CBF) is required for Epstein-Barr virus EBNA3 proteins to regulate target gene expression. Nucleic Acids Res. 2017;45:2368–2383. doi: 10.1093/nar/gkw1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, Varga SM, Taniuchi I, Harty JT, Peng W, Badovinac VP, Xue HH. The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol. 2017;18:931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo B, Kavishwar A, Ross A, Wang P, Tabassum DP, Polyak K, Barteneva N, Petkova V, Pantazopoulos P, Tena A, Moore A, Medarova Z. Combining miR-10b-targeted nanotherapy with low-dose doxorubicin elicits durable regressions of metastatic breast cancer. Cancer Res. 2015;75:4407–4415. doi: 10.1158/0008-5472.CAN-15-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.