Abstract
A variety of Good Manufacturing Practice (GMP) compliant processes have been reported for production of non-replicating adenovirus vectors, but important challenges remain. Most clinical development of adenovirus vectors now uses simian adenoviruses or rare human serotypes, whereas reported manufacturing processes mainly use serotypes such as AdHu5 which are of questionable relevance for clinical vaccine development. Many clinically relevant vaccine transgenes interfere with adenovirus replication, whereas most reported process development uses selected antigens or even model transgenes such as fluorescent proteins which cause little such interference. Processes are typically developed for a single adenovirus serotype – transgene combination, requiring extensive further optimization for each new vaccine.
There is a need for rapid production platforms for small GMP batches of non-replicating adenovirus vectors for early-phase vaccine trials, particularly in preparation for response to emerging pathogen outbreaks. Such platforms must be robust to variation in the transgene, and ideally also capable of producing adenoviruses of more than one serotype. It is also highly desirable for such processes to be readily implemented in new facilities using commercially available single-use materials, avoiding the need for development of bespoke tools or cleaning validation, and for them to be readily scalable for later-stage studies.
Here we report the development of such a process, using single-use stirred-tank bioreactors, a transgene-repressing HEK293 cell – promoter combination, and fully single-use filtration and ion exchange components. We demonstrate applicability of the process to candidate vaccines against rabies, malaria and Rift Valley fever, each based on a different adenovirus serotype. We compare performance of a range of commercially available ion exchange media, including what we believe to be the first published use of a novel media for adenovirus purification (NatriFlo® HD-Q, Merck). We demonstrate the need for minimal process individualization for each vaccine, and that the product fulfils regulatory quality expectations. Cell-specific yields are at the upper end of those previously reported in the literature, and volumetric yields are in the range 1 × 1013 – 5 × 1013 purified virus particles per litre of culture, such that a 2–4 L process is comfortably adequate to produce vaccine for early-phase trials. The process is readily transferable to any GMP facility with the capability for mammalian cell culture and aseptic filling of sterile products.
Keywords: Simian adenovirus, GMP, Clinical trials, Single-use, Biomanufacturing, Bioreactor, Purification
1. Introduction
Historically, development of novel vaccines has been a painstaking process taking many years and involving the development of entirely bespoke manufacturing processes. This approach is incapable of delivering a rapid response to emerging pathogens. It is also unsuited to the development of ‘niche’ vaccines for diseases with small commercial markets- for example human diseases which are significant problems only within a restricted geographical area, livestock diseases predominantly affecting low-income countries, or even ‘personalized’ cancer vaccines. In contrast ‘platform technologies’ for subunit vaccines allow the application of a single vaccine production method to the induction of immune responses against any protein antigen of interest. A key group of such ‘platforms’ are viral vector vaccines, which use a replication-deficient virus to deliver a transgene encoding the antigen of interest for expression in the recipient’s cells. In principle, manufacture of such vectors should be similar regardless of the encoded antigen.
Among the major classes of viral vectors, adenoviruses are arguably the most versatile and potent in inducing a combined antibody and T cell response [1], [2]. Viruses lacking E1 and E3 regions are replication-incompetent outside helper cells and a high level of assurance against the generation of replication-competent virus can be achieved [3], [4]. Pre-existing immunity to human adenoviruses in the human population may inhibit their effectiveness as vaccine vectors but can be overcome using simian adenoviruses. These have been shown to be safe and to have excellent immunogenicity in various human populations, including African children and adults, without any patient selection on the basis of pre-existing anti-vector immunity [5], [6]. Rare human serotypes may have similar properties [7], [8]. As a result, there has been significant investment by large pharmaceutical companies in the development of high-yielding and cost-effective large-scale adenovirus production processes [4]. An adenovirus-vectored vaccine against Ebola has been licensed in China [9] and numerous candidates are in clinical trials and advanced pre-clinical development, targeting diseases including malaria, Middle East respiratory syndrome, respiratory syncytial virus and a variety of cancers [10].
In keeping with this potential – and also the fact that adenovirus-based gene therapy has been intensively investigated – there is extensive literature regarding GMP-compliant adenovirus production processes. Suspension HEK293 cells are commonly used in batch-mode stirred-tank upstream processes [11], although the highest volumetric yields have been reported in perfusion-based processes in PERC6 cells [4], [12]. Downstream processes have commonly made use of bead-based anion exchange resins, often with an additional polish step such as size exclusion, hydrophobic interaction or multi-modal core-shell (eg CaptoCore®) chromatography [13], [14], [15], [16], [17]. More recently, membrane-based and monolith-based chromatography have been used [18], [19].
Most previous publications describing adenovirus production processes have reported careful optimization of the manufacture of a single vector, often using the AdHu5 serotype and/or model transgenes such as green fluorescent protein (both lacking relevance to most clinical vaccine development) [11]. Marked differences between the growth characteristics of different viruses are observed when varying adenovirus serotypes and pathogen-derived transgenes are used. This necessitates optimization of production processes for each individual virus, which is clearly undesirable for low-cost manufacture or outbreak response. In extreme cases, inhibition of viral growth by ‘transgene toxicity’ can result in failure of viral rescue or genetic instability [20], [21].
Partly as a result of this variability, production of an initial Good Manufacturing Practice (GMP)-compliant batch of virus for early-phase clinical trials remains a bottleneck in the adenovirus-vectored vaccine development pipeline. Our own Clinical Biomanufacturing Facility (CBF) is among the most experienced facilities in manufacture of such batches, having produced GMP adenovirus for numerous clinical trials in the past decade. This has been accomplished using a simple method involving a shake-flask-based upstream process and ultracentrifugation-based downstream process. This process is robust but is labour-intensive, not scalable, requires a preceding period of product-specific process development, and limits cleanroom throughput to around three batches per year.
Here, we set out to develop a GMP-suitable process which is sufficiently robust to be used across multiple adenovirus serotypes carrying a variety of pathogen transgenes with minimal product-specific process development. Although our principal focus has been the production of an initial GMP batch for early-phase clinical trials, we have aimed to use processing techniques suitable for subsequent scale-up. Throughout, we have sought to make the process as simple as possible, using entirely off-the-shelf materials and disposable product-contact parts, with the aim of facilitating adoption by facilities with little or no experience of adenovirus production.
2. Methods
2.1. Note regarding materials
For the avoidance of doubt, throughout this manuscript ‘Merck’ refers to the European entity ‘Merck KGaA’, known as ‘MilliporeSigma’ in North America.
2.2. Viruses
Derivation of the ChAdOx2 RabG and ChAd63 ME-TRAP vaccines has previously been described [22], [23]. Briefly, ChAdOx2 RabG is a vector expressing the rabies virus glycoprotein, based upon the AdC68 chimpanzee adenovirus serotype but with modifications to the E4 region; ChAd63 ME-TRAP is a chimpanzee adenovirus serotype 63 vector expressing a multi-epitope string and thrombospondin related adhesive protein from Plasmodium falciparum. The ChAdOx1 RVF vaccine used here was similar to that previously reported [24], with the exception of modified codon usage in the GnGc transgene sequence, designed to avoid repetitive sequence: it is a vector based upon the Y25 chimpanzee adenovirus serotype, with modifications to the E4 region [25], expressing Rift Valley Fever virus glycoproteins Gn and Gc. The purified virus inocula used to infect cultures, for small-scale gradient-elution ion exchange experiments and as standards in quality control assays were produced by caesium chloride density-gradient ultracentrifugation by the Jenner Institute Viral Vector Core Facility.
2.3. Cells and bioreactors
A master cell bank of HEK293 T-rex cells (Invitrogen) was produced under GMP conditions and subjected to testing compliant with pharmacopoeial requirements. Cells were adapted to suspension growth in CD293 media with 4 mM L-glutamine (both from ThermoFisher), 10 µg/mL blasticidin (Melford Laboratories) and gradually reducing concentrations of foetal bovine serum (5% to 0.5%), in non-baffled polycarbonate Erlenmeyer flasks (Corning) in an ISF1-X shaking incubator (Kuhner) at 37 °C, 8% CO2, 100 RPM with 50 mm orbit. Media with 0.5% serum is henceforth referred to as complete medium.
For small-scale upstream process experiments (virus growth kinetics, Fig. 1), 15 mL of culture at the specified density was inoculated at the specified multiplicity of infection (MOI) in an Erlenmeyer flask; 3 h later, a feed of 1.5 volumes of fresh complete medium was added.
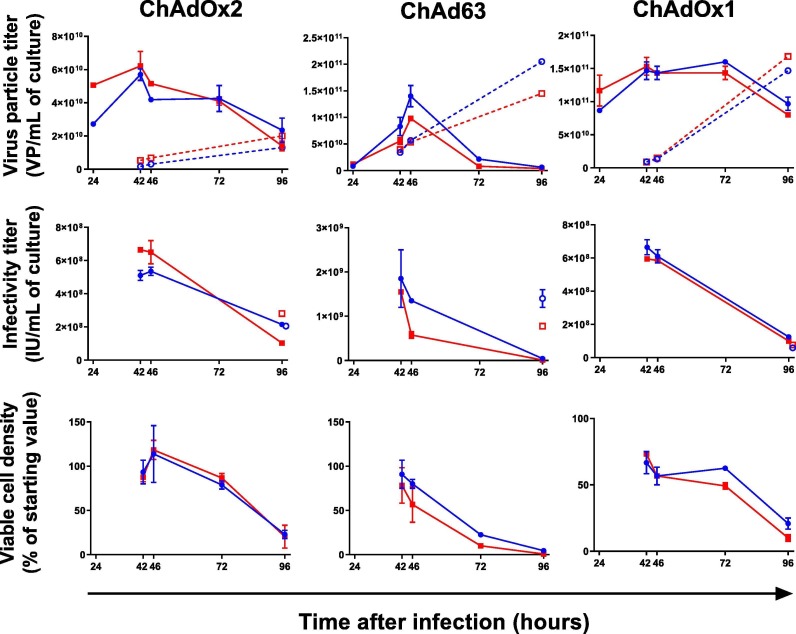
Fig. 1.
Growth kinetics of three virus serotypes in suspension-adapted HEK293 T-rex cells.
Each column of graphs represents a different serotype (as indicated by column captions), and each row a different measured parameter (as indicated by row captions: virus particle titer, assayed by qPCR; infectious unit titer; and viable cell density respectively). Each parameter was measured at multiple timepoints after infection. Blue lines indicate MOI of 3; red lines indicate MOI of 10. Solid lines and filled symbols indicate cell-associated virus [assays using cell pellets]; dashed lines and open symbols, where present, indicate virus in supernatant. Points and error bars indicate the median and range of two independent flasks within an experiment; absence of a visible error bar indicates close inter-replicate agreement. For each flask the plotted results are the mean of triplicate qPCR measurements and duplicate infectivity titers. Results are representative of two independent experiments for ChAdOx2 and ChAd63. In the case of ChAdOx1, a second experiment performed using a different transgene insert (a different viral glycoprotein) also gave similar results (data not shown).
Mobius® 3L (Merck) and BioBlu 3c (Eppendorf) single-use bioreactor vessels were used in accordance with the manufacturers’ instructions. A GX bioreactor controller unit and C-BIO software (both from Global Process Control) were used to control both vessel types. Dissolved oxygen (DO) was regulated at a setpoint of 50% air saturation by addition of medical air via microsparger. pH was regulated in the range 7.2–7.3 by addition of CO2 or 7.5% sodium bicarbonate solution (ThermoFisher) as required. Vessels were seeded with 800 mL (Mobius®) or 1.2 L (BioBlu) of culture at 1.5–2.0 × 106 cells/mL, then inoculated with virus at an MOI of 3. Three hours later, 1.5 volumes of fresh complete medium was added.
In the case of the BioBlu vessels, an OP-76 optical pH transducer (Eppendorf) was used for non-invasive pH sensing. One-point calibration of the optical pH sensor was performed by taking a sample three hours after vessel set-up (allowing time for equilibration of the sensor, but before addition of the feed).
2.4. Lysis, nucleic acid digestion and clarification
Lysis was initiated 42 h after infection by addition of 1/9 culture volume of buffer containing 10% v/v polysorbate 20, 50% w/v sucrose, 20 mM MgCl2, 500 mM Tris pH 8.0, plus Benzonase® (all from Merck) to a final in-culture concentration of 60 units/mL. DO and pH control of the bioreactors was de-activated, but agitation and heating to 37 °C continued.
Two hours after addition of lysis buffer, clarification was initiated. Except for the initial small-scale comparison of filters, for which a variety of 23 cm2 units were used (Fig. 2), 140 cm2 Millistak+® HC Pro depth filters with C0SP media filters were used (Merck). A flow rate of 3.3 L/min/m2 was used in early experiments (Fig. 2); this was subsequently accelerated to 6.6 L/min/m2 without detriment to turbidity reduction or virus recovery. Prior to use, filters were equilibrated with 1% v/v polysorbate 20, 5% w/v sucrose, 2 mM MgCl2, 50 mM Tris pH 8.0; after completion of filtration of lysate, 140 cm2 filters were flushed with 750 mL of the same buffer.
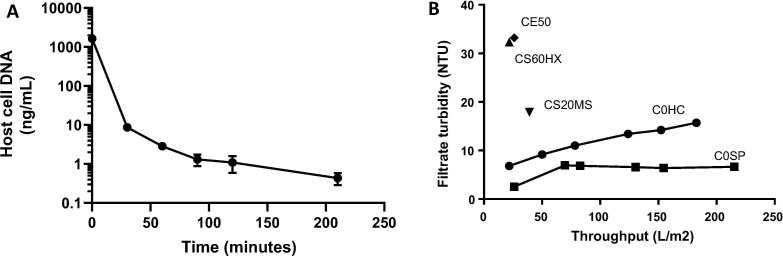
Fig. 2.
DNA lysis and clarification.
Panel A shows kinetic of host cell DNA reduction, as measured by qPCR, during cell lysis in the presence of 60 U/mL Benzonase®. Data shown is the mean (points) and range (error bars, if range large enough to render them visible) of duplicate samples. The result for each sample was itself the mean of triplicate assay wells.
Panel B shows comparison of five depth filters (indicated by annotation on the graph) on the basis of the relationship of filtrate turbidity versus cumulative throughput. The same cell lysate, with a pre-filtration turbidity of 70 NTU, was passed through each filter. Initial filtrate measurements obtained with Millistak+® CE50, Clarisolve® CS60HX and Clarisolve® CS20MS filters were clearly inferior to those with Millistak+® HC Pro C0SP and so these were not evaluated beyond the initial filtrate sample collection.
2.5. Tangential flow and sterile filtration
Unless otherwise specified in the Results section, tangential flow filtration (TFF) was performed using Pellicon® 2 Mini cassettes with BioMax® polyethersulfone membranes and C-screens (Merck), in conjunction with Spectrum process reservoirs (Spectrum Laboratories), Masterflex tubing (ColeParmer) and single-use pressure sensors (Pendotech).
For concentration and diafiltration of clarified lysate (the first TFF step, ‘TFF1′), feed flow rates of 6 L/min/m2 were used, with minimal transmembrane pressure (no pinch valve restriction). Diafiltration buffer feed rate was manually adjusted to match the permeate flow rate by maintaining a constant mass of the process reservoir. Where conversion rates (permeate flow as proportion of feed flow) exceeded 20%, a third pump was used for permeate control [26]. After 6 to 10-fold concentration, clarified lysate was diafiltered into ion exchange loading buffer comprising 100 mM NaCl, 1 mM MgCl2, 0.1% v/v polysorbate 20, 5% w/v sucrose, 50 mM bis-tris, pH 6.5.
Following ion exchange chromatography (IEX), eluate was buffer exchanged with 6–8 diavolumes of A438 formulation buffer [27] by means of a second TFF step (‘TFF2′). This step was performed using either PES flat sheet modules (Pellicon® 2 Mini as above, or in one case a smaller Pellicon® XL50 module) or using 300 kDa MWCO PES MidiKros hollow fibre units (Spectrum Laboratories). Whichever module was used, feed flow rates for this second TFF step were 2.4 L/min/m2. A retentate pinch valve was used to target a permeate flow rate of 0.83 L/min/m2, which was matched by the rate of addition of fresh A438 buffer to the system by a transfer pump. This was typically achieved with transmembrane pressure of c. 0.5 bar. Permeate control was not used for the second TFF. After the second TFF, the product was sterilized by passage through successive 0.45 µm and 0.2 µm filters (Millipak-20, 100 cm2 membrane area, Merck).
2.6. Anion exchange chromatography
Small-scale anion exchange to guide media selection was performed using an Akta Purifier (GE) and scale-down columns/capsules: 0.18 mL bed volume Mustang Q Acrodiscs (Pall); 1 mL CIMmultus QA-1 monoliths (BIA Separations); and 0.2 mL bed volume NatriFlo® HD-Q Recon Mini discs (Merck). Buffers used were ‘loading buffer’ (as described above) and ‘Buffer B’ (1 M NaCl, 1 mM MgCl2, 0.1% v/v polysorbate 20, 5% w/v sucrose, 50 mM trisHCl, pH 8.0). In the case of the NatriFlo® HD-Q device, virus in the above bis-tris-based loading buffer was diluted 5-fold into a phosphate-based loading buffer prepared by titrating 50 mM orthophosphoric acid to pH 6.5 with sodium hydroxide.
Dynamic binding capacity (DBC) was assessed by loading excess of a pre-purified ChAdOx2 RabG sample in loading buffer, at the lower end of manufacturers’ recommended flow rate ranges for each ion exchange medium, until breakthrough was observed (assessed by A280). DBC was defined as the load at which breakthrough A280 exceeded that observed early during loading (i.e. flowthrough A280) by 10% of the difference between the A280 of the loaded sample and the flowthrough A280. After wash of >10 media bed volumes with loading buffer, a linear gradient of increasing proportion of buffer B in loading buffer was applied. Subsequently, elution characteristics of ChAd63 ME-TRAP and ChAdOx1 RVF were investigated in similar linear gradient elution experiments using Mustang Q Acrodisc capsules.
To assess performance of the media under step elution conditions, diafiltered lysate containing ChAdOx2 RabG was loaded at manufacturers’ recommended flow rates, up to a load of 5 × 1012 virus particles (VP) for the Mustang Q and NatriFlo® HD-Q capsules, and 1x1013 VP for the larger-volume CIM-QA column. Wash and elution buffers were prepared by mixing loading buffer with buffer B until the desired conductivity was obtained. Buffer conductivity was selected on the basis of the conductivity value at the centre of the virus elution peak in the above linear gradient experiments, such that wash buffer conductivity was 3–4 mS/cm lower than this value, and elution buffer conductivity 2–4 mS/cm higher. After completion of loading, loading buffer, wash buffer, elution buffer and buffer B were applied in sequence, with 10 bed volumes or 10 mL (whichever was greater) of each buffer being applied. Eluted virus was collected during application of elution buffer, then analysed by quantitative PCR (qPCR) for VP quantification and by ELISA for residual host cell protein (see below).
For at-scale chromatography, Mustang Q capsules with 10 mL bed volume (Pall) were used in peristaltic pump-driven rigs comprising Masterflex tubing (Cole Parmer). Single-use UV absorbance, conductivity and pressure sensors (Pendotech) were used unless otherwise stated in results/figure legend. Capsules were pre-equilibrated with loading buffer (as above). Diafiltered lysate was loaded at a flow rate of 40–45 mL/min and followed by a wash with 10–15 column volumes of loading buffer. For ChAdOx2, wash and elution conditions were as described above for small-scale experiments. For ChAdOx1, wash conductivity was identical to that of ChAdOx2 and elution conductivity was higher (37 mS/cm). For ChAd63, due to the lower conductivity at which this vaccine eluted, only the loading buffer was used for washing, and elution buffer conductivity was adjusted to 26 mS/cm.
2.7. Product yield quantification
Purified genome-containing virus particles were quantified by UV spectrophotometry as previously described [28]; all results are based upon the mean of triplicate absorbance measurements. Infectious units were quantified by a cell-based infectivity assay which has also been described previously [25].
For in-process samples, genome-containing virus particles were quantified by TaqMan quantitative PCR (qPCR) using a probe set designed to bind a non-coding region of the adenoviral genome common to ChAdOx1, ChAdOx2 and ChAd63. Forward and reverse primers were GTGGGAAAAGTGACGTCAAACGAG and TGCATCCGCCTAGAAACACCTCA respectively. A probe with sequence GAGAGCGCGGGAAAATTGAGTATT was labelled with TAMRA/FAM (ThermoFisher). All samples were pre-treated with Benzonase®, to remove unencapsidated viral DNA, followed by proteinase K treatment to release encapsidated genomes. Quantification was performed relative to a standard curve of purified, spectrophotometrically-quantified ChAdOx2 RabG virus.
2.8. Assessment of product quality
SDS-PAGE was performed using 4–12% Bis-Tris gels, MOPS running buffer and NuPAGE sample buffer supplemented with DTT (all from ThermoFisher), stained with Quick Coomassie (Generon) and imaged using a ChemiDoc system (Bio-Rad).
For quantification of empty viral capsids, purified virus was subjected to isopycnic ultracentrifugation in 1.35 g/mL CsCl for 18 h at 160,000 g. The relative protein content of the upper (empty capsid) and lower (genome-containing virus particle) bands were then quantified by running SDS-PAGE gels (as above) followed by staining using Silver Express (ThermoFisher) and densitometry, as previously reported [29].
Residual host-cell protein (HCP) was quantified using the HEK293 HCP ELISA kit (Cygnus Technologies), according to the manufacturer’s instructions. In some cases an in-house version of the same assay was used, employing the same standards and goat anti-HEK293 HCP antibody (both from Cygnus Technologies), with the antibody used in unconjugated and horseradish-peroxidase-conjugated formats (produced using Lynx Rapid kit [Bio-Rad]) as capture and detection antibodies respectively.
Residual benzonase was quantified using the Benzonase ELISA kit II (Merck), according to the manufacturer’s instructions.
Residual host cell DNA was quantified using a previously reported quantitative PCR method targeting a 94 base pair amplicon within the Alu repeats (which comprise 11% of the human genome and have a copy number >1 million/genome) [30], [31]. The lower limit of our standard curve was 100 pg/mL for intact HEK293 DNA.
3. Results
3.1. Vectors of three serotypes carrying three antigen transgenes have similar growth characteristics in tet-repressing suspension cells
A major cause of the need for product-specific optimisation of adenovirus vector production processes is the fact that some transgenes have a marked impact upon the growth of adenovirus vectors [20]. We and others have previously reported that viruses carrying such ‘toxic’ transgenes can be rescued by incorporating ‘tet operator’ elements in the transgene promoter and growing the virus in cells such as HEK293 T-Rex (ThermoFisher) and Procell 92 which repress expression from such modified promoters [20], [21], [32], [33]. Unfortunately however, such cell lines have not been widely accessible for GMP manufacture in suspension culture. We and most other adenovirus manufacturing facilities have thus continued to use non-tet-repressing cells for GMP manufacture, requiring empirical determination of optimal time of harvest (ToH) and multiplicity of infection (MOI) for individual viruses prior to manufacture.
We have now produced and tested a HEK293 T-rex cell bank which is suitable for GMP use. We sought to investigate growth of vectors of three simian serotypes in these cells: ChAdOx1, a derivative of the Y25 serotype [25]; ChAdOx2, a derivative of AdC68 [3]; and ChAd63 [23]. We selected vectors encoding a variety of transgenes: ‘ChAdOx1 RVF’ encoding the GnGc antigen of Rift Valley fever virus (RVF) [24]; ‘ChAdOx2 RabG’ encoding rabies virus glycoprotein [22], and ‘ChAd63 ME-TRAP’ encoding a previously described malaria antigen [23]. We examined virus yield over time in HEK293 T-rex cells in small-scale shake flasks (maximum culture volume 40 mL), comparing MOIs of 3 and 10. Cell density was 1.5 × 106 cells/mL at the time of infection, reduced to 0.6 × 106 cells/mL by addition of a 1.5 volume feed 3 h after infection. Our aim was not to determine strictly optimal conditions (which would require a wider range of MOIs and cell densities), but instead to assess whether we could identify ‘standard’ conditions providing acceptable yields. Previous work suggests cell-specific productivity is reduced at cell densities above those we tested unless more complex medium-exchange or perfusion methods are used [11]. Given our aim to develop a simple and transferable method, we restricted ourselves to lower cell densities and simply sought to establish a readily-achieved window of densities achieving acceptable yield.
For all viruses, yields exceeding 4 × 1010 virus particles and 5 × 108 infectious units per mL of culture were achieved between 42 and 46 h post-infection at either cell density (Fig. 1 ). ‘Super-infecting’ cells with an MOI of 10 achieved little additional yield versus an MOI of 3 (in fact reducing the amplification factor [output yield per input IU]). Yields were similar at cell densities of 2.0 × 106 cells/mL during infection (data not shown). At 42–46 h, the majority of virus remains cell associated; by 96 h, much of the virus is in supernatant.
We therefore proceeded to use similar conditions in all subsequent work: ToH of 42–46 h; MOI of 3; cell density of 1.5–2.0 × 106 cells/mL at point of infection.
It is important to note that, although repression of transgene expression substantially reduces variability between viruses of a single serotype encoding different antigens, it does not affect the fundamental growth characteristics of the different serotypes. Notably, ChAd63 infection consistently causes more rapid cell lysis than ChAdOx1 or ChAdOx2- an effect seen here, and also repeatedly in pre-clinical work (Jenner Institute Viral Vector Core Facility, unpublished observations). Nonetheless, up to 42–46 h, kinetics are sufficiently similar to allow use of this as a single ToH.
3.2. Small-scale development of a simple lysis and clarification process
We next sought to implement a straightforward procedure for cell lysis, host cell DNA reduction and clarification. Starting from a culture infected with ChAdOx2 RabG (parameters as above), we observed rapid reduction in intact host cell DNA levels (as assessed by qPCR) during lysis with a detergent-based buffer in the presence of 60 units/mL Benzonase® during incubation with stirring at 37 °C (Fig. 2 A).
We compared a range of depth filters, seeking to identify a candidate which might be suitable for a single-step clarification process. At 23 cm2 scale, the fully synthetic Millistak+® HC Pro C0SP filter (MilliporeSigma) was found to achieve 90% turbidity reduction (70 to 7 nephelometric turbidity units [NTU]) with no evidence of turbidity breakthrough at 200 L/m2 and back-pressure < 3 psi (Fig. 2B). There was little difference between the filters in efficiency of product recovery (data not shown); subsequent experience with the Millistak+® HC Pro C0SP filter demonstrated recovery consistently > 90% (median 95%, range 91–100, n = 4, based upon infectivity unit titrations). Additional flushing with 1 M NaCl recovered minimal extra virus (<2% of load).
3.3. A standardized upstream process using culture volumes of 2-4L achieves yields adequate for early-phase clinical trials
Having selected upstream process parameters in the above small-scale shake flask experiments, we proceeded to transfer the process to single-use stirred tank bioreactors. For our initial work with ChAdOx2 RabG, we explored two single-use bioreactor systems. We obtained satisfactory results with both 2L cultures in the Mobius® 3L (MilliporeSigma; maximum working volume 2.4L; we occasionally used two such vessels in parallel for a 4L total culture volume), and 3.2L cultures in the BioBlu 3c (Eppendorf; maximum working volume 3.8L). Subsequent work with ChAdOx1 RVF and ChAd63 ME-TRAP was performed using Mobius® vessels. Typical process parameter logging data is shown in Fig. 3 . Clarification using 140 cm2 Millistak+® HC Pro depth filters with C0SP media filters resulted in post-clarification turbidity of ≤6 NTU for all runs with all viruses. Maximal throughput was 285 L/m2 (4L), with no turbidity breakthrough observed.
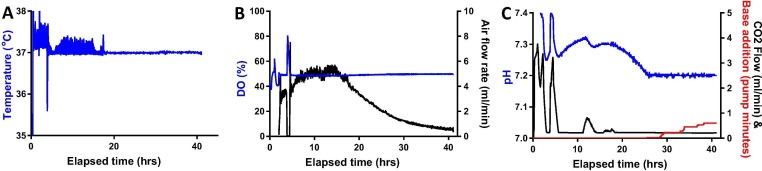
Fig. 3.
Typical upstream process logging data.
Panel A shows temperature; Panel B shows control of oxygenation (i.e. DO and air flow); Panel C shows pH control (i.e. measured pH, CO2 flow and base addition). In each case, the controlled parameter is shown in blue and plotted on the left-hand axis. For Panels B and C, actuator responses are plotted on the right-hand axes and shown in black or, in the case of base addition on Panel C, in red. Data shown is from a process run with ChAdOx1 RVF in Mobius® 3L vessels. Similar results were obtained with other viruses using Mobius® 3L vessels, and for ChAdOx2 RabG in BioBlu 3c vessels.
Yields are shown on a volumetric basis in Table 1 . Upstream process yields per bioreactor vessel ranged from 7.2 × 1013 to 2.5 × 1014 VP [by qPCR] across four runs with ChAdOx2 RabG, two runs with ChAdOx1 RVF, and two runs with ChAd63 ME-TRAP, i.e. >2500 human doses of c. 2.5 × 1010 VP.
Table 1.
Product yield and quality.
| Virus | Number of runs and vessel type | Culture volume (litres) | USP yield (VP [by qPCR], per litre of culture) | DSP yield (VP [by spectrophotometry], per litre of culture) | Host-cell protein (ng/mL) | Host-cell DNA (ng/mL) | Residual Benzonase (ng/mL) | A260:A280 ratio | Empty capsid : VP ratio (n = 1 per serotype) | Particle: infectivity ratio |
|---|---|---|---|---|---|---|---|---|---|---|
| ChAdOx2 RabG | 1 (2x Mobius vessels) | 4 | 1.2 × 1014 | 3.60 × 1013 | 5.2 (3.3–8.6) | <0.1 | <0.3 | 1.3 (1.28–1.34) | 0.17 | 103 (85–122) |
| 3 (BioBlu vessels) | 3.2 | 8.3 × 1013 | 4.3 × 1013 | |||||||
| (7.9 × 1013–1.4 × 1014) | (1.7 × 1013–6.4 × 1013) | |||||||||
| ChAd63 ME-TRAP | 2 (Mobius vessels) | 2 | 6.4 × 1013 | 3.1 × 1013 | 3 | <0.1 | <0.3 | 1.39 | <0.1 | 56 (23–88) |
| (3.7 × 1013–9.0 × 1013) | (1.3 × 1013–5 × 1013) | (2.9–3.1) | (1.35–1.42) | |||||||
| ChAdOx1 RVF | 2 (Mobius vessels) | 2 | 5.5 × 1013 | 1.3 × 1013 | 4.9 | <0.1 | <0.3 | 1.38 | <0.1 | 146 (99–192) |
| (3.6 × 1013–7.0 × 1013) | (1.1 × 1013–1.5 × 1013) | (3.1–7.9) | (1.37–1.39) | |||||||
Results of a series of process runs with each virus are presented. Results shown are the median of the indicated number of runs, with the range of results in brackets, with the exception of empty capsid : VP ratio, which was assessed once for each serotype. USP yields were quantified by qPCR at the stage of clarified lysate; for each run, yields values were calculated means of triplicate qPCR reactions. Note that yields shown are per litre of culture (to standardize for the varying volumes of the different bioreactor vessels). Approximate yields per infected cell can be calculated by dividing these values by 109 (based upon cell density of c. 109 cells per litre, after the addition of the feed).
3.4. Selection of downstream process media
Consistent with previous reports of TFF of adenovirus products using various devices with polyethersulfone (PES) membranes with 300 kDa molecular weight cut-off (MWCO), we obtained satisfactory results using 300 kDa MWCO BioMax® PES C-screen Pellicon® XL50 or Pellicon® 2 Mini devices (Merck) [14], [18], [34]. With Pellicon® 2 Mini devices, recovery was typically c. 80% for all viruses after 10-fold concentration and 6 diavolumes (DV) of buffer exchange into anion exchange loading buffer. This achieved reduction of >95% in residual host-cell protein and reduced host-cell DNA to undetectable levels. Further diafiltration beyond 6 DV achieved no further reduction in residuals.
We next sought to compare a range of commercially available anion exchange media for use in bind - wash – elute mode in small-scale experiments using up to 1 mL of media. We focused our efforts upon membrane and monolith matrices with quaternary amine chemistry. As compared to bead-based resins such matrices provide much improved convective access for large species such as viruses to the binding groups, higher binding capacities for such species, and substantially higher flow rate: matrix volume ratios. For the purposes of manufacture of small GMP batches, they also have the substantial advantage of the off-the-shelf availability of GMP-suitable disposable capsules in sizes suitable for purification of ∼1 × 1014 VP, avoiding the need for packing of custom-sized columns with bead-based resins.
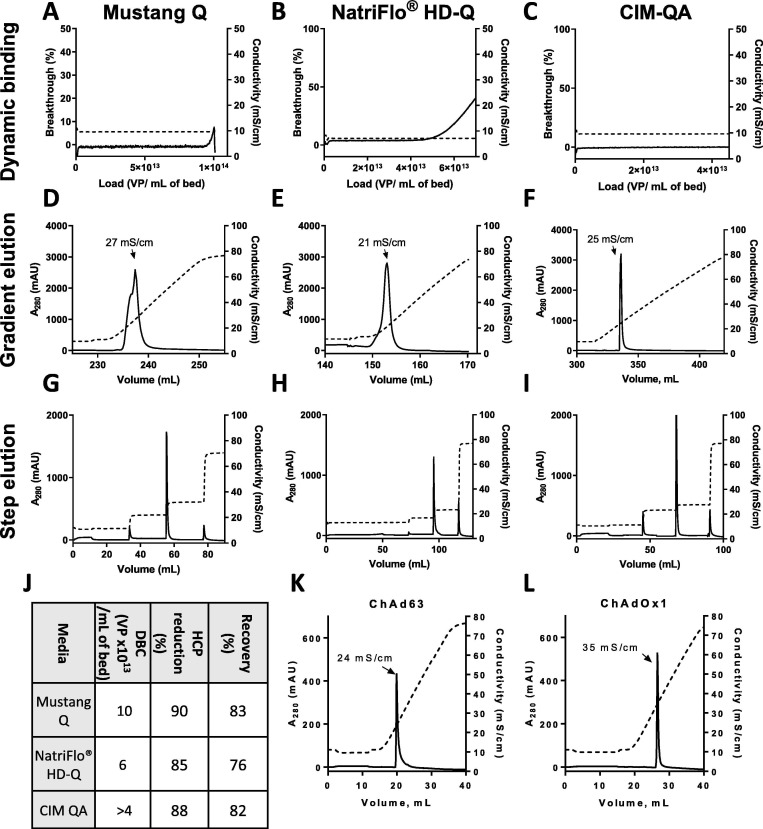
We observed satisfactory results with three such matrices: Mustang Q membrane capsules; NatriFlo® HD-Q hydrogel membrane capsules; and CIM-QA monoliths (Fig. 4 A–J). Satisfactory separation with the NatriFlo® HD-Q capsule required transfer of the virus into phosphate buffer prior to application to the column. Binding capacity, virus recovery, and host cell protein reduction are shown in Fig. 4J. It is likely that a scaled-up process could have been designed using any of these media, but for further work, we selected Mustang Q capsules on the basis of binding capacity, product recovery and host cell protein reduction.
Fig. 4.
Comparison of anion exchange media.
Chromatograms from small-scale anion exchange experiments are shown. In all cases, solid lines indicate A280 (left axis) and dashed lines indicate conductivity (right axis). A260 data was also collected, with results paralleling the A280 data, but is omitted from graphs for clarity.
In panels A-I, each column of graphs represents data from a single type of media, as per column captions. Panels A-F show chromatograms from experiments in which previously purified ChAdOx2 RabG was loaded on Mustang Q Acrodisc, NatriFlo® HD-Q Recon Mini, and CIM-QA 1 mL capsules. In panels A-C. breakthrough was observed and hence dynamic binding capacity was calculated for the Mustang Q and NatriFlo® capsules; breakthrough was not observed after loading 4x1013 VP on the (larger) CIM-QA column. Panels D-F show the continuations of the above experiments, in which virus was eluted using a linear gradient of increasing conductivity to allow estimation of elution conditions; the conductivity at the peak of virus elution is shown.
Panels G-I show chromatograms from experiments in which diafiltered lysate containing ChAdOx2 RabG was loaded on Mustang Q Acrodisc, NatriFlo® HD-Q Recon Mini, and CIM-QA 1 mL capsules, followed by step elution.
Panel J presents DBC, virus recovery and HCP reduction data from the experiments shown in Panels A-F. In the case of the CIM-QA column, breakthrough was not reached despite loading 3x1013 VP on the 1 mL device.
Panels K-L show chromatograms from experiments in which previously purified ChAd63 ME-TRAP and ChAdOx1 RVF were loaded on Mustang Q Acrodisc capsules and eluted with a linear conductivity gradient, as above.
To ascertain elution conditions for the ChAd63 and ChAdOx1 serotypes, we performed small-scale gradient elution from Mustang Q membranes. ChAd63 ME-TRAP eluted at 24 mS/cm and ChAdOx1 RVF at 35 mS/cm (Fig. 4K and L).
3.5. 2-4L downstream process yields product suitable for early-phase clinical trials
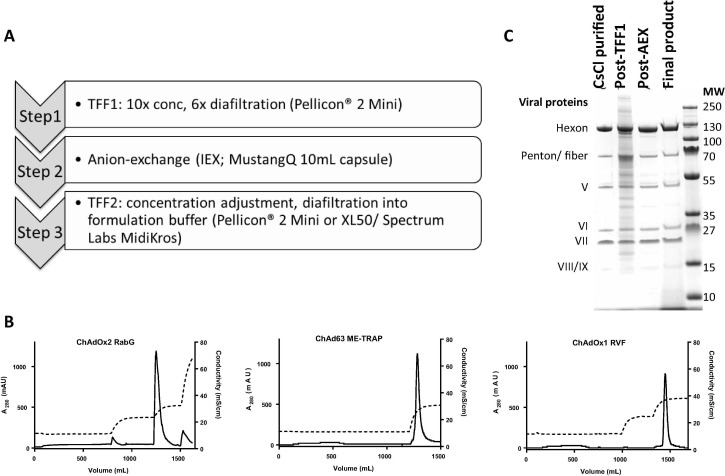
Drawing upon the experience from the above scaled-down experiments, we implemented a downstream process as shown in Fig. 5 A. To ensure that all product-contact components were disposable, single-use liners (Merck) were used with the Pellicon® 2 Mini TFF cassettes, and chromatography monitoring employed single-use pressure, conductivity and UV spectrometry sensor flow cells (Pendotech) (data shown in Fig. 5B).
Fig. 5.
Downstream process overview and results.
Panel A shows an overview of the downstream process steps.
Panel B shows conductivity and A280 traces obtained for at-scale chromatography for each of the three vaccines, as indicated in captions for each plot. In each case, loading occurs over the first ∼ 600 mL (a degree of A280 deflection from impurity flow-through is visible) and the largest A280 peak represents the eluted virus which was collected. Note for ChAdOx2 RabG that the pre-elution wash and the post-elution high-salt wash produce relatively small A280 peaks (predominantly impurity, with some virus, as assessed by SDS-PAGE); this data was obtained using UV and conductivity sensors of the Akta Purifier. For ChAd63 and ChAdOx1, data was obtained using the complete single-use chromatography rig (i.e. with single-use UV and conductivity sensors [Pendotech]) and no post-elution wash was performed. For ChAd63, due to the lower conductivity at which this vaccine eluted, no pre-elution wash was used.
Panel C shows samples from each stage of the downstream process on a Coomassie-stained SDS-PAGE gel. Samples shown are from a single process run with the ChAdOx2 RabG vaccine, but are typical of runs with all three investigated vaccines.
The only difference between the processes for the different viruses was the choice of anion exchange wash and elution conductivity (shown in Fig. 5B), based upon the differing elution characteristics observed in small-scale experiments (Fig. 4K and L). For ChAd63 ME-TRAP washing was performed with loading buffer but a higher-conductivity wash was omitted in view of the lower elution conductivity and the modest amount of impurity eluted by the wash step with the other serotypes.
For each virus, we obtained >800 human doses from a 2–5 L culture (Table 1). Recovery at each process step is shown in Table 2 ; substantial losses were noted during IEX for ChAdOx1, and at TFF2 in the single run using the Pellicon XL50 unit. Product quality was within specifications accepted for clinical use by local regulators (particle:infectivity [P:I] ratio, A260:A280 ratio, residual host-cell protein, host-cell DNA, and benzonase), and results of further assays performed for information were also consistent with high quality product (empty capsid:VP ratio quantification and Coomassie-stained SDS-PAGE) (Table 1 and Fig. 5C). P:I ratios of these simian adenovirus serotypes are commonly higher than those usually obtained with AdHu5, even when purification is by ultracentrifugation: values in the range 50–120 are commonly obtained after lab-scale ultracentrifugation (based upon experience of >1000 virus preparations by the Jenner Institute Viral Vector Core Facility), and batches prepared for clinical trials at our Clinical Biomanufacturing Facility have had P:I ratios in the range 50–200.
Table 2.
Downstream process step recovery.
| Run | TFF1 | IEX | TFF2 device type | TFF2 & sterile filtration, by qPCR | Sterile filtration, by spectrophotometry | |
|---|---|---|---|---|---|---|
| ChAdOx2 RabG | 1 | 85 | 44 | Pellicon 2 Mini | 53 | 98 |
| 2 | 79 | 72 | Pellicon 2 Mini | 94 | 96 | |
| 3 | 113 | 65 | MidiKros | 119 | 93 | |
| ChAd63 ME-TRAP | 1 | 80 | 100 | Pellicon 2 Mini | 59 | 100 |
| 2 | Not done | 48 | Pellicon XL50 | 39 | 48 | |
| ChAdOx1 RVF | 1 | 101 | 39 | MidiKros | 77 | 76 |
| 2 | 107 | 36 | MidiKros | 54 | 90 | |
| OVERALL | 93 (79–113) | 48 (36–100) | Pellicon 2 Mini | 59 (53–94) | 98 (96–100) | |
| MidiKros | 77 (54–119) | 90 (76–93) | ||||
Percentage product recovery at each step was calculated for consecutive individual runs with each virus. All calculations were based upon viral genome qPCR, with the exception of recovery after sterile filtration which is based upon spectrophotometry (as there is no change in buffer composition at this point).
qPCR-based results are the means of results obtained from three replicate samples, with three replicate qPCR reactions per sample. All calculations were based upon comparisons made within a single qPCR run. Apparent recoveries in excess of 100% reflect residual qPCR assay variability.
The qPCR-based results presented for TFF2 recovery were produced by comparing samples taken prior to TFF2 and from the final product (after TFF2 and sterile filtration), on the basis that product loss upon sterile filtration typically reflects aggregation as a result of poor performance of the TFF2 process. Summary results are presented as medians with ranges in brackets, and are broken down by device type for TFF2 (excluding the single run performed using a Pellicon XL50 unit).
4. Discussion
Methods for GMP production of a variety of adenovirus vectors at a variety of scales have previously been reported [4], [11]. Here, we focused upon developing a process which could be rapidly adapted to any adenovirus serotype/transgene combination to produce a small GMP batch suitable for early-phase clinical trials. We have aimed for simplicity and transferability in view of increasing interest in the production of adenovirus vectors in diverse contexts including – though not limited to – use in low and middle-income countries, in the response to outbreaks of emerging pathogens, and veterinary use. The resulting process should be suitable for adoption by any facility with competence in routine bioprocess procedures (mammalian cell culture, bioreactor batch processes, tangential flow filtration and chromatography). Key elements are the use of a cell line which minimizes the need for virus/antigen specific growth optimization, and the use of entirely ‘off-the-shelf’ equipment and materials, with single-use product-contact materials throughout.
Of the two bioreactor vessels we used for the upstream process, the BioBlu vessel (Eppendorf) has the advantage of non-invasive pH and DO sensing; nonetheless, given the relatively low cost of invasive pH and DO sensors compatible with the Mobius® vessel (c. USD 2000), these could be regarded as single-use items for a GMP production run. These vessels could readily be used in any cleanroom with existing capability to grow mammalian cells in suspension in shake flasks: at this scale, the only gas required in addition to CO2 is a 640L (under-bench) medical air cylinder. Both manufacturers offer a family of larger vessels for scaled-up production (with somewhat more choice at the lower end in the BioBlu range). Due to our desire for simplicity and satisfactory results with this process, we deliberately did not pursue intensification of the upstream process (with more modern media formulations, fed batch or perfusion processes), but clearly there are a variety of options available to increase the volumetric yield of the upstream process.
Similarly, we prioritized simplicity over consumable costs (which are a relatively small component of the total cost of a GMP run of this scale). We were therefore satisfied with the host cell DNA reduction achieved with Benzonase® at 60 U/mL (and the ease of quantifying residual Benzonase® with an off-the-shelf ELISA kit), despite its relatively high cost as compared to alternative methods such as precipitation with domiphen [35]. Pharmaceutical-grade Benzonase® at this concentration costs c. USD 150 per liter, or c. USD 0.15 per vaccine dose. This is trivial in the context of manufacture of a Phase I clinical trial batch, and indeed might be tolerable for marketed products in certain contexts. Nonetheless our data (Fig. 2A) suggest that, if scaling up this process, the concentration of Benzonase® used, or the digestion time, could be considerably reduced while maintaining acceptable host cell DNA levels and without requiring any other change to the method. The conditions we used reduced host cell DNA to beneath regulatorily-accepted levels of 10 ng per human dose prior to clarification; a considerably higher level at this stage would be expected to be reduced within acceptable limits by the downstream process.
A variety of commercial anion exchange products were shown to provide acceptable results; notably, our results are, to our knowledge, the first published report of the use of the NatriFlo® HD-Q hydrogel membrane device for viral purification. All are available in a range of sizes suitable not only for processes of this scale, but for scaled-down process development and scaled-up production. We observed substantially higher dynamic binding capacities than some previous reports for similar media- substantially in excess of 1 × 1013 VP per mL of bed for all three media. Our primary measurements of binding capacity were made using pre-purified virus, which will have favoured high capacity (as compared to using a load including impurities). Nonetheless, we routinely observed total process yields of 1 × 1014 – 2 × 1014 VP using the 10 mL Mustang Q capsule in our full-scale process. Such capacities have clear beneficial implications for process scalability and cost-effectiveness; Mustang Q units are available with bed volumes up to 780 mL, which our data suggests would be adequate for purification of nearly a million human doses.
We observed slightly different elution characteristics for the three tested serotypes. Although the salt concentration required to elute various adenovirus serotypes from anion exchange media has previously been reported to correlate with charge of the hexon, the major capsid protein [36], we did not observe such a pattern. At pH 7, predicted hexon net charges are −17.7 for ChAdOx2, −18.5 for ChAd63, and −19.7 for ChAdOx1. The report correlating elution characteristics with charge found this relationship when comparing viruses with hexon charge spanning the range −14 to −28, while the relationship was imperfect when comparing viruses with smaller differences. It may be that chromatographic differences between viruses sharing similar hexon charges are attributable to differing charges on other structural proteins. The serotypes we tested are representative of the diversity of major structural protein (hexon and fiber) sequences within adenovirus species E [25]. It would be of interest to extend the use of this process to clinically relevant serotypes of other adenovirus species. Encouraging such work, there are several reports of the application of TFF- and anion-exchange-based downstream processes to the species B adenovirus AdHu35 and the species C virus AdHu5 [37], and anion exchange has also been successfully applied to species D viruses [36]. Given that residual host cell protein levels were within acceptable limits after single-step chromatography, we did not require or explore a ‘polish’ step (although we note the possibility of use of core–shell octylamine resins (such as Captocore 700 [GE]) to provide such a polish in flow-through mode [16]). Our downstream process is thus strikingly straightforward and accessible to any GMP facility with minimal equipment investment: the only requirements are peristaltic pumps and the Pendotech sensor monitoring apparatus (total one-off cost < USD 30,000).
Our final process provided USP yields consistently > 2500 doses per litre. Yield per infected cell ranged from 5.5 × 104–1.4 × 105 VP, favourably comparable to values previously reported in a review encompassing a variety of cell types, media, and process types (HEK293 and PER.C6; adherent and suspension; batch, fed batch and perfusion) [11], and also comparable also to those achieved in more recent perfusion-based processes using PER.C6 cells [12]. Overall DSP recovery was in some cases as low as 20%, offering clear scope for further improvement in the overall process yield; analysis of step recovery from individual runs indicated particular potential for improvement of the ChAdOx1 RVF IEX step and the consistency of performance of the TFF2 step. Nonetheless, final yields were consistently more than adequate for early-phase clinical trials.
This method can be implemented for a new vaccine with minimal or even no vaccine-specific process development. Given one to two weeks, it is possible to perform a shake flask study to confirm optimal time of harvest, and a small-scale gradient-elution anion exchange experiment to confirm suitable chromatography elution +/- wash conditions. Among vaccines of a single adenovirus serotype, however, transgene repression should minimize differences in growth kinetics, and chromatographic characteristics are believed to be determined by the (constant) structural proteins rather than the (varying) transgene [36]. It is likely that conditions pre-established to be suitable for a given serotype (i.e. for ChAdOx2, ChAdOx1 and ChAd63, those conditions used here) will be applicable regardless of the transgene- and hence even those simple optimization experiments may not be necessary to assure satisfactory yields for early-phase clinical trials. The key rate-limiting step in the production of GMP adenovirus in the outbreak-response context is now the rescue of virus and production of a suitable inoculum (c. 2 × 1010 IU) to initiate a process such as ours. Ongoing work in our Institute aims to address this issue.
Funding
This work was supported by Merck KGaA, the UK Medical Research Council (grant MR/P017339/1), and the UK Engineering and Physical Sciences Research Council (grant EP/R013756/1). ADD is supported by the Wellcome Trust [grants 201477/Z/16/Z and 204826/Z/16/Z] and is a Jenner Investigator. The study was performed in collaboration between the University of Oxford and Merck KGaA; both partners reviewed the manuscript prior to submission. The other funders had no input to the design of the study or decision to publish. Correspondence and requests for materials can be addressed to sandy.douglas@ndm.ox.ac.uk.
Acknowledgments
Acknowledgements
We are grateful to Ali Turner, Claire Powers and the Jenner Institute Viral Vector Core Facility for skilled assistance; to Cath Green, Emma Bolam, Elena Boland and the Clinical Biomanufacturing Facility for HEK293-Trex cells; and to Adrian Hill for provision of the ChAd63 ME-TRAP vaccine. We thank Youness Cherradi, Lenaig Savary and Josselyn Haas from MilliporeSigma/Merck for their consultancy.
Competing Interests
ADD, SJM, and SCG are named inventors on patent filings relating to the use of simian adenoviruses, but not directly related to the work described here. SCG is a founder of Vaccitech Ltd, which develops adenovirus-vectored vaccines.
References
- 1.Gilbert S.C., Warimwe G.M. Rapid development of vaccines against emerging pathogens: the replication-deficient simian adenovirus platform technology. Vaccine. 2017;35:4461–4464. doi: 10.1016/j.vaccine.2017.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewer K.J., Lambe T., Rollier C.S., Spencer A.J., Hill A.V., Dorrell L. Viral vectors as vaccine platforms: from immunogenicity to impact. Curr Opin Immunol. 2016;41:47–54. doi: 10.1016/j.coi.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Morris SJS, Sarah; Spencer, Alexandra J.; Gilbert, Sarah C. Simian adenoviruses as vaccine vectors. Future Virology. 2016;11(9):649-59. [DOI] [PMC free article] [PubMed]
- 4.Vellinga J., Smith J.P., Lipiec A., Majhen D., Lemckert A., van Ooij M. Challenges in manufacturing adenoviral vectors for global vaccine product deployment. Hum Gene Ther. 2014;25(4):318–327. doi: 10.1089/hum.2014.007. [DOI] [PubMed] [Google Scholar]
- 5.Afolabi M.O., Tiono A.B., Adetifa U.J., Yaro J.B., Drammeh A., Nebie I. Safety and Immunogenicity of ChAd63 and MVA ME-TRAP in West African children and infants. Mol Ther. 2016;24(8):1470–1477. doi: 10.1038/mt.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogwang C., Afolabi M., Kimani D., Jagne Y.J., Sheehy S.H., Bliss C.M. Safety and immunogenicity of heterologous prime-boost immunisation with Plasmodium falciparum malaria candidate vaccines, ChAd63 ME-TRAP and MVA ME-TRAP, in healthy Gambian and Kenyan adults. PLoS One. 2013;8(3):e57726. doi: 10.1371/journal.pone.0057726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baden L.R., Walsh S.R., Seaman M.S., Tucker R.P., Krause K.H., Patel A. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001) J Infect Dis. 2013;207(2):240–247. doi: 10.1093/infdis/jis670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch D.H., Kik S.V., Weverling G.J., Dilan R., King S.L., Maxfield L.F. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29(32):5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CanSinoBIO. Ad5-EBOV 2017 [Available from: <http://www.cansinotech.com/homes/article/show/45/26.html> Accessed: [10/12/2018].
- 10.Ewer K., Sebastian S., Spencer A.J., Gilbert S., Hill A.V.S., Lambe T. Chimpanzee adenoviral vectors as vaccines for outbreak pathogens. Human Vacc Immunotherapeut. 2017;13(12):3020–3032. doi: 10.1080/21645515.2017.1383575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadeau I., Kamen A. Production of adenovirus vector for gene therapy. Biotechnol Adv. 2003;20(7–8):475–489. doi: 10.1016/s0734-9750(02)00030-7. [DOI] [PubMed] [Google Scholar]
- 12.Luitjens A, Lewis JA, inventors; Crucell Holland BV, assignee. Method for the production of adenoviral vectors. US patent number 10,041,049. 2011.
- 13.Kamen A., Henry O. Development and optimization of an adenovirus production process. J Gene Med. 2004;6(Suppl 1):S184–S192. doi: 10.1002/jgm.503. [DOI] [PubMed] [Google Scholar]
- 14.Green A.P., Huang J.J., Scott M.O., Kierstead T.D., Beaupre I., Gao G.P. A new scalable method for the purification of recombinant adenovirus vectors. Hum Gene Ther. 2002;13(16):1921–1934. doi: 10.1089/10430340260355338. [DOI] [PubMed] [Google Scholar]
- 15.Konz J.O., Pitts L.R., Sagar S.L. Scaleable purification of adenovirus vectors. Methods Mol Biol. 2008;434:13–23. doi: 10.1007/978-1-60327-248-3_2. [DOI] [PubMed] [Google Scholar]
- 16.Nestola P., Peixoto C., Villain L., Alves P.M., Carrondo M.J., Mota J.P. Rational development of two flowthrough purification strategies for adenovirus type 5 and retro virus-like particles. J Chromatogr A. 2015;1426:91–101. doi: 10.1016/j.chroma.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 17.Riske F., Berard N., Albee K., Pan P., Henderson M., Adams K. Development of a platform process for adenovirus purification that removes human SET and nucleolin and provides high purity vector for gene delivery. Biotechnol Bioeng. 2013;110(3):848–856. doi: 10.1002/bit.24742. [DOI] [PubMed] [Google Scholar]
- 18.Peixoto C., Ferreira T.B., Sousa M.F., Carrondo M.J., Alves P.M. Towards purification of adenoviral vectors based on membrane technology. Biotechnol Prog. 2008;24(6):1290–1296. doi: 10.1002/btpr.25. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes P., Peixoto C., Santiago V.M., Kremer E.J., Coroadinha A.S., Alves P.M. Bioprocess development for canine adenovirus type 2 vectors. GeneTher. 2013;20(4):353–360. doi: 10.1038/gt.2012.52. [DOI] [PubMed] [Google Scholar]
- 20.Cottingham M.G., Carroll F., Morris S.J., Turner A.V., Vaughan A.M., Kapulu M.C. Preventing spontaneous genetic rearrangements in the transgene cassettes of adenovirus vectors. Biotechnol Bioeng. 2012;109(3):719–728. doi: 10.1002/bit.24342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanton R.J., McSharry B.P., Armstrong M., Tomasec P., Wilkinson G.W. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques. 2008;45(6):659–662. doi: 10.2144/000112993. 64–8. [DOI] [PubMed] [Google Scholar]
- 22.Wang C., Dulal P., Zhou X., Xiang Z., Goharriz H., Banyard A. A simian-adenovirus-vectored rabies vaccine suitable for thermostabilisation and clinical development for low-cost single-dose pre-exposure prophylaxis. PLoS NeglTrop Dis. 2018;12(10):e0006870. doi: 10.1371/journal.pntd.0006870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes-Sandoval A., Berthoud T., Alder N., Siani L., Gilbert S.C., Nicosia A. Prime-boost immunization with adenoviral and modified vaccinia virus Ankara vectors enhances the durability and polyfunctionality of protective malaria CD8+ T-cell responses. InfectImmun. 2010;78(1):145–153. doi: 10.1128/IAI.00740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warimwe G.M., Lorenzo G., Lopez-Gil E., Reyes-Sandoval A., Cottingham M.G., Spencer A.J. Immunogenicity and efficacy of a chimpanzee adenovirus-vectored Rift Valley fever vaccine in mice. Virol J. 2013;10:349. doi: 10.1186/1743-422X-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dicks M.D., Spencer A.J., Edwards N.J., Wadell G., Bojang K., Gilbert S.C. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7(7):e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghunath B.B.W., Pattanaik P., Janssens J. Best practices for optimization and scale-up of microfiltration TFF processes. BioProcessing J. 2012;11(1):30–40. [Google Scholar]
- 27.Colloca S., Barnes E., Folgori A., Ammendola V., Capone S., Cirillo A. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med. 2012;4(115):115ra2. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maizel J.V., Jr., White D.O., Scharff M.D. The polypeptides of adenovirus. I. evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36(1):115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi E., Cohen S.L., Tsai P.K., Sweeney J.A. Quantitation of adenovirus type 5 empty capsids. Anal Biochem. 2006;349(2):208–217. doi: 10.1016/j.ab.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Wu M., Menesale E., Lu T., Magliola A., Bergelson S. Development and qualification of a high sensitivity, high throughput Q-PCR assay for quantitation of residual host cell DNA in purification process intermediate and drug substance samples. J Pharm Biomed Anal. 2014;100:145–149. doi: 10.1016/j.jpba.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 31.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12(12):236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas A.D., Williams A.R., Illingworth J.J., Kamuyu G., Biswas S., Goodman A.L. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat Commun. 2011;2:601. doi: 10.1038/ncomms1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitelli A., Quirion M.R., Lo C.Y., Misplon J.A., Grabowska A.K., Pierantoni A. Vaccination to conserved influenza antigens in mice using a novel Simian adenovirus vector, PanAd3, derived from the bonobo Pan paniscus. PLoS One. 2013;8(3):e55435. doi: 10.1371/journal.pone.0055435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne R.O., Silk S.E., Elias S.C., Miura K., Diouf A., Galaway F. Human vaccination against RH5 induces neutralizing antimalarial antibodies that inhibit RH5 invasion complex interactions. JCI Insight. 2017;2(21):e96381. doi: 10.1172/jci.insight.96381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goerke A.R., To B.C., Lee A.L., Sagar S.L., Konz J.O. Development of a novel adenovirus purification process utilizing selective precipitation of cellular DNA. Biotechnol Bioeng. 2005;91(1):12–21. doi: 10.1002/bit.20406. [DOI] [PubMed] [Google Scholar]
- 36.Konz J.O., Livingood R.C., Bett A.J., Goerke A.R., Laska M.E., Sagar S.L. Serotype specificity of adenovirus purification using anion-exchange chromatography. Hum Gene Ther. 2005;16(11):1346–1353. doi: 10.1089/hum.2005.16.1346. [DOI] [PubMed] [Google Scholar]
- 37.Weggeman M, Van Corven EJJM, inventors; Crucell Holland B.V. (Leiden, NL), assignee. Virus purification methods. US patent number 8,574,595. 2012