Abstract
Hepatotoxic microcystins (MCs) are the most widespread class of cyanotoxins and the one that has most often been implicated in cyanobacterial toxicosis. One of the main challenges in studying and monitoring MCs is the great structural diversity within the class. The full chemical structure of the first MC was elucidated in the early 1980s and since then, the number of reported structural analogues has grown steadily and continues to do so, thanks largely to advances in analytical methodology. The structures of some of these analogues have been definitively elucidated after chemical isolation using a combination of techniques including nuclear magnetic resonance, amino acid analysis, and tandem mass spectrometry (MS/MS). Others have only been tentatively identified using liquid chromatography-MS/MS without chemical isolation. An understanding of the structural diversity of MCs, the genetic and environmental controls for this diversity and the impact of structure on toxicity are all essential to the ongoing study of MCs across several scientific disciplines. However, because of the diversity of MCs and the range of approaches that have been taken for characterizing them, comprehensive information on the state of knowledge in each of these areas can be challenging to gather. We have conducted an in-depth review of the literature surrounding the identification and toxicity of known MCs and present here a concise review of these topics. At present, at least 279 MCs have been reported and are tabulated here. Among these, about 20% (55 of 279) appear to be the result of chemical or biochemical transformations of MCs that can occur in the environment or during sample handling and extraction of cyanobacteria, including oxidation products, methyl esters, or post-biosynthetic metabolites. The toxicity of many MCs has also been studied using a range of different approaches and a great deal of variability can be observed between reported toxicities, even for the same congener. This review will help clarify the current state of knowledge on the structural diversity of MCs as a class and the impacts of structure on toxicity, as well as to identify gaps in knowledge that should be addressed in future research.
Keywords: microcystin, cyanobacteria, cyanotoxin, structural elucidation, toxicology
1. Introduction
There are an increasing number of warnings about toxic cyanobacterial blooms observed worldwide and global warming is thought to stimulate their development in eutrophic waters [1,2,3,4,5]. These blooms are often accompanied by production of a variety of cyanotoxins generally classified according to the target organs: hepatotoxins (liver), neurotoxins (nervous system), and dermatotoxins (skin). Among these cyanotoxins, it appears that hepatotoxic and tumor promoting microcystins (MCs) are more commonly found in cyanobacterial blooms and considered to be one of the most hazardous groups throughout the world [6,7,8,9,10]. Despite the significant amount of available information, interest in MCs continues to increase due to their well-known hazards to farm animals, fisheries, aquaculture, human health, and wildlife through exposure via drinking, environmental, and recreational waters [7,8,10,11,12].
Louw and Smit [13] were among the first scientists who isolated and attempted to characterize a cyanotoxin from a cyanobacterial bloom, dominated by Microcystis toxica, which occurred in the Vaal Dam reservoir in South Africa in 1942–1943. They concluded that it was an alkaloid of undetermined structure with acute and chronic hepatotoxic properties. Initial attempts to characterize the structures of MCs started in the late 1950s [14]. However, full structural identification of the first MC congeners was achieved in 1984, when a combination of amino acid analysis, nuclear magnetic resonance (NMR) spectroscopy, and mass spectrometry (MS) were used [15,16]. These and subsequent studies showed the chemical structure of these toxins to consist of a cyclic heptapeptide composed of five relatively conserved amino acids plus two variable L-amino acids [14,15,16,17,18,19,20].
Two decades later, studies have shown that these toxins are biosynthesized nonribosomally via an MC synthetase gene cluster (mcy), consisting of a combination of polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs) and tailoring enzymes [21,22,23,24]. Recent studies have reported the bulk of this structural diversity is the result of genetic and/or environmental factors, which have an impact on the functioning of enzymes encoded in the mcy gene cluster [25,26,27,28,29,30,31,32]. This mcy gene cluster is often spontaneously modified through point mutations, deletions and insertions, or a series of genetic recombinations, which affect the functioning of the MC peptide synthetases and result in the chemical diversity observed in nature [33,34,35,36,37].
As a result of the large number of literature reports spanning over four decades of research, it can be difficult to glean accurate information on the total number of identified MCs. The phrase ‘more than 100 microcystin congeners’ is still often used in the literature [12,38,39,40,41] however estimates as high as 248 known MCs have recently been published [42]. In this review, we update this number to 279 and describe the methods by which this structural elucidation was carried out. In addition, we provide an in-depth review of their toxic potential and a discussion of the structure–activity relationships this information provides.
2. Nomenclature and General Chemical Structure of Microcystins
Hughes et al. [43] first described a hepatotoxic ‘fast death factor’ in an extract of the isolated strain Microcystis aeruginosa NRC-1. This hepatotoxic factor was later renamed microcystin, derived from the genus Microcystis [44,45,46]. Since, they have also been referred to in the literature as cyanoginosin, with prefix ‘cyano’ from the term cyanobacteria and ‘ginosin’ derived from aeruginosa [19,47]; and cyanoviridin, with the root ‘viridin’ from the species M. viridis [48].
After almost two decades of structural analysis of toxic peptides from the colonial bloom-forming cyanobacterium M. aeruginosa, Botes et al. [15,16,17,18,19] and Santikarn et al. [20] provided structural details of these toxins with the general structure of: cyclo-(D-Ala1-X2-D-Masp3-Z4-Adda5-D-γ-Glu6-Mdha7) (Figure 1) in which X and Z are variable L-amino acids, D-Masp3 is D-erythro-β-methyl-isoaspartic acid, and Mdha is N-methyldehydroalanine. The β-amino acid Adda, (2S,3S,4E,6E,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid, is the most unusual substructure, and has not been reported elsewhere in nature apart from in the closely related group of toxic pentapeptides, the nodularins. After a time, leading researchers in the field warned that the continued use of multiple naming systems could generate confusion as the number of publications on these cyclic peptides increased [49]. Therefore, a universal system of nomenclature was proposed based on the original term microcystin-XZ, where X and Z denote the variable amino acid residues in positions 2 and 4 [50].
Figure 1.
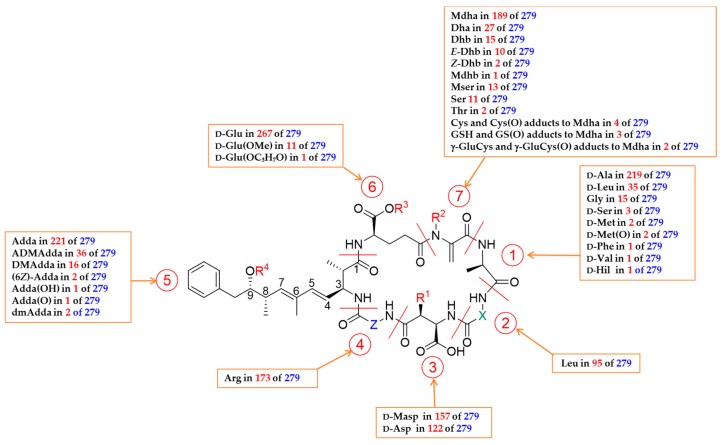
General structure of microcystins (MCs) and an overview of their observed structural diversity. R1 = H or CH3; R2 = H or CH3; R3 = H, CH3 or C3H6OH; R4 = H, CH3 or COCH3; X and Z = variable L-amino acids.
The number of fully and partially characterized MCs increased significantly throughout the 1990s and structural variations have been reported in all seven amino acids, but most frequently with substitution of the two variable L-amino acids X and Z at positions 2 and 4, respectively, and demethylation of amino acids at position 3 and/or 7. Multiple combinations of the variable L-amino acids (X and Z), and modifications to the other amino acids, lead to the high levels of structural diversity observed in MCs which have been characterized to date from bloom samples and isolated strains of Microcystis, Oscillatoria, Planktothrix, Anabaena (syn. Dolichospermum), and Nostoc spp. [51]. While Carmichael et al. [50] originally proposed abbreviation in the format MCYST-XZ, in recent decades the abbreviation MC-XZ has come in to general use, where X and Z are the one-letter amino acid abbreviations (where these exist) and with any variations at positions 1, 3, and 5–7 (relative to D-Ala, D-Masp, Adda, D-Glu, and Mdha, respectively) shown in square brackets, using 3–7-letter amino acid abbreviations [52], in numerical order with the position indicated by a superscript, and separated by commas without spaces in square brackets immediately prior to MC. For example, [D-Leu1,D-Asp3,Dha7]MC-LR contains D-Leu, D-Asp and Dha at positions 1, 3, and 7, and L-Leu and L-Arg at positions 2 and 4, respectively, with Adda and D-Glu assumed by default at positions 5 and 6 (see Figure 1). If an amino acid residue at positions 2 or 4 is not one of the 20 standard amino acids, the three-letter (or more, where necessary) abbreviation is used; e.g., the congener containing Leu at position 2 and homoarginine in position 4 is named MC-LHar. Ring opened MCs are designated with the prefix [seco-a/b], where a and b are the residue numbers between which the amide bond has been hydrolysed. For example, [seco-4/5]MC-LR indicates MC-LR hydrolysed between the Arg4- and Adda5-residues. A MC name-generator is included in version 16 of the MC mass calculator tab of a publicly available toxin mass list [53]. To facilitate ongoing efforts to maintain comprehensive lists and databases of toxins and/or cyanobacterial metabolites, it is recommended that researchers use these naming conventions going forward when reporting the identification of new MCs.
3. Biosynthesis of Microcystins
The nonribosomal biosynthesis of MCs has been described in detail in Microcystis [21,22,23,54], Planktothrix [55], and Anabaena [56]. This MC synthetase system is encoded by two transcribed operons in M. aeruginosa, mcyABC, and mcyDEFGHIJ, from a central bidirectional promoter located between mcyA and mcyD [23,54]. Insertional gene knockout experiments have demonstrated that all MC congeners produced by a strain are synthesized by a single enzyme complex encoded by a 55 kb gene cluster [21,55,57,58]. Therefore, strains that do not produce MCs can result from inactivation either by point mutation, or by partial or complete deletion of mcy genes [36,57,59,60]. Recently, Shishido et al. [61] reported that the mcy gene cluster did not encode all enzymes for the synthesis of rare variants of MC that contain a selection of homo-amino acids by the benthic cyanobacterium Phormidium sp. LP904c, but may instead activate homo-amino acids produced during the synthesis of anabaenopeptins.
Several studies have assessed the impact of environmental factors, including nutrient availability, light, iron limitation, temperature, and pH on cellular MC content [25,31,32,62,63,64,65,66,67,68]. Some of these factors are involved in enhancing or suppressing the expression of the mcy gene, leading to an increase or decrease in MC production. For example, Kaebernick et al. [62] reported that light quality has a significant effect on the transcription levels of the genes mcyB and mcyD. Recently, Yang and Kong [69] reported that the exposure of M. aeruginosa cells to UV-B not only remarkably inhibited the growth of Microcystis, but also led to a reduction in MC concentration by decreasing transcription of the gene mcyD. Sevilla et al. [64] demonstrated that iron starvation causes an increase in mcyD transcription and MC levels. Moreover, it was also shown that stress conditions caused by nutrient deprivation increased mcyD transcription and MC production [68]. Horst et al. [70] combined laboratory and field experiments and observed a correlation between nitrogen limitation and lower MC cell quotas in a field survey of Lake Erie (situated at the international boundary between Canada and the USA). Since MC is a nitrogen-rich metabolite, Van de Waal et al. [30] have observed an increase in MC cell quotas after excess nitrogen supply, the nitrogen-rich variant MC-RR being the most significantly affected. Similarly, Puddick et al. [32] recently reported that when Microcystis strain CAWBG11 was grown in batch culture until nitrogen-depletion, the relative abundance of the MC congeners shifted and the relative abundance of arginine-containing MCs decreased, whilst the congeners which did not contain arginine increased. These changes coincided with a large decrease in nitrate concentration to 0.35% of initial levels.
Therefore, the biosynthesis of MCs is strain dependent and their diversity is due to variability in the coding of the MC synthetase genes among cyanobacterial strains [71] as well as some environmental factors such as nitrogen concentration [29,30,32,72,73]. The large variety of amino acids that can be incorporated and further modified by tailoring enzymes, having activities such as the epimerization, cyclisation, N-methylation, formylation, and reduction of amino acids, accounts for the production of highly complex peptides [74,75]. In the field, it is important to consider that multiple strains of cyanobacteria co-exist, often leading to more complex profiles of MCs. Typically, cyanobacterial strains in the field and in controlled conditions produce up to twenty MC congeners but usually only one or two congeners dominate in any single strain [12,40,76,77,78,79,80]. However, the number of congeners produced in a single strain can exceed 47 when all minor MC analogues are considered [33] but in many other cases MC profiles reportedly contain only a small number of congeners. This highlights the fact that there are still new MCs being constantly identified, with varying degrees of certainty, in the very recent literature [81,82]. However, gaps remain in the understanding of how environmental factors can modulate the relative abundance of MC congeners in a bloom, requiring further research on the topic.
4. Structural Elucidation of Microcystins
Definitive structural elucidation of new MCs can only be achieved by purifying individual MCs using preparative isolation techniques followed by comprehensive NMR, amino acid analysis and tandem MS (MS/MS) experiments.
Proper purification, structural elucidation, and quantitation are also of critical importance to toxicity studies, where the knowledge of structure, purity, and amount of each MC tested for toxicity are required to ensure the validity and comparability of toxicology results. In this section we give a brief overview of the techniques available for structural elucidation of MCs and the structural information that each technique can provide. This will be useful in choosing appropriate methods for structural elucidation as well as in the critical evaluation of published MC identifications and toxicities.
4.1. Mass Spectrometry for Structural Elucidation
In addition to offering sensitive and selective targeted quantitative analysis of known MCs, MS is an invaluable tool for tentative structural elucidation of new MCs, providing information about their molecular formula (full scan) and structure (MS/MS). The types of MS techniques that have been used for the identification of MCs have evolved as mass spectrometers suitable for intact analysis of peptides have become commercially available. Early work relied heavily on direct analysis of MCs purified off-line, using fast atom bombardment MS in a variety of low and high resolution as well as tandem MS configurations [15,75,83,84,85,86]. Similarly, the usually high-resolution MS information from matrix assisted laser desorption ionization-time of flight MS with and without post-source decay has been invaluable in identifying new purified MCs over the years [87,88,89,90,91]. The widespread availability of electrospray ionization (ESI) has made it commonplace to couple analytical liquid chromatography (LC) separations directly to high resolution and/or tandem mass spectrometers. This has offered the possibility for structural characterization and tentative identification of many MCs in complex mixtures without the need for isolation of milligram amounts of toxin needed for full structural elucidation. LC with sequential multi-stage MS dissociation (LC-MSn), while usually offering only unit mass resolution, provides a great deal of information for the determination of structure and connectivity and has been widely used for MC identification [33,75,77,90,92,93]. Alternatively, LC with high resolution MS/MS (LC-HRMS/MS) provides accurate mass data on precursor and product ions suitable for determining their molecular formulae [81,82,94,95,96,97].
LC-MS alone is usually not considered definitive proof of chemical structure, but a great deal of structural information can nevertheless be obtained. LC separation of MCs is typically carried out in reverse phase using C18 or similar stationary phases and acidic mobile phases, resulting in protonation of carboxyl groups, with acetonitrile as the organic modifier. The hydrophobicity and therefore elution order of MCs in reverse-phase LC is governed primarily by the number of arginine residues in their structure, and the relative elution order of new congeners can provide useful information about structure. MCs with two arginine or homoarginines (e.g., MC-RR) are the most polar and elute first, followed by MCs with a single Arg at position four (e.g., MC-LR), then those with a single Arg at position 2 (e.g., MC-RY), and finally those with no Arg residues (e.g., MC-LA). Though sometimes difficult to separate chromatographically [98], the relative elution order of desmethylated MCs can also be useful for identitication of minor analogues with [DMAdda5]-variants eluting much earlier than [D-Asp3]-variants, which are followed closely by [Dha7]-variants [82].
The nominal molecular weights for known parent MCs range from 882 Da to 1101 Da, although the theoretical range using the known amino acids identified to date in natural MCs is significantly wider than this [53]. These masses are too large for definitive molecular formula elucidation by HRMS when all possible elements are considered (e.g., F, P, Cl, etc.), but when the possible elemental composition and ring or double bond equivalents (RDBE) are limited to a reasonable range similar to known MCs, in most cases a candidate formula can be determined. Careful evaluation of isotopic patterns in HRMS can further help narrow down possible molecular formulae, relying less on pre-determined limits in elements, as reported recently in Mallia et al. [99]. Known parent (unconjugated) MCs have between 44 and 57 C atoms, 63 and 84 H atoms, 7 and 13 N atoms, 12 and 17 O atoms, and up to one S atom in methionine-containing MCs [53]. It can also be useful to limit possible molecular formulas by the number of RDBEs in the structure, which ranges from 17 to 27. Within these limits there are currently no cases among known MC where [M+H]+ or [M+2H]2+ ions are within 5 ppm mass accuracy from one another. However, there are >120 cases reported of isomeric MCs reported, significantly limiting the utility of accurate mass measurements alone for MC identification. It should be noted that conjugates between MCs and thiol-containing biomolecules (e.g., cysteine, glutathione) have been reported [81,100,101] and that mass ranges and molecular formula tolerances need to be widened accordingly when characterizing these compounds.
The ionization and MS/MS fragmentation of MCs are also generally similar between MCs with the same number of Arg/Har residues. In positive ESI mode, MCs with two Arg/Har residues, or conjugated to Cys or GSH, appear primarily as doubly charged ions after ESI while those with one or no Arg/Har are predominantly singly charged. MCs with no Arg/Har ionize comparatively poorly and therefore tend to form adducts with other cations present in the sample (e.g., [M+Na]+ or [M+NH4]+). In negative ESI all MCs are observed predominantly as singly charged [M-H]- ions. The MS/MS dissociation of MCs has been studied extensively in both positive [102,103], and negative [104,105] ionization modes. In most cases, MCs fragment to produce sensitive class-characteristic fragments at m/z 135.0804 originating from the Adda5–residue in positive mode, and m/z 128.0353 originating from the Glu6-residue in negative mode [82,105,106]. Since MCs are cyclic peptides, a large body of previous work on the collision induced dissociation (CID) of non-microcystin peptides also exists [107] that can be useful in interpretation of microcystin MS/MS spectra. In addition to the class-characteristic fragments, various “sequence ions” are formed during CID following ring opening that represent cleavages of individual amino acids. These can be used to effectively reconstruct the MC structure one amino acid at a time. For example, in negative mode, the B-ion series is useful for determining the mass of the amino acid at position 4 since ions represent successive cleavages from the B1 ion [C5H4O3-X4-Adda-(Glu-H2O)-Mdha-Ala]− [105]. Similarly, in positive mode, fragments associated with each individual amino acid can be readily identified from HRMS/MS data. This allows the amino acid at which small structural variations, such as demethylation have occurred, to be determined but not always the definitive structure of amino acid side chains, particularly for low resolution MS data [102].
There are two different types of CID that provide different and often complementary information for structural elucidation; quadrupole and ion trap CID. Quadrupole CID, sometimes termed higher energy collisional dissociation (HCD), is a single-stage process involving a smaller number of more energetic collisions. The main advantage of this approach is that the full m/z range of product ions can be detected simultaneously, allowing for sensitive detection of both class-specific and structurally informative product ions in a single experiment. However, it can be more difficult to trace the origin of lower mass product ions. Ion trap CID involves longer time scales and a larger number of lower energy collisions, as well as offering the potential for multiple stages of MS/MS (MSn). Here, structural elucidation is done by constructing a fragmentation tree where the origin of lower mass product ions can be established in higher order experiments (e.g., MS3 or MS4) in relation to higher-mass product ions observed in MS2. The principal limitation of ion trap dissociation is the 1/3 rule, stating that product ions of less than 1/3 the m/z of a selected precursor ion cannot be isolated for detection. This means that class-specific low mass product ions cannot be detected in MS2 in an ion trap regardless of the collision energy used.
Use of established chemical degradation or modification reactions to probe the structure of unknown chemicals is a classical approach that pre-dates the availability of modern analytical instrumentation such as NMR or MS. This approach was an important part of the first structural elucidation studies on MCs, where partial hydrolysis reactions followed by Edman degradation reactions and mass spectrometric methods of characterizing the linearized products were utilised [15,16]. However, when combined with modern LC-MS, similar approaches are even more powerful due to the ease with which multiple products of reactions with multiple reactants can be selectively detected. The reactivity of the double bond in Dha7 and Mdha7 of MCs towards thiols has emerged as being particularly useful for characterizing MCs congeners [81,82,99]. In particular, mercaptoethanol derivatization has been exploited in such a manner both to identify new MCs from complex samples [77,108] or to quickly differentiate Mdha7- from Dhb7-containing MCs [109], which is not possible using LC-MS alone unless authentic standards are available. Another useful reaction is methyl esterification that, together with LC-MS, can be used to count the number of reactive carboxylic acid (the CO2H group of Glu6 is reactive, but not that of Masp3/Asp3) and phenolic groups in MCs [96,99]. Selective oxidation with mild oxidants can also be combined with LC-MS to identify MCs containing sulfide and sulfoxides, such as methionine (e.g., MC-MR) [81,93]. This approach has also proved useful in helping to identify sulfide (e.g., Cys, GSH) conjugates of MCs, and their autoxidation products, in natural samples [81]. These conjugates are also readily semi-synthesised to produce authentic standards for confirmation of their presence in samples and cultures by LC-MS (e.g., [81,100]) (see Section 5.7). In addition, the identity of S-conjugates at position-7 of MCs can be confirmed through a range of deconjugation reactions [101,110,111].
It should be noted that stereochemical information is not generally accessible from LC-MS/MS analyses. It is therefore generally assumed in the literature that the stereochemistry of the amino acids in MCs whose structures are established using LC-MS methods are the same as those in well-established structures such as MC-LR unless there is evidence to the contrary, and the same assumption is made throughout this review.
4.2. Preparative Isolation of Microsystins
While a great deal of structural information is available from LC-MS without the need for additional purification of MCs, stereochemistry is not accessible by LC-MS. Definitive structural elucidation by NMR currently requires isolation of 50–100 μg or more of a pure MC. This is most often done by scaling up production of laboratory cultured cyanobacteria, or directly from naturally occurring bloom samples. Dried cyanobacterial biomass is usually first extracted under acidic conditions. Aqueous extracts can then be applied to a reverse phase (e.g., C18) open column or large solid phase extraction (SPE) cartridge, depending on the complexity of the toxin profile. Elution is then carried out in a stepwise fashion, usually with aqueous methanol. In some cases, size exclusion chromatography has been used [83] but is often not required. For more complex toxin mixtures, a final semi-preparative isolation step is required, again using a reverse phase column. Additional selectivity at each step can be achieved by varying the pH of the eluent with various buffers. These then need to be removed with an additional SPE step. Long-term storage of MCs in acidic methanol should be avoided because of the potential for formation of methyl esters [81,112] (see Section 5.7).
4.3. Amino Acid Analysis
One of the classical approaches to structural characterization of MCs after preparative isolation is to hydrolytically digest them to their constituent amino acids, which are then identified chromatographically based on retention time comparison to amino acid standards. Typically, hydrolysis is done in 6 M HCl at 105 °C for 24 h. It should be noted that under these conditions, Mdha and Adda were not detected [18] and some amino acids (e.g., tryptophan, glutamine, serine, threonine, tyrosine, methionine, and cysteine) are altered [113] and their presence must be inferred. Amino acids can then be derivatized with a UV absorbent tag and analyzed by reverse phase LC-UV [83]. Alternatively, they can be derivatized with trifluoroacetic anhydride to make them volatile and then analyzed by gas chromatography [114]. When chiral stationary phases or derivatizing agents are used, it is possible to determine the absolute configuration of the amino acids, which is not possible by MS or NMR alone.
4.4. Structural Elucidation by Nuclear Magnetic Resonance Spectroscopy
Although it is most often used in combination with other approaches mentioned earlier, NMR is usually required for definitive structural identification of MCs including relative stereochemistry. Often CD3OD and sometimes D2O were used in the past as solvents for NMR spectroscopy of MCs to reduce 1H signals arising from the solvent signals in 1- and 2-D spectra e.g., [115]. The disadvantage is that exchangeable amide protons, and thus their correlations, cannot be detected. It is therefore advantageous to use solvents that do not exchange the amide protons, such as (CD3)2SO or CD3OH, and with modern NMR spectrometers and solvent suppression techniques, the use of partially protonated solvents such as CD3OH is not usually problematic. However, NMR spectroscopy is a relatively insensitive technique, and significant amounts of purified compound are generally required. With modern high-field NMR spectrometers, full structural elucidations can be performed on less than 50 μg of an MC in CD3OH in 5 mm NMR tubes.
A comprehensive approach to assigning all protons and carbons in MCs by NMR alone was presented nearly 3 decades ago that relied on a combination of three 2-D NMR techniques: double-quantum filtered correlation spectroscopy (DQF-COSY), lH-detected multiple quantum coherence (HMQC), and heteronuclear multiple bond correlation (HMBC) NMR spectroscopy [115]. DQF-COSY was first used to assign H-2 of each constituent amino acid along with several other proton assignments. HMQC was then used to assign C-2 of each constituent amino acid based on the H-2 resonances already assigned. Finally, HMBC NMR spectra, that can detect 2J- and 3J-couplings between carbon and hydrogen atoms, were used to assign the carbonyl carbon from each amino acid and ultimately to determine the overall amino acid sequence. Nowadays, total correlation spectroscopy (TOCSY) or decoupling in the presence of scalar interactions (DIPSI-2) NMR spectra would typically also be acquired to identify the individual amino acid spin systems, and edited heteronuclear single quantum correlation (HSQC) spectra to identify methyl, methylene, and methine 13C and 1H resonances. Rotating frame nuclear overhauser effect spectroscopy (ROESY) NMR spectra are generally better suited for detection of through-space correlations for molecules of the size of MCs than are nuclear overhauser effect spectroscopy (NOESY) NMR spectra. In addition to detecting through-space correlations and thus potentially defining the relative stereochemistry of the molecule, ROESY NMR correlations can be useful for establishing the connectivity of the amino acids as a supplement to HMBC NMR correlations. Selective 1D-TOCSY or –DIPSI-2 NMR spectra can often be used to obtain 1H signal multiplicities and coupling constants in regions of spectral overlap, which can be useful for chemical shift and stereochemical assignments. The higher resolution afforded in the 13C axis by band-selective HMBC and HSQC spectra is sometimes useful for resolving crowded regions of the HSQC (e.g., methylene region) and HMBC (e.g., amide carbonyl region) spectra, and data acquisition times for 2-D spectra can be substantially reduced by use of non-uniform sampling and NMR by ordered acquisition using 1H detection techniques.
NMR spectroscopy can also be used quantitatively to measure the concentrations of algal toxin solutions relative to reference solutions [116]. This approach has been applied to produce quantitative certified reference materials (CRMs) for MC-RR, MC-LR, and [Dha7]MC-LR [117] with CRMs for other MCs including MC-LA and [D-Leu1]MC-LY in preparation (P. McCarron, personal communication).
4.5. 3-Dimensional Structures of Microcystins
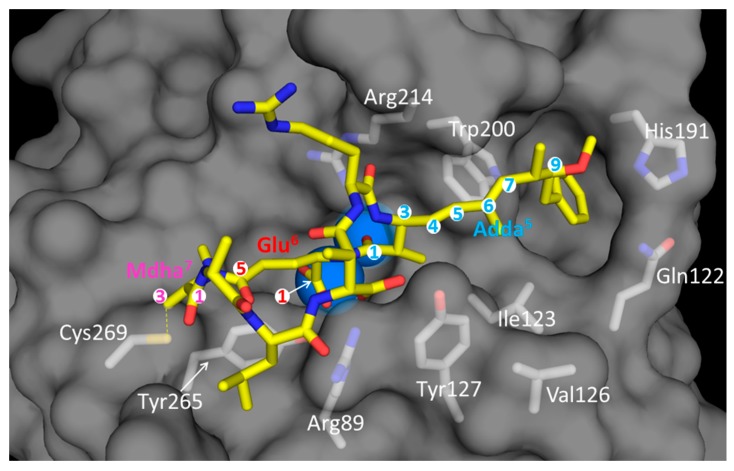
Two methods have been used for determining the 3-dimensional structures of MCs. X-ray crystallography has been used for nearly a century to reveal the 3-dimensional relationships of heavy atoms in crystallized organic molecules. Unfortunately, MCs have not so far yielded suitable crystals for such studies. However, in recent decades the 3-dimensional structures of MCs bound to protein phosphatases (PPs) have been determined [118,119,120,121,122,123,124,125,126] (see Section 6.2). In addition to providing key information about the mode of action and structure–activity relationships of the MCs, this also provides 3-dimensional structural information about the toxins themselves. The other approach that has been used is NMR spectroscopy, using through-space NOESY and ROESY correlations to estimate the relative intramolecular distances between hydrogen atoms in the MCs in solution [127,128,129,130]. Such measurements can be supplemented with measurements of 1H–1H coupling constants, which provide information about the dihedral angles between spin-coupled hydrogen atoms within the molecule. Such studies have led to several published 3-dimensional NMR-based solution structures, something that is not available from X-ray crystal structures because the structures are based on the solid-state structures that do not necessarily reflect the structure in solution.
Although not in and of itself proof of structure or stereochemistry, observation of high affinity of a purified MC for PPs or MC-specific antibodies can be taken as strong supporting evidence that the absolute stereochemistry of the MC is largely the same as that of other MCs. The reason is that recognition of MCs by receptor biomolecules is dependent on the 3-dimensional structure of the ligand (see Section 6.2), which is controlled by amino acids present in the MC and their stereochemistries. Biosynthetic reasoning also suggests that most MCs likely share the same stereochemistry, since they are assembled by closely related synthetases produced from genetically similar gene clusters, even in separate MC-producing genera (see Section 3).
5. Diversity of Characterized Microcystin Congeners
To date, the identification of at least 279 different MC congeners have been reported in the literature (Table 1) using various combinations of the techniques reviewed in Section 4. These congeners include MCs biosynthesized with structural variations at every amino acid position as well as the products of several chemical and biochemical transformations that can occur in the environment or the laboratory, all of which are reviewed in this section.
Table 1.
Microcystin congeners reported in the literature as identified from cyanobacterial cultures and field samples.
| Entry | Microcystin | Molecular Formula | Exact Mass a |
Characterization | Reference |
|---|---|---|---|---|---|
| 1 | [D-Asp³,DMAdda5]MC-LA | C44H63N7O12 | 881.4535 | LC-MS/MS | [33] |
| 2 | [D-Asp³]MC-VA | C44H63N7O12 | 881.4535 | LC-MS/MS | [33] |
| 3 | [D-Asp3]MC-LA | C45H65N7O12 | 895.4691 | LC-HRMS/MS | [94] |
| 4 | [Dha7]MC-LA | C45H65N7O12 | 895.4691 | LC-MS/MS | [33] |
| 5 | [DMAdda5]MC-LA | C45H65N7O12 | 895.4691 | LC–MS/MS, thiol | [131] |
| 6 | MC-VA | C45H65N7O12 | 895.4691 | LC-MS/MS | [33] |
| 7 | MC-LA | C46H67N7O12 | 909.4848 | MS, NMR, AA, LC-HRMS/MS |
[15,94] |
| 8 | MC-LAbu | C47H69N7O12 | 923.5004 | HRFABMS, AA, NMR | [132] |
| 9 | [D-Asp3,D-Glu(OMe)6]MC-LAbu b | C47H69N7O12 | 923.5004 | LC-HRMS/MS | [38] |
| 10 | [D-Asp³]MC-LV | C47H69N7O12 | 923.5004 | LC-MS/MS | [33] |
| 11 | [D-Asp³]MC-FA | C48H63N7O12 | 929.4535 | LC-MS/MS, thiol | [12] |
| 12 | [D-Asp³,Dha7]MC-YA | C47H61N7O13 | 931.4327 | MALDI-TOF MS | [89] |
| 13 | [D-Asp3]MC-LL | C48H71N7O12 | 937.5161 | LC-HRMS/MS | [94] |
| 14 | MC-LV | C48H71N7O12 | 937.5161 | AA, LC-MS/MS | [33,133] |
| 15 | [D-Asp³]MC-RA | C45H66N10O12 | 938.4862 | LC-MS/MS, thiol | [12,33] |
| 16 | MC-FA | C49H65N7O12 | 943.4691 | NMR, LC-MS/MS, AA, thiol | [134] |
| 17 | MC-LL | C49H73N7O12 | 951.5317 | LC-HRMS/MS, AA | [94,133] |
| 18 | MC-AR | C46H68N10O12 | 952.5018 | AA, HRMS, NMR | [76] |
| 19 | MC-RA | C46H68N10O12 | 952.5018 | LC-MS/MS, NMR, thiol | [77,135] |
| 20 | [D-Asp³]MC-RAbu | C46H68N10O12 | 952.5018 | LC-MS/MS, thiol | [12] |
| 21 | MC-FAbu | C50H67N7O12 | 957.4848 | LC-MS/MS, thiol | [12] |
| 22 | MC-YA | C49H65N7O13 | 959.4640 | AA, NMR, MS, LC-MS/MS, thiol |
[16,77] |
| 23 | MC-AHar | C47H70N10O12 | 966.5175 | LC-MS/MS | [136] |
| 24 | [Gly1,D-Asp3]MC-LR | C47H70N10O12 | 966.5175 | LC-MS/MS | [92] |
| 25 | [D-Asp3,Dha7]MC-LR | C47H70N10O12 | 966.5175 | AA, FABMS/MS | [84] |
| 26 | [D-Asp³,DMAdda5]MC-LR | C47H70N10O12 | 966.5175 | LC-MS/MS | [137] |
| 27 | [D-Asp³,DMAdda5,Dhb7]MC-LR | C47H70N10O12 | 966.5175 | LC-MS/MS, thiol | [109] |
| 28 | [Gly¹,D-Asp3,Dhb7]MC-LR | C47H70N10O12 | 966.5175 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 29 | MC-RAbu | C47H70N10O12 | 966.5175 | LC-MS/MS, thiol | [77] |
| 30 | [D-Asp3]MC-HarAbu | C47H70N10O12 | 966.5175 | LC-MS/MS | [40] |
| 31 | [D-Asp³]MC-WA | C50H64N8O12 | 968.4644 | LC-MS/MS, thiol | [12] |
| 32 | [D-Asp3,Dha7]MC-EE(OMe) b | C46H63N7O16 | 969.4331 | HRMS, MS/MS | [138] |
| 33 | [D-Asp³]MC-EE | C46H63N7O16 | 969.4331 | LC-HRMS/MS, thiol, esterification, 15N-label | [99] |
| 34 | MC-LM | C48H71N7O12S | 969.4881 | AA, MS | [133] |
| 35 | [D-Asp3]MC-LF | C51H69N7O12 | 971.5004 | LC-MS, MS/MS, LC-MS/MS, thiol |
[108,139] |
| 36 | MC-VF | C51H69N7O12 | 971.5004 | LC-MS/MS, 15N-enrichment |
[140] |
| 37 | [D-Asp3,Dha7]MC-LY | C50H67N7O13 | 973.4797 | LC-MS/MS, thiol | [108] |
| 38 | MC-YAbu | C50H67N7O13 | 973.4797 | LC-MS/MS, thiol | [77] |
| 39 | [D-Asp3]MC-LR | C48H72N10O12 | 980.5331 | MS/MS, HRMS, AA | [141] |
| 40 | [D-Asp3,(E)-Dhb7]MC-LR | C48H72N10O12 | 980.5331 | NMR, AA, HRMS | [142] |
| 41 | [D-Asp3,(Z)-Dhb7]MC-LR | C48H72N10O12 | 980.5331 | NMR, AA, HRMS | [142] |
| 42 | [Dha7]MC-LR | C48H72N10O12 | 980.5331 | AA, FABMS/MS | [84] |
| 43 | [DMAdda5]MC-LR | C48H72N10O12 | 980.5331 | AA, HRMS, NMR | [76] |
| 44 | [Gly1,D-Asp3,Dhb7]MC-LHar | C48H72N10O12 | 980.5331 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 45 | MC-RApa | C48H72N10O12 | 980.5331 | LC-MS/MS, thiol | [77] |
| 46 | MC-VR | C48H72N10O12 | 980.5331 | LC-MS/MS | [143] |
| 47 | MC-WA | C51H66N8O12 | 982.4800 | NMR, LC-MS/MS, AA, thiol |
[134] |
| 48 | [D-Ser1,D-Asp³,Dha7]MC-LR | C47H70N10O13 | 982.5124 | LC-MS/MS | [33] |
| 49 | [Dha7]MC-EE(OMe) b | C47H65N7O16 | 983.4488 | HRMS, MS/MS | [138] |
| 50 | [D-Asp3,Dha7]MCE(OMe)E(OMe) b | C47H65N7O16 | 983.4488 | HRMS, MS/MS | [138] |
| 51 | MC-FL | C52H71N7O12 | 985.5161 | LC-MS/MS, thiol | [12] |
| 52 | MC-LF | C52H71N7O12 | 985.5161 | AA, MS | [133] |
| 53 | MC-KynA b | C50H66N8O13 | 986.4749 | LC-MS/MS, HRMS, thiol, semisynthesis | [90] |
| 54 | [D-Asp³]MC-LY | C51H69N7O13 | 987.4953 | LC-MS/MS, thiol | [108] |
| 55 | [D-Asp3,(E)-Dhb7]MC-LY | C51H69N7O13 | 987.4953 | NMR, LC-HRMS/MS, thiol | [109] |
| 56 | [Gly1,D-Asp3,ADMAdda5]MC-LR | C48H70N10O13 | 994.5124 | LC-MS/MS | [92] |
| 57 | [Gly1,D-Asp³,ADMAdda5,Dhb7]MC-LR | C48H70N10O13 | 994.5124 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 58 | [D-Asp³,ADMAdda5]MC-VR | C48H70N10O13 | 994.5124 | LC-MS/MS | [33] |
| 59 | [D-Asp³,Dhb7]MC-AhaR | C49H74N10O12 | 994.5488 | LC-MS/MS, thiol | [109] |
| 60 | [D-Asp³]MC-Hil/HleR | C49H74N10O12 | 994.5488 | LC-MS/MS | [33] |
| 61 | [D-Asp³,(E)-Dhb7]MC-HilR | C49H74N10O12 | 994.5488 | NMR, HRMS, AA | [144] |
| 62 | [Dha7]MC-HilR | C49H74N10O12 | 994.5488 | HRMS, NMR, AA | [145] |
| 63 | [DMAdda5]MC-HilR | C49H74N10O12 | 994.5488 | LC-MS/MS | [33] |
| 64 | [DMAdda5]MC-LHar | C49H74N10O12 | 994.5488 | LC-MS/MS | [33] |
| 65 | [D-Asp3,D-Glu(OMe)6]MC-LR b | C49H74N10O12 | 994.5488 | HRMS, MS/MS, AA | [85] |
| 66 | MC-LR | C49H74N10O12 | 994.5488 | AA, NMR, HRMS, LC-MS/MS |
[16,141,146] |
| 67 | [D-Asp3]MC-ER | C47H68N10O14 | 996.4916 | LC-HRMS/MS, thiol, esterification, 15N-label | [99] |
| 68 | [(6Z)-Adda5]MC-LR b | C49H74N10O12 | 994.5488 | NMR, AA, MS | [115,147] |
| 69 | MC-RL | C49H74N10O12 | 994.5488 | LC-MS/MS, thiol | [77] |
| 70 | MC-WAbu | C52H68N8O12 | 996.4957 | LC-MS/MS, thiol | [90] |
| 71 | [Dha7]MC-E(OMe)E(OMe) b | C48H67N7O16 | 997.4644 | HRMS, MS/MS | [138] |
| 72 | MC-OiaA b | C51H66N8O13 | 998.4749 | LC-MS/MS, HRMS, thiol, semisynthesis | [90] |
| 73 | [D-Asp³]MC-MR | C47H70N10O12S | 998.4895 | LC-MS/MS, thiol, S-oxidation |
[93] |
| 74 | [seco-4/5][D-Asp³]MC-LR b | C48H74N10O13 | 998.5437 | LC-MS/MS, thiol | [93] |
| 75 | [D-Asp³,Mser7]MC-LR | C48H74N10O13 | 998.5437 | LC-MS/MS, MS/MS, thiol | [88,93] |
| 76 | [Ser7]MC-LR | C48H74N10O13 | 998.5437 | AA, HRMS, MS/MS | [84] |
| 77 | MC-LHph | C53H73N7O12 | 999.5317 | LC-MS/MS | [75] |
| 78 | MC-KynAbu b | C51H68N8O13 | 1000.4906 | LC-MS/MS, thiol | [90] |
| 79 | [D-Asp³,Dha7]MC-FR | C50H68N10O12 | 1000.5018 | LC-MS/MS | [33] |
| 80 | [Ser7]MC-EE(OMe) b | C47H67N7O17 | 1001.4593 | HRMS, MS/MS | [138] |
| 81 | [D-Asp3,Ser7]MC-E(OMe)E(OMe) b | C47H67N7O17 | 1001.4593 | HRMS, MS/MS | [138] |
| 82 | [D-Asp³]MC-HilY | C52H71N7O13 | 1001.5110 | LC-MS/MS, thiol | [108] |
| 83 | MC-LY | C52H71N7O13 | 1001.5110 | LC-MS/MS, NMR | [38,148] |
| 84 | MC-YL | C52H71N7O13 | 1001.5110 | LC-MS/MS | [38] |
| 85 | [D-Asp³,Mser7]MC-LY | C51H71N7O14 | 1005.5059 | LC-MS/MS, thiol | [108] |
| 86 | [D-Asp³,ADMAdda5,Dha7]MC-HilR | C49H72N10O13 | 1008.5280 | LC-MS/MS | [33] |
| 87 | [Gly1,D-Asp3,ADMAdda5,Dhb7]MC-LHar | C49H72N10O13 | 1008.5280 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 88 | [Gly1,D-Asp3,ADMAdda5]MC-LHar | C49H72N10O13 | 1008.5280 | LC-MS/MS | [92] |
| 89 | [D-Asp3,ADMAdda5]MC-LR | C49H72N10O13 | 1008.5280 | HRMS, NMR, AA, MS/MS, LC-MS/MS |
[78,79,149] |
| 90 | [ADMAdda5,Dha7]MC-LR | C49H72N10O13 | 1008.5280 | LC-MS/MS | [33] |
| 91 | [D-Asp³,ADMAdda5,Dhb7]MC-LR | C49H72N10O13 | 1008.5280 | NMR, HRMS, AA | [150] |
| 92 | MC-HilR | C50H76N10O12 | 1008.5644 | MS/MS, HRMS, NMR, AA | [80] |
| 93 | MC-LHar | C50H76N10O12 | 1008.5644 | AA, MS/MS, HRMS, NMR | [151] |
| 94 | [D-Glu(OMe)6]MC-LR b | C50H76N10O12 | 1008.5644 | HRMS, MS/MS, AA, HRMS/MS | [85,152] |
| 95 | [Mdhb7]MC-LR | C50H76N10O12 | 1008.5644 | AA, MS | [79] |
| 96 | [D-Leu1,D-Asp³,DMAdda5]MC-LR | C50H76N10O12 | 1008.5644 | LC-MS/MS | [153] |
| 97 | [D-Asp3,Dha7]MC-RR | C47H71N13O12 | 1009.5345 | AA, NMR, HRMS | [76] |
| 98 | [D-Asp³,DMAdda5]MC-RR | C47H71N13O12 | 1009.5345 | MS, MS/MS | [55] |
| 99 | [Gly1,D-Asp3]MC-RR | C47H71N13O12 | 1009.5345 | LC-MS/MS | [92] |
| 100 | [Gly1,D-Asp3,Dhb7]MC-RR | C47H71N13O12 | 1009.5345 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 101 | [D-Asp3]MC-LW | C53H70N8O12 | 1010.5113 | LC-MS, MS/MS | [139] |
| 102 | [D-Asp3,(E)-Dhb7]MC-LW | C53H70N8O12 | 1010.5113 | NMR, LC-HRMS/MS | [95] |
| 103 | MC-OiaAbu b | C52H68N8O13 | 1012.4906 | LC-MS/MS, thiol | [90] |
| 104 | MC-MR | C48H72N10O12S | 1012.5052 | LC-MS/MS, thiol, S-oxidation |
[93] |
| 105 | [Mser7]MC-LR | C49H76N10O13 | 1012.5593 | LC-HRMS | [80,94] |
| 106 | [seco-4/5]MC-LR b | C49H76N10O13 | 1012.5593 | LC-MS/MS, HRMS, thiol, NMR | [93,154] |
| 107 | [seco-1/2]MC-LR b | C49H76N10O13 | 1012.5593 | MS/MS, HRMS, NMR | [154] |
| 108 | MC-NfkA b | C51H66N8O14 | 1014.4698 | NMR, LC-MS/MS, HRMS, thiol, semisynthesis | [90] |
| 109 | [D-Asp³]MC-M(O)R b | C47H70N10O13S | 1014.4845 | LC-MS/MS, thiol, S-oxidation |
[93] |
| 110 | [D-Asp³,Dha7]MC-HphR | C51H70N10O12 | 1014.5175 | LC-MS/MS | [33] |
| 111 | [D-Asp3]MC-FR | C51H70N10O12 | 1014.5175 | AA, MS, NMR | [87] |
| 112 | [Dha7]MC-FR | C51H70N10O12 | 1014.5175 | AA, HRMS, MS/MS | [155] |
| 113 | [DMAdda5]MC-FR | C51H70N10O12 | 1014.5175 | LC-MS/MS | [137] |
| 114 | [D-Asp³]MC-RF | C51H70N10O12 | 1014.5175 | LC-MS/MS, thiol | [108] |
| 115 | [Ser7]MC-E(OMe)E(OMe) b | C48H69N7O17 | 1015.4750 | HRMS, MS/MS | [138] |
| 116 | MC-LHty | C53H73N7O13 | 1015.5266 | LC-MS/MS | [75] |
| 117 | [D-Asp³,Dha7]MC-RY | C50H68N10O13 | 1016.4967 | LC-MS/MS, thiol | [108] |
| 118 | [D-Asp³,DMAdda5]MC-RY | C50H68N10O13 | 1016.4967 | LC-MS/MS, thiol | [109] |
| 119 | MC-YM | C51H69N7O13S | 1019.4674 | AA, NMR, MS | [16] |
| 120 | [D-Asp3,ADMAdda5]MC-LHar | C50H74N10O13 | 1022.5437 | HRMS, MS/MS, AA | [86] |
| 121 | [ADMAdda5]MC-LR | C50H74N10O13 | 1022.5437 | HRMSNMR, AA, MS/MS | [78,79] |
| 122 | [D-Leu1,DMAdda5]MC-LR | C51H78N10O12 | 1022.5801 | LC-HRMS/MS, thiol | [81] |
| 123 | [D-Leu1,dmAdda5]MC-LR (isomer 1) c | C51H78N10O12 | 1022.5801 | LC-HRMS/MS, thiol | [81] |
| 124 | [D-Leu1,dmAdda5]MC-LR (isomer 2) c | C51H78N10O12 | 1022.5801 | LC-HRMS/MS, thiol | [81] |
| 125 | [D-Leu1,D-Asp3]MC-LR | C51H78N10O12 | 1022.5801 | LC-MS/MS, HRMS/MS | [152,153] |
| 126 | [D-Leu1,Dha7]MC-LR | C51H78N10O12 | 1022.5801 | LC-MS/MS | [75] |
| 127 | [D-Val1]MC-LR | C51H78N10O12 | 1022.5801 | LC-MS/MS | [75] |
| 128 | [Gly1,D-Asp³,Dhb7]MC-RHar | C48H73N13O12 | 1023.5502 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 129 | [D-Asp3]MC-RR | C48H73N13O12 | 1023.5502 | AA, HRMS, NMR | [76,156] |
| 130 | [Dha7]MC-RR | C48H73N13O12 | 1023.5502 | AA, HRMS, MS/MS, NMR | [157] |
| 131 | [D-Asp3,(E)-Dhb7]MC-RR | C48H73N13O12 | 1023.5502 | NMR, HRMS | [158] |
| 132 | [Gly1,D-Asp3]MC-RHar | C48H73N13O12 | 1023.5502 | LC-MS/MS | [92] |
| 133 | [DMAdda5]MC-RR | C48H73N13O12 | 1023.5502 | LC-HRMS/MS, thiol | [82] |
| 134 | MC-WL | C54H72N8O12 | 1024.5270 | LC-MS/MS, thiol | [90] |
| 135 | MC-LW | C54H72N8O12 | 1024.5270 | LC-MS/MS, 15N-enrichment |
[140] |
| 136 | [D-Asp3]MC-RCit | C48H72N12O13 | 1024.5342 | LC-HRMS/MS, thiol, 15N-label | [99] |
| 137 | [D-Asp3,ADMAdda5,Thr7]MC-LR | C49H74N10O14 | 1026.5386 | LC-MS/MS, thiol | [159] |
| 138 | [Seco-1/2]MC-HilR b | C50H78N10O13 | 1026.5750 | MS/MS, HRMS | [80] |
| 139 | [D-Asp3,Ser7]MC-RR | C47H73N13O13 | 1027.5451 | LC-MS/MS, thiol | [159] |
| 140 | MC-NfkAbu b | C52H68N8O14 | 1028.4855 | LC-MS/MS, thiol | [90] |
| 141 | MC-M(O)R b | C48H72N10O13S | 1028.5001 | AA, HRMS, NMR | [76] |
| 142 | MC-FR | C52H72N10O12 | 1028.5331 | AA, HRMS, NMR | [76,160] |
| 143 | MC-RF | C52H72N10O12 | 1028.5331 | LC-MS/MS, thiol | [77] |
| 144 | [D-Asp³]MC-HphR | C52H72N10O12 | 1028.5331 | MS/MS | [88] |
| 145 | [Dha7]MC-HphR | C52H72N10O12 | 1028.5331 | AA, HRMS, MS/MS, 1H-NMR |
[161] |
| 146 | [D-Asp³]MC-M(O2)R b | C47H70N10O14S | 1030.4794 | LC-MS/MS, thiol, S-oxidation |
[93] |
| 147 | [D-Asp3,Dha7]MC-HtyR | C51H70N10O13 | 1030.5124 | AA, HRMS, MS/MS, NMR | [161] |
| 148 | [D-Asp³,DMAdda5]MC-HtyR | C51H70N10O13 | 1030.5124 | MS/MS | [88] |
| 149 | [Dha7]MC-YR | C51H70N10O13 | 1030.5124 | HRMS, MS/MS, AA | [162] |
| 150 | [D-Asp3]MC-RY | C51H70N10O13 | 1030.5124 | HRMS, LC-MS/MS, thiol |
[108,163,164] |
| 151 | [Dha7]MC-RY | C51H70N10O13 | 1030.5124 | LC-MS/MS, thiol | [77] |
| 152 | [D-Asp³,Dhb7]MC-RY | C51H70N10O13 | 1030.5124 | LC-MS/MS, thiol | [109] |
| 153 | [D-Asp3]MC-YR | C51H70N10O13 | 1030.5124 | AA, HRMS, MS/MS | [165] |
| 154 | [D-Asp3,(E)-Dhb7]MC-YR | C51H70N10O13 | 1030.5124 | NMR, LC-HRMS/MS | [95] |
| 155 | [DMAdda5]MC-YR | C51H70N10O13 | 1030.5124 | LC–MS/MS, thiol | [131] |
| 156 | MC-LY(OMe) | C53H73N7O14 | 1031.5216 | LC-MS/MS, thiol | [108] |
| 157 | [D-Asp³]MC-(H4)YR | C51H74N10O13 | 1034.5437 | LC–MS/MS, thiol | [131] |
| 158 | [Dha7]MC-(H4)YR | C51H74N10O13 | 1034.5437 | HRMS, NMR, AA | [145] |
| 159 | [DMAdda5]MC-(H4)YR | C51H74N10O13 | 1034.5437 | LC-MS/MS | [137] |
| 160 | MC-YM(O) b | C51H69N7O14S | 1035.4623 | AA, NMR, MS | [166] |
| 161 | [ADMAdda5]MC-HilR | C51H76N10O13 | 1036.5593 | LC-MS/MS | [33] |
| 162 | [ADMAdda5]MC-LHar | C51H76N10O13 | 1036.5593 | HRMS, NMR, AA, MS/MS | [78,79] |
| 163 | MC-AnaR | C52H80N10O12 | 1036.5957 | LC–MS/MS, thiol | [131] |
| 164 | [D-Leu1]MC-LR | C52H80N10O12 | 1036.5957 | NMR, HRMS, MS/MS, AA | [167,168] |
| 165 | [D-Asp³]MC-YY | C54H67N7O14 | 1037.4746 | LC-MS/MS, thiol | [108] |
| 166 | [Gly1,D-Asp3,ADMAdda5,Dhb7]MC-RR | C48H71N13O13 | 1037.5294 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 167 | [Gly1,D-Asp3,ADMAdda5]MC-RR | C49H71N13O13 | 1037.5294 | LC-MS/MS | [92] |
| 168 | MC-RR | C49H75N13O12 | 1037.5658 | NMR, AA, MS | [48] |
| 169 | [(6Z)-Adda5]MC-RR b | C49H75N13O12 | 1037.5658 | NMR, AA, MS | [115,147] |
| 170 | [D-Asp3,D-Glu(OMe)6]MC-RR b | C49H75N13O12 | 1037.5658 | NMR, AA, MS/MS | [169] |
| 171 | [D-Asp3]MC-RHar | C49H75N13O12 | 1037.5658 | LC-HRMS/MS | [170] |
| 172 | [D-Ser1,ADMAdda5]MC-LR | C50H74N10O14 | 1038.5386 | HRMS, MS/MS, AA | [86] |
| 173 | [D-Met1,D-Asp3]MC-LR | C50H76N10O12S | 1040.5365 | LC-MS/MS | [75] |
| 174 | [ADMAdda5,Mser7]MC-LR | C50H76N10O14 | 1040.5542 | HRMS, MS/MS, AA | [86] |
| 175 | [Ser7]MC-RR | C48H75N13O13 | 1041.5607 | AA, HRMS, MS/MS | [84] |
| 176 | [D-Asp³,Mser7]MC-RR | C48H75N13O13 | 1041.5607 | AA, HRMS, MS/MS | [114] |
| 177 | [D-Asp³,Thr7]MC-RR | C48H75N13O13 | 1041.5607 | MS/MS | [171] |
| 178 | [seco-1/6][D-Asp3]MC-RR b | C48H75N13O13 | 1041.5607 | NMR, AA, MS/MS | [169] |
| 179 | MC-HphR | C53H74N10O12 | 1042.5488 | LC-MS/MS, thiol | [34,93] |
| 180 | [D-Glu(OMe)6]MC-FR b | C53H74N10O12 | 1042.5488 | LC-MS/MS | [40] |
| 181 | [D-Leu1]MC-LY | C55H77N7O13 | 1043.5579 | LC-HRMS/MS, LC-MS/MS |
[97,172] |
| 182 | MC-M(O2)R b | C48H72N10O14S | 1044.4950 | LC-MS/MS, thiol, S-oxidation |
[93] |
| 183 | [D-Asp3]MC-HtyR | C52H72N10O13 | 1044.5280 | AA, MS, NMR | [173] |
| 184 | [Dha7]MC-HtyR | C52H72N10O13 | 1044.5280 | AA, HRMS, MS/MS, 1H-NMR |
[161] |
| 185 | [D-Asp3,(E)-Dhb7]MC-HtyR | C52H72N10O13 | 1044.5280 | NMR, AA, HRMS | [142] |
| 186 | [D-Asp3,(Z)-Dhb7]MC-HtyR | C52H72N10O13 | 1044.5280 | NMR, AA, HRMS | [142] |
| 187 | MC-RY | C52H72N10O13 | 1044.5280 | LC-MS/MS, NMR, thiol | [77,163] |
| 188 | MC-YR | C52H72N10O13 | 1044.5280 | AA, NMR, MS | [16] |
| 189 | [Dhb7]MC-YR | C52H72N10O13 | 1044.5280 | LC-HRMS/MS, thiol | [174] |
| 190 | [seco-1/2]MC-FR b | C52H74N10O13 | 1046.5437 | HRMS, MS/MS | [80,154] |
| 191 | MC-(H2)YR | C52H74N10O13 | 1046.5437 | LC-HRMS/MS, thiol | [82] |
| 192 | MC-HphHph | C57H73N7O12 | 1047.5317 | LC-MS/MS | [75] |
| 193 | [D-Asp³,Ser7]MC-HtyR | C51H72N10O14 | 1048.5229 | LC-MS/MS | [33] |
| 194 | [D-Asp³,Mser7]MC-RY | C51H72N10O14 | 1048.5229 | LC-MS/MS, thiol | [108] |
| 195 | [Ser7]MC-YR | C51H72N10O14 | 1048.5229 | LC–MS/MS | [102] |
| 196 | MC-(H4)YR | C52H76N10O13 | 1048.5593 | HRMS, MS/MS, NMR | [80] |
| 197 | [ADMAdda5]MC-HilHar | C52H78N10O13 | 1050.5750 | LC-MS/MS | [33] |
| 198 | [D-Leu1, D-Asp³,ADMAdda5]MC-LR | C52H78N10O13 | 1050.5750 | LC-MS/MS | [153] |
| 199 | [D-Leu1,Adda(O)5]MC-LR b | C52H78N10O13 | 1050.5750 | LC-MS/MS, thiol | [81] |
| 200 | [D-Leu1]MC-HilR | C53H82N10O12 | 1050.6114 | LC-MS/MS | [75] |
| 201 | [D-Leu1]MC-LHar | C53H82N10O12 | 1050.6114 | LC-MS/MS | [75] |
| 202 | [D-Leu1,Glu(OMe)6]MC-LR b | C53H82N10O12 | 1050.6114 | HRMS/MS | [152] |
| 203 | [Hil1]MC-LR | C53H82N10O12 | 1050.6114 | LC-MS/MS, thiol | [81] |
| 204 | [D-Asp³,(E)-Dhb7]MC-HtyY | C55H69N7O14 | 1051.4902 | AA, NMR, HRMS | [175] |
| 205 | MC-YY | C55H69N7O14 | 1051.4902 | NMR, LC-HRMS/MS | [95] |
| 206 | [Gly1,D-Asp3,ADMAdda5]MC-RHar | C49H73N13O13 | 1051.5451 | LC-MS/MS | [92] |
| 207 | [Gly1,D-Asp3,ADMAdda5,Dhb7]MC-RHar | C49H73N13O13 | 1051.5451 | LC-MS/MS, HRMS, AA, thiol |
[91] |
| 208 | [D-Asp³,ADMAdda5]MC-RR | C49H73N13O13 | 1051.5451 | LC-MS/MS | [153] |
| 209 | [D-Asp3,ADMAdda5,Dhb7]MC-RR | C49H73N13O13 | 1051.5451 | NMR, HRMS, AA, LC-MS/MS, thiol |
[150,176] |
| 210 | [D-Glu(OC3H6O)6]MC-LR b | C52H80N10O13 | 1052.5906 | AA, HRMS, NMR | [76] |
| 211 | [D-Leu1,Adda(OH)5]MC-LR b | C52H80N10O13 | 1052.5906 | LC-MS/MS, thiol | [81] |
| 212 | [D-Asp3]MC-WR | C53H71N11O12 | 1053.5284 | AA, NMR, MS | [87] |
| 213 | [Dha7]MC-WR | C53H71N11O12 | 1053.5284 | LC–MS/MS, thiol | [131] |
| 214 | [DMAdda5]MC-WR | C53H71N11O12 | 1053.5284 | LC–MS/MS, thiol | [131] |
| 215 | [D-Asp3]MC-RW | C53H71N11O12 | 1053.5284 | LC-HRMS/MS, thiol, 15N-label | [99] |
| 216 | [D-Met1]MC-LR | C51H78N10O12S | 1054.5521 | LC-MS/MS | [75] |
| 217 | [D-Leu1]MC-MR | C51H78N10O12S | 1054.5521 | LC-HRMS/MS, thiol, S-oxidation |
[81] |
| 218 | [D-Leu1,Mser7]MC-LR | C52H82N10O13 | 1054.6063 | HRMS/MS | [152] |
| 219 | [Mser7]MC-RR | C49H77N13O13 | 1055.5764 | LC-HRMS/MS, thiol | [82] |
| 220 | [ADMAdda5]MC-FR | C53H72N10O13 | 1056.5280 | LC-MS/MS | [137] |
| 221 | [D-Asp³,ADMAdda5]MC-HphR | C53H72N10O13 | 1056.5280 | LC-MS/MS | [153] |
| 222 | MC-HtyR | C53H74N10O13 | 1058.5437 | AA, MS, NMR | [173] |
| 223 | [D-Asp³,D-Glu(OMe)6]MC-HtyR b | C53H74N10O13 | 1058.5437 | MS/MS | [88] |
| 224 | [D-Glu(OMe)6]MC-YR b | C53H74N10O13 | 1058.5437 | NMR, LC-HRMS/MS | [95] |
| 225 | [D-Ser1,D-Asp³]MC-HtyR | C52H72N10O14 | 1060.5229 | LC-MS/MS, thiol | [93] |
| 226 | [D-Asp³]MC-Y(OMe)R | C52H72N10O14 | 1060.5229 | LC–MS/MS, thiol | [131] |
| 227 | [DMAdda5]MC-Y(OMe)R | C52H72N10O14 | 1060.5229 | LC–MS/MS, thiol | [131] |
| 228 | [seco-4/5][D-Asp³]MC-HtyR b | C52H74N10O14 | 1062.5386 | LC-MS/MS, thiol | [93] |
| 229 | [Ser7]MC-HtyR | C52H74N10O14 | 1062.5386 | AA, HRMS, MS/MS, NMR | [161] |
| 230 | [D-Asp³,Mser7]MC-HtyR | C52H74N10O14 | 1062.5386 | MALDI-TOF MS | [88] |
| 231 | [D-Asp³,ADMAdda5]MC-(H4)YR | C52H74N10O14 | 1062.5386 | LC-MS/MS | [153] |
| 232 | [Mser7]MC-RY | C52H74N10O14 | 1062.5386 | LC-MS/MS, thiol | [77] |
| 233 | [D-Asp³,Mser7]MC-YHar | C52H74N10O14 | 1062.5386 | NMR, LC-HRMS/MS | [95] |
| 234 | [Mser7]MC-YR | C52H74N10O14 | 1062.5386 | LC–MS/MS, NMR, thiol, HRMS | [131] |
| 235 | MC-HphHty | C57H73N7O13 | 1063.5266 | LC-MS/MS | [75] |
| 236 | [D-Leu1,ADMAdda5]MC-LR | C53H80N10O13 | 1064.5906 | LC –MS/MS | [153] |
| 237 | [D-Leu1,Glu(OMe)6]MC-HilR b | C54H84N10O12 | 1064.6270 | HRMS/MS | [152] |
| 238 | [D-Asp³,(E)-Dhb7]MC-HtyHty | C56H71N7O14 | 1065.5059 | AA, NMR, HRMS | [175] |
| 239 | [ADMAdda5]MC-RR | C50H75N13O13 | 1065.5607 | LC-MS/MS, LC-HRMS/MS |
[104,153] |
| 240 | MC-HarHar | C51H79N13O12 | 1065.5971 | LC–MS/MS | [177] |
| 241 | [D-Leu1,D-Asp3]MC-RR | C51H79N13O12 | 1065.5971 | LC-MS/MS | [75] |
| 242 | MC-WR | C54H73N11O12 | 1067.5440 | AA, HRMS, NMR | [76] |
| 243 | [D-Leu1,D-Glu(OMe)6,Mser7]MC-LR b | C53H84N10O13 | 1068.6219 | HRMS/MS | [152] |
| 244 | [D-Met(O)1]MC-LR b | C51H78N10O13S | 1070.5471 | HRMS/MS | [152] |
| 245 | [D-Leu1]MC-M(O)R b | C51H78N10O13S | 1070.5471 | LC-HRMS/MS, thiol, S-oxidation |
[81,97] |
| 246 | [D-Leu1,D-Asp³]MC-HphR | C55H78N10O12 | 1070.5801 | LC-MS/MS | [153] |
| 247 | [D-Phe1]MC-LR | C55H78N10O12 | 1070.5801 | LC-MS/MS | [75] |
| 248 | [D-Leu1]MC-FR | C55H78N10O12 | 1070.5801 | LC-MS/MS, thiol | [81] |
| 249 | MC-KynR b | C53H73N11O13 | 1071.5389 | LC-MS/MS, thiol | [90] |
| 250 | [D-Asp3,ADMAdda5]MC-HtyR | C53H72N10O14 | 1072.5229 | AA, HRMS, MS/MS | [178] |
| 251 | [D-Asp3,ADMAdda5,Dhb7]MC-HtyR | C53H72N10O14 | 1072.5229 | NMR, HRMS, AA | [150] |
| 252 | [ADMAdda5]MC-YR | C53H72N10O14 | 1072.5229 | LC-MS/MS | [33] |
| 253 | MC-MhtyR | C54H76N10O13 | 1072.5593 | LC-HRMS/MS, thiol, esterification | [96] |
| 254 | [D-Asp3,(E)-Dhb7]MC-HtyW | C57H70N8O13 | 1074.5062 | NMR, LC-HRMS/MS | [95] |
| 255 | MC-RY(OMe) | C53H74N10O14 | 1074.5386 | LC-MS/MS, thiol | [77] |
| 256 | [D-Asp³]MC-Hty(OMe)R | C53H74N10O14 | 1074.5386 | LC-MS/MS, thiol | [93] |
| 257 | MC-Y(OMe)R | C53H74N10O14 | 1074.5386 | LC–MS/MS, thiol | [131] |
| 258 | [seco-4/5]MC-HtyR b | C53H76N10O14 | 1076.5542 | LC-MS/MS, thiol | [93] |
| 259 | [ADMAdda5]MC-(H4)YR | C53H76N10O14 | 1076.5542 | LC-MS/MS | [137] |
| 260 | [Mser7]MC-HtyR | C53H76N10O14 | 1076.5542 | LC-MS/MS, thiol | [93] |
| 261 | [D-Leu1,ADMAdda5]MC-LHar | C54H82N10O13 | 1078.6063 | LC-MS/MS | [153] |
| 262 | [ADMAdda5]MC-RHar | C51H77N13O13 | 1079.5764 | LC-MS/MS | [153] |
| 263 | [D-Leu1]MC-RR | C52H81N13O12 | 1079.6128 | LC-MS/MS | [75] |
| 264 | MC-OiaR b | C54H73N11O13 | 1083.5389 | LC-MS/MS, thiol | [90] |
| 265 | [D-Met(O)1,Glu(OMe)6]MC-LR b | C52H80N10O13S | 1084.5627 | HRMS/MS | [152] |
| 266 | [D-Leu1]MC-HphR | C56H80N10O12 | 1084.5957 | LC-MS/MS | [75] |
| 267 | [D-Leu1]MC-M(O2)R b | C51H78N10O14S | 1086.5420 | LC-HRMS/MS, thiol, S-oxidation |
[81] |
| 268 | [ADMAdda5]MC-(H4)YHar | C54H78N10O14 | 1090.5699 | LC-MS/MS | [137] |
| 269 | MC-NfkR b | C54H73N11O14 | 1099.5338 | LC-MS/MS, thiol | [90] |
| 270 | [D-Leu1]MC-HtyR | C56H80N10O13 | 1100.5906 | LC-MS/MS | [153] |
| 271 | [D-Leu1,Ser7]MC-HtyR | C55H80N10O14 | 1104.5855 | LC-HRMS/MS | [170] |
| 272 | MC-LR Cys conjugate b | C52H81N11O14S | 1115.5685 | HRMS, MS/MS, semisynthesis | [80] |
| 273 | [D-Leu1]MC-LR Cys conjugate b | C55H87N11O14S | 1157.6155 | LC-HRMS/MS, semisynthesis | [81] |
| 274 | [D-Leu1]MC-LR Cys sulfoxide conjugate b | C55H87N11O15S | 1173.6104 | LC-HRMS/MS, semisynthesis, S-oxidation |
[81] |
| 275 | [D-Leu1]MC-LR γ-GluCys conjugate b | C60H94N12O17S | 1286.6581 | LC-HRMS/MS, semisynthesis | [81] |
| 276 | [D-Leu1]MC-LR γ-GluCys sulfoxide conjugate b | C60H94N12O18S | 1302.6530 | LC-HRMS/MS, semisynthesis, S-oxidation |
[81] |
| 277 | [D-Asp³]MC-RRGSH conjugate b | C58H90N16O18S | 1330.6340 | LC-HRMS/MS, thiol, 15N-label, semisynthesis | [99] |
| 278 | [D-Leu1]MC-LR GSH conjugate b | C62H97N13O18S | 1343.6795 | LC-HRMS/MS, semisynthesis | [81] |
| 279 | [D-Leu1]MC-LR GSH sulfoxide conjugate b |
C62H97N13O19S | 1359.6744 | LC-HRMS/MS, semisynthesis, S-oxidation |
[81] |
a Exact monoisotopic neutral mass. For protonated/deprotonated ions, m/z = (monoisotopic neutral mass +/− (z × 1.0073))/z. b Congeners expected to form through chemical or biochemical transformations of other MCs rather than through biosynthesis by cyanobacteria, see Section 5.7 for additional details. c Entries 123 and 124 are a pair of congeners demethylated at different positions somewhere between C-2 and C-8 of their Adda moieties. Characterization Techniques. AA, amino acid analysis; MS, mass spectrometry (low resolution MS analysis of isolated MCs implied); NMR, nuclear magnetic resonance(including 1H, 13C or 2-dimensional experiments); MS/MS, tandem mass spectrometry (direct analysis of isolated fractions using either ESI or MALDI ionization); HRMS, high resolution mass spectrometry (direct analysis of isolated fractions); LC-MS/MS, liquid chromatography-tandem mass spectrometry (without isolation); LC-HRMS/MS, liquid chromatography-high resolution tandem mass spectrometry (without isolation); thiol, reactivity with a thiol-containing reagent (e.g., mercaptoethanol) to confirm presence of an α,β-unsaturated amide such as in Mdha7; S-oxidation, selective oxidation with mild oxidant (e.g., periodate) to confirm presence of methionine residues and thiol conjugates as well as their corresponding sulfones and sulfoxides; semisynthesis, chemical conversion of a known congener to the unknown one (e.g., by derivatization with GSH, or oxidation of Trp residues); esterification, derivatization of carboxylic acid groups as esters or phenolic groups as ethers; 15N-labelling, culturing in 15N-labelled medium to count N-atoms.
5.1. Congeners with Variable Amino Acid at Position 1
In the first ever report of the structural elucidation of a MC, MC-LA, produced by the South African M. aeruginosa strain WR70 (UV-0l0), the amino acid residue in position 1 (Figure 1) was found to be D-Ala [15,17,18,20]. This amino acid is incorporated by the NRPS module of mcyA [23] and was initially considered as relatively conserved in MCs [6,94]. However, subsequent characterization of new congeners has shown that the amino acid residue in position 1 is also variable, although not as diverse as those in positions 2 and 4. D-Ala is found in position 1 in 219 of the 279 reported MCs (Table 1). However, other amino acid residues reported in position 1 include D-Ser, D-Leu, Gly, D-Met, D-Phe, D-Val, D-Hil, and D-Met S-oxide. Among these congeners, the first one reported was [D-Ser1,ADMAdda5]MC-LR isolated from Nostoc sp. strain 152 by Sivonen et al. [86]. Subsequentlty, [D-Leu1]MC-LR was identified as the most abundant MC from a laboratory strain RST 9501 of a Microcystis sp. isolated from a hepatotoxic Microcystis bloom from brackish waters in the Patos Lagoon estuary, southern Brazil [167], and from a cyanobacterial bloom dominated by M. aeruginosa collected in Pakowki Lake, AB, Canada [168]. A decade later, Shishido et al. [75] identified [D-Met1]MC-LR and [D-Met1,D-Asp3]MC-LR from M. aeruginosa strains NPLJ-4 and RST 9501. Furthermore, the authors reported that the M. aeruginosa strain NPLJ-4 produced a larger diversity of MC variants, but most of them in trace amounts, such as [D-Val1]MC-LR and [D-Phe1]MC-LR. Recently, Puddick et al. [91] described eight new congeners with Gly at position-1 in two hydro-terrestrial cyanobacterial mat samples from the McMurdo Dry Valleys of Eastern Antarctica. Recently, Foss et al. [81] identified a new congener by LC-MS/MS with homoisoleucine (D-Hil) at position 1, from a Microcystis bloom at Poplar Island, MD, USA.
5.2. Congeners with Variable Amino Acid at Positions 2 and 4
The two L-amino acids in positions 2 and 4 are the most variable of the seven amino acids in MCs. The adenylation domains of mcyB and mcyC are responsible for the activation of the amino acids that appear in positions 2 and 4, respectively [23,54,55,56]. Therefore, the variable L-amino acids incorporated into MCs are dependent upon the substrate specificity of the adenylation domain responsible for loading the amino acid onto the MC synthetase module [32,179]. To date, 21 different amino acids have been identified at position 2 and 42 at position 4 (Table 1). Leu is present in position 2 in about one third of reported MCs (95 of the 279) and Arg is present in position 4 in more than half of the reported MCs (173 of the 279). For example, Puddick et al. [12], when conducting an analysis of the relevant literature on 49 isolated strains of cyanobacteria, reported that most of the strains assessed were only able to incorporate one or two amino acids into position 4, usually including Arg. Consequently, the congener MC-LR is the most commonly observed MC in most cyanobacterial blooms worldwide [6,7,8,9,180]. However, in many other cases, different congeners with differing residues at positions 2 and 4 can dominate a bloom and methods targeting MC-LR exclusively are known to significantly underestimate total MC content in many cases.
5.3. Congeners with Variable Amino Acid at Position 3
Position 3 is highly conserved in MCs, as only D-erythro-β-methyl isoaspartic acid (D-Masp3) and D-erythro-isoaspartic acid (D-Asp3) have been reported in MCs. D-Masp is most commonly reported (157 of 279 MCs), with D-Asp reported in all remaining MCs (Table 1). The amino acid at position-3 is incorporated by the activity of the NRPS module of mcyB [23]. Furthermore, Pearson et al. [181] and Sielaff et al. [182] reported that one of the moieties, D-erythro-β-methyl-aspartic acid, required the activity of two additional enzymes prior to incorporation in the peptide chain: the 2-hydroxy acid dehydrogenase mcyI and the aspartate racemase mcyF. Both dicarboxylic acids, D-Masp/Asp at position 3 and D-Glu in position 6, are inserted into the cyclic peptide structure in the iso-configuration [15,16].
5.4. Congeners with Variable Amino Acid at Position 5
The most characteristic twenty-carbon β-amino acid in position 5 of MCs, and which plays an important role in their biological activity, is Adda (reported in 220 of 279 MCs). Four PKS modules, mcyG, J, D, and E are responsible for the synthesis of the Adda moiety of MCs [24]. The natural structural diversity of the Adda moiety is largely limited to the group on the C-9 oxygen, that can be present as 9-O-desmethylAdda (DMAdda), the 9-hydroxy derivative of Adda, or as 9-acetoxy-9-O-desmethylAdda (ADMAdda). The O-methylation on the C-9 oxygen of MCs is carried out by the mcyJ O-methyltransferase [55]. Recently, Fewer et al. [183] reported that the production of O-acylated variants of MC in Nostoc is the result of specific enzymatic O-acetylation of the MC catalyzed by the mcyL O-acetyltransferase. In fact, several earlier studies reported that O-acylated MCs are produced almost exclusively by strains of the genus Nostoc isolated from a range of aquatic and terrestrial habitats in free-living and lichen-associated growth states [79,86,137,153]. However, O-acetylated MCs have also been reported in a single strain of Planktothrix agardhii PH123 [178].
The first DMAdda microcystin congener, [DMAdda5]MC-LR, was identified from a cyanobacterial bloom containing M. aeruginosa (dominant), M. viridis and M. wesenbergii collected from Homer Lake, IL, USA [76,80]. Furthermore, 15 different DMAdda MC congeners have been described (Table 1), that could originate from either incomplete methylation by the mcyJ gene during biosynthesis or from hydrolysis of the 9-methoxy or 9-acetoxy group in the [Adda5] and [ADMAdda5] MC variants, respectively. However, since most reports of DMAdda5-microcystins have occurred in strains or blooms where the corresponding ADMAdda-congeners were not present, and ethers are relatively resistant to hydrolysis, the origin of DMAdda5-microcystins is likely to be due primarily to incomplete methylation during biosynthesis. LC-MS studies recently indicated the existence of small amounts of incompletely characterized MC congeners containing oxidized Adda (hydroxyAdda and ketoAdda) and Adda demethylated (dmAdda) at two positions other than at the 9-O-methyl group [81]. Furthermore, [epoxyAdda5]MC-LR was tentatively identified in a field sample [81]. These oxidized congeners are likely to arise through metabolism or autoxidation, while the two dmAdda variants presumably arise through biosynthesis.
5.5. Congeners With Variable Amino Acid at Position 6
D-Glu is incorporated in position 6 by the activity of the NRPS module adjacent to the PKS module of mcyE [23] and is highly conserved in this position. The esterification of the α-carboxyl group of the D-Glu residue is rare, and only 11 such MC congeners have been characterized in the literature (Table 1). Namikoshi et al. [76,80] characterized the only non-methyl (-C3H6OH) monoester of the α-carboxyl on the D-Glu6 unit of MC-LR ([D-Glu(OC2H3(CH3)OH)6]MC-LR) from a toxic bloom containing M. aeruginosa (dominant), M. viridis, and M. wesenbergii collected from Homer Lake (Illinois) in 1988. However, the mono-methyl ester derivatives at the D-Glu6 unit [D-Glu(OMe)] were more frequent and 11 different congeners have been characterized (Table 1). These methyl esters appear to be artefacts produced by exposure to methanol (Section 5.7).
5.6. Congeners With Variable Amino Acid at Position 7
The amino acid residue in position 7 is also variable; however, it is not as diverse as those in positions 2 and 4. N-methyldehydroalanine (Mdha) is found in position 7 in 189 of the 279 reported MCs (Table 1). However, other amino acids have been reported in this position, including dehydroalanine (Dha) [33,83,108,109,138,161], dehydrobutyrine (Dhb) [75,77,79,91,142], N-methyldehydrobutyrine (Mdhb) [78], Ser [33,84,102,138,152,161], N-methylserine (Mser) [80,82,86,88,93,108,109], Thr [171], and conjugates formed by addition of Cys, γ-GluCys and glutathione (GSH) with the reactive double bond of Mdha7 [80,81]. Chemical and metabolic transformations of MCs are discussed in Section 5.7.
The N-terminal adenylation domain of mcyA catalyzes the N-methylation of MCs [21,23,55,56]. The N-methyltransferase (NMT) domain recognizes and activates Ser as an AMP-adenylate, causing the Ser residue to undergo N-methylation and dehydration to form Mdha [21,23,34,55,56]. Thus, the NMT domain catalyzes the transfer of the methyl group from S-adenosylmethionine (SAM) to the α-amino group of the thioesterified amino acid [184]. However, Fewer et al. [34] reported that the order and timing of the N-methylation, dehydration, and condensation reactions have not yet been determined. For example, deletion of the entire NMT domain of mcyA in Anabaena [34] and Planktothrix [185] or point mutations in this gene in Microcystis [37], were associated with the production of Dha7-microcystins by these genera. Therefore, simultaneous production of Mdha- and Dha-containing MCs in Anabaena could be the result of the inactivation of the NMT domain during the assembly of Dha-containing MCs [34]. However, Marahiel et al. [186] suggested that the concomitant biosynthesis of Dha- and Mdha-containing MC variants could also be the result of the limitation of SAM, the source of the N-methyl group. Such mixtures of MCs containing both Mdha and Dha are common in Planktothrix [114] and Microcystis [139], and partial inactivation of the NMT domain may be a widespread feature of MC biosynthesis.
MCs-containing Dhb instead of Dha in Planktothrix are thought to be the result of a homologous recombination event in the first module of mcyA that replaces the adenylation domain, leading to a change in substrate specificity (from Ser to Thr) and presumably the concomitant deletion of the NMT domain [185]. For the biosynthesis of nodularin, a microcystin-like pentapeptide containing Mdhb instead of Mdha, the nodularin synthetase gene cluster is proposed to have arisen by deletion of two NRPS modules and a change in substrate specificity from Ser to Thr in the C-terminal adenylation domain of NdaA [187]. Kurmayer et al. [185] reported a significant correlation between mcyAAd1 genotypes with NMT and mcyAAd1 genotypes without NMT and the occurrence of either Mdha or Dhb in position 7 of the MC structures in Planktothrix strains. Therefore, strains of cyanobacteria producing MC variants containing either Dha or Dhb, exclusively, are known from all major MC producing genera (Microcystis, Nostoc, Anabaena, and Planktothrix) [6,33,91,109,138,144]. Furthermore, Sano and Kaya [142] reported that the configuration of the Dhb7 unit of Dhb-microcystins isolated from the CCAP strains of O. agardhii was E, while the Dhb of nodularins from Nodularia spumigena has been determined as Z [188], suggesting that the Dhb unit in Dhb-microcystins is biosynthesized via a dehydration of Thr or allo-threonine.
The MC congener [D-Asp3,Thr7]MC-RR was identified from an environmental bloom of P. rubescens collected from Austrian alpine lakes [171]. Additionally, 11 MC analogues containing Ser and 13 containing Mser instead of Mdha in position 7 have been reported (Table 1). According to Tillett et al. [23], the module mcyA is responsible for the incorporation of Ser into the growing molecule. Ser is further N-methylated by the corresponding NMT domain and transformed by dehydration into Mdha prior to or following the condensation reaction. Analogous to Ser, Thr is transformed by dehydration into Dhb [189]. Therefore, the production of the unique congener with Thr residue and the congeners with Ser or Mser residues in position 7 could be the result of the partial inactivation of the NMT domain during the dehydration and N-methylation reactions of Thr- and Ser-containing MCs. In fact, Fewer et al. [34] reported that there is a 10–500-fold increase in the amount of Ser incorporated into the MCs in strains of Anabaena lacking a functional NMT domain, suggesting that there may be a functional coupling of the dehydration and N-methylation reactions.
5.7. Chemical and Biochemical Transformations of Microcystins
Several chemical and biochemical reactions have been reported to occur in the cell, the environment, or during sample extraction and storage in the laboratory that lead to the formation of new MC analogues beyond those that are biosynthetic products of cyanobacteria (Figure 2). Even though these are not of biosynthetic origin, their occurrence can still be important to measure for studies of MC occurrence and toxicity. It is recommended that authors describing the identification of new MC variants try to clearly differentiate between those that are metabolites of cyanobacteria, those that form through conjugation or metabolism after biosynthesis, and those that form artefactually during sample handling or storage.
Figure 2.
Examples of chemical reactions of MCs demonstrated using the hypothetical congener [ADMdda5]MC-RM. The numbers in circles indicate the amino acid residue-number.
Eight MCs containing sulfur oxidation in their Met moiety have been reported (Table 1) that are likely artefacts produced by autoxidation. This is consistent with the findings of Miles et al. [93] who reported that a fresh culture of Dolichospermum flos-aquae contained only traces of sulfoxides [D-Asp3]MC-M(O)R and MC-M(O)R, but that these increased markedly during storage or sample extraction and preparation. This suggests that MCs containing methionine sulfoxide are primarily post-extraction oxidation artefacts, rather than being produced by biosynthesis in cyanobacteria. These can then be further oxidized to the corresponding sulfones (e.g., MC-M(O2)R) through an additional step of autoxidation [93]. In addition, Puddick et al. [90] reported MC congeners containing the oxidized tryptophan moieties kynurenine (Kyn), oxindolylalanine (Oia), and N-formylkynurenine (Nfk) that are also considered as autoxidation artefacts.
Isomerization of the C-6 double bond in the Adda5 group is rarely reported and is thought to be due to ultraviolet (UV) irradiation rather than arising through biosynthesis [190,191,192]. In early studies, Harada et al. [115] and Namikoshi et al. [76] isolated geometrical isomers at C-6 of [(6Z)-Adda5]MC-LR and -RR, as minor components with along with the corresponding normal (6E)-Adda5 MCs (MC-LR and -RR) from Microcystis spp. The DMAdda derivative at the Adda unit could be also an artefact produced during extraction of cyanobacterial samples dominated by ADMAdda derivatives. In fact, Beattie et al. [150] observed that [ADMAdda5]MCs are unstable and hydrolyze at room temperature in aqueous solution and/or in methanol, and that [D-Asp3,ADMAdda5,Dhb7]MC-RR isolated from a Nostoc strain DUN901 was converted to [D-Asp3,DMAdda5,Dhb7]MC-RR within one week. Similarly, Ballot et al. [131] reported the half-life for hydrolysis of several [ADMadda5]MCs (to form the corresponding [DMadda5]MCs) to be ca 30 h at pH 9.7 and 30 °C.
In addition, MCs containing methyl esters in their Glu6 moiety are almost certainly artefacts produced by esterification in methanolic solutions, possibly due to traces of residual acid from extraction or isolation steps. For example, [D-Glu(OMe)6]MC-LR and [D-Glu(OMe)6]MC-FR characterized recently by Bouhaddada et al. [40] were identified in the presence of their much more abundant non-esterified congeners MC-LR and MC-FR, respectively. In addition, the same results were reported for the eight other described congeners with methyl esters on the α-carboxyl group of the D-Glu residue in position 6 [76,80,85,95,152,169]. Harada et al. [112] demonstrated formation of Glu6 methyl esters of MCs in acidic methanolic solutions, and Foss et al. [81] observed substantial esterification of [Leu1]MC-LR that was purified with an acidic eluent then evaporated to dryness before storage in MeOH at −20 °C. In fact, del Campo and Ouahid [38] identified the presence of [D-Asp3,D-Glu(OMe)6]MC-LAba from a cultured Microcystis strain UTEX-2666, but not its non-esterified residue [D-Asp3]MC-LAba, suggesting that near-complete esterification is possible under some conditions. Therefore, the technique described by Fastner et al. [193] consisting of analyzing of a single colony of Microcystis directly, without methanol extraction may circumvent this probem. As a precaution, use of both acids and methanol during sample preparation or storage should be avoided where possible. Esters of MCs containing L-Glu at position-2 and -4 have also been reported [138] and given the facile esterification of D-Glu6 it seems probable that these two are esterification artefacts as well.
MCs with either Dha or Mdha at position 7 possess an α,β-unsaturated carbonyl group that is reactive toward thiols (e.g., cysteine), and to a lesser extent hydroxyl groups, through the Michael addition reaction [100,101]. Consequently, the unusual MC congener [Mlan7]MC-LR that was characterized from a bloom of Microcystis spp. collected from Homer Lake (IL, USA) can be considered the result of a chemical reaction between the major constituent (MC-LR) biosynthesized in the cyanobacterium and cysteine [80]. On the other hand, the biosynthesis of lanthionine has been proposed to involve addition of the thiol group of a Cys residue to the double bond of Dha, generated by dehydration from Ser, forming a sulfide bridge [80]. It has also been proposed that glutathione-S-transferase catalyzes the addition of glutathione (GSH) to MCs, but also that the reaction proceeds at a similar rate with or without the enzyme under physiological conditions [101,194], suggesting that the reaction may be primarily chemical rather than purely biochemical. Recently, 3 new MCs containing cysteine (Cys), glutathione (GSH), and γ-glutamylcysteine (γ-GluCys) conjugated to the Mdha7 olefinic bond of [D-Leu1]MC-LR were identified in extracts of freeze-dried Microcystis bloom material from Poplar Island (MD, USA) [81]. Small amounts of the corresponding sulfoxides of the conjugates (Cys(O), cysteine sulfoxide; GS(O), glutathione sulfoxide; and γ-GluCys(O), γ-glutamylcysteine sulfoxide), probably produced by autoxidation, were also detected. Conjugated MCs present in the bloom material were characterized for the first time, but they could nonetheless be metabolites produced during detoxification of MC congeners in aquatic organisms. The principal pathway of the metabolism and the excretion of MCs in mammals and aquatic organisms has been found to be as MC–GSH and MC–Cys conjugates [8,195,196,197]. However, cyanobacteria are known to produce both Cys and GSH [198], and low levels of GSH conjugates of MCs have been detected in several cyanobacterial cultures using untargeted LC-HRMS methods (D. G. Beach and C. O. Miles, unpublished observations; [99]). GSH conjugates can be further metabolized to γ-GluCys and Cys conjugates [199], so it is possible that the γ-GluCys and Cys conjugates observed from cyanobacteria are produced via this route or by direct conjugation of MCs to these smaller peptides. Nucleophilic addition of water (to produce D,L-Ser7 and -Mser7 congeners) and methanol (to produce the corresponding O-methoxyserine congeners), and intramolecular addition of the 2-arginyl group, have all also been observed at the reactive double bond of Dha7 and Mdha7 of MCs under basic conditions during studies of their derivatization with thiols [101], as shown in Figure 2.
Eight linear MCs (Table 1) have been reported to date that could well be the products of bacterial degradation of MCs, since many studies have shown that the first step of biodegradation of MCs by multiple different species of bacteria is through the cleavage and opening of the macrocycle (reviewed by Li et al. [200]).
MC congeners containing oxidized Adda (an epoxide, ketone, and hydroxylated derivative) have also been reported recently in small amounts in a bloom, and partially characterized by LC-HRMS/MS and chemical derivatizations [81]. At this stage it is unclear whether these are the result of enzyme-catalyzed oxidation or are products of autoxidation.
Researchers should consider the detection in a sample of a potentially artefactual MC congener, such as an oxidized, esterified, or conjugated variant, as possible evidence for the presence in the original sample of its unmodified precursor MC.
6. Toxicity of Microcystin Congeners
6.1. Toxicity In Vivo
It should be borne in mind when comparing LD50 values (the dose of toxin that kills 50% of exposed animals) that toxicological measurements are inherently subject to uncertainty caused by biological variability, and the possibility that in some cases isolated MCs may not have been adequately characterized with respect to purity and amount of toxin prior to toxicity assessment. For this reason, only large differences in toxicity should be considered significant when comparing toxicities from different experiments and different MC congeners.
The toxic effects of MCs on animals were first studied with unpurified cyanobacterial extracts. Several studies reported enlargement and congestion of the liver, with necrosis of the hepatic cells in laboratory and domestic animals after oral administration or parenteral injection of toxic algal extracts in which M. aeruginosa was the predominant species [44,201,202]. A decade later, Elleman et al. [166] confirmed these observations and reported that the lethal dose (LD100) in mice by intraperitoneal (i.p.) injection of an M. aeruginosa (then Anacystis cyanea) bloom extract was 15–30 mg of lyophilized algal bloom per kilogram body weight (b.w.). The LD50 of the purified toxin from this extract was estimated to be 56 µg kg−1 in mice, and the approximate LD100 was 70 µg kg−1. Acute toxicity by parenteral administration of the purified toxin to mice produced extensive liver lobular hemorrhage and death within 1–3 h. However, repeated administration of sublethal doses daily over some weeks produced progressive hepatocyte degeneration and necrosis and the development of fine hepatic fibrosis.
Since then and following the identification of the toxin responsible, toxicological research has focused on purified MCs. The most extensive toxicological information is available for MC-LR. The acute toxicity of MC-LR has been tested in several studies on mice, with i.p. LD50 values ranging from 25 to 150 µg kg−1 b.w. [203,204]. An LD50 value of 50 µg kg−1 b.w. is generally accepted (Table 2) (for a review, see [205]). The inhalation toxicity (intratracheal and intranasal) in mice is similar, with an LD50 of 36 to 122 µg kg−1 b.w. [206,207,208,209]. However, MC-LR is 100 times less toxic by oral ingestion than it is by intra-peritoneal injection, with an LD50 of 5000 µg kg−1 b.w. [203,210]. Although some studies indicate that MCs are stable to proteolytic enzymes in vitro [211], Freitas et al. [212] suggested that a decrease in the bioaccessibility and thus bioavailability of free MC-LR after proteolytic digestion of contaminated shellfish by the pancreatic enzymes, trypsin and chymotrypsin, may be an important factor that can decrease its toxicity by oral route. Although the toxicity of all characterized MC congeners has not been determined, there are some variations within the MC family with i.p. LD50 values ranging from 50 to >1200 µg kg−1 b.w. in mice (see Table 2). Firstly, the cyclic structure of MCs is very important for the toxicity and opening of the macrocycle at any position makes the molecule much less toxic. For example, the two congeners [seco-1/2]MC-LR and [seco-4/5]MC-LR, corresponding to the opening of the cyclic structure of the congener MC-LR between the amino acid residues at positions 1/2 and 4/5, respectively, showed no toxicity to mice at 22,500 and 1100 µg kg−1 b.w., respectively [80]. Secondly, two of the most conserved residues, at position 5 (Adda, present in 220 of 279 identified MC congeners) and position 6 (D-Glu, present in 267 of 279 identified MC congeners) are also very important for the toxicological potential. Adda group stereochemistry and its double bond configuration (6E) are crucial for the toxicity since [(6Z)-Adda5]MC-LR showed an LD50 in mice >1200 µg kg−1 b.w. [115,147]. However, the toxicity appears to be only slightly affected by 9-O-demethylation (DMAdda) or 9-O-acetylation (ADMAdda) of the Adda group. [DMAdda5]MC-LR and [ADMAdda5]MC-LR were found to have LD50 values of 90–100 and 60 µg kg−1 b.w., respectively, by i.p. injection in mice. The free carboxylic acid on the D-Glu6 residue seems to be essential for toxicity because the esters ([D-Glu(OC3H7O)6]MC-LR [76] and [D-Glu(OMe)6]MC-LR) [85,140,189] of the MC-LR showed no toxicity in the i.p. mouse bioassay at 1000 µg kg−1 b.w. These results are consistent with the results of studies with synthetic fragments containing elements of the MC structure, where the presence of Adda and a suitably placed carboxylic acid group mimicking the carboxyl group of Glu6 was enough to elicit PP inhibition, albeit at potencies far below that of MC-LR [213,214]. However, substitution of amino acids in positions 1, 3, and 7 only had a small impact on the toxic potential of MCs in vivo. For example, MC toxicity seems to be barely affected by substitution of the moderately conserved D-Ala in position 1 by other D-amino acids. Matthiensen et al. [167] and Park et al. [168] reported that the LD50 of [D-Leu1]MC-LR by i.p injection in mice was 100 µg kg-1 b.w. Likewise, MC toxicity was only slightly affected by demethylation of the amino acid at position 3 (D-Asp3 instead of D-Masp3) or by demethylation or amino acid substitution at position 7. For example, demethylated forms of MCs containing D-Asp3 or Dha7 residues are two to five times less or more toxic, respectively, compared to their methylated forms [83,139,215,216]. Similarly, Sano and Kaya [142] reported that the toxicity is slightly affected by the addition of a methyl group to the double bond of the Dha7 unit to form the Dhb7, whereas, conjugation of the Mdha7 residue with Cys residue (to give [L-Mlan7] instead of Mdha7)) decreased the toxicity 20-fold compared to MC-LR by i.p. injection in mice [80]. Nevertheless, the acute toxicity is significantly affected by substitution at the two most variable amino acid residues in positions 2 and 4. Most of the structural MC congeners containing a hydrophobic L-amino acid in position 2 or 4, such as MC-LA, MC-YR, MC-YA, and MC-YM(O), have similar LD50 values to MC-LR (Table 2), whereas the LD50 of the hydrophilic MC-RR is about ten times higher (Table 2).
Table 2.
Lethal dose (LD50) values (µg kg-1 b.w., i.p. mouse) of MC congeners.
| Entry a | Microcystin | LD50 | Reference |
|---|---|---|---|
| 7 | MC-LA | 50 | [226] |
| 18 | MC-AR | 250 | [76] |
| 22 | MC-YA | 60–70 | [227] |
| 25 | [D-Asp3,Dha7]MC-LR | 160–300 | [83] |
| 39 | [D-Asp3]MC-LR | 200–500 | [139,215] |
| 40 | [D-Asp3,(E)-Dhb7]MC-LR | 70 | [142] |
| 42 | [Dha7]MC-LR | 250 | [83] |
| 43 | [DMAdda5]MC-LR | 90–100 | [76,149] |
| 66 | MC-LR | 50 | [205] |
| 68 | [(6Z)-Adda5]MC-LR | >1200 | [115,147] |
| 83 | MC-LY | 90 | [206] |
| 89 | [D-Asp3,ADMAdda5]MC-LR | 160 | [78,79] |
| 92 | MC-HilR | 100 | [80] |
| 94 | [D-Glu(OMe)6]MC-LR | >1000 | [85,140,189] |
| 105 | [Mser7]MC-LR | 150 | [80] |
| 111 | [D-Asp3]MC-FR | 90 ± 10 | [216] |
| 121 | [ADMAdda5]MC-LR | 60 | [78,79] |
| 129 | [D-Asp3]MC-RR | 350 ± 10 | [216] |
| 130 | [Dha7]MC-RR | 180 | [157] |
| 131 | [D-Asp3,(E)-Dhb7]MC-RR | 250 | [142] |
| 141 | MC-M(O)R | 700–800 | [76] |
| 142 | MC-FR | 250 | [76] |
| 160 | MC-YM(O) | 56 | [166] |
| 162 | [ADMAdda5]MC-LHar | 60 | [78,79] |
| 164 | [D-Leu1]MC-LR | 100 | [167,168] |
| 168 | MC-RR | 500–800 | [48,76] |
| 169 | [(6Z)-Adda5]MC-RR | >1200 | [115,147] |
| 183 | [D-Asp3]MC-HtyR | 80–100 | [147] |
| 185 | [D-Asp3,(E)-Dhb7]MC-HtyR | 70 | [142] |
| 188 | MC-YR | 70 | [16] |
| 209 | [D-Asp3,ADMAdda5,Dhb7]MC-RR | 200 | [150] |
| 210 | [D-Glu(OC3H6O)6]MC-LR | >1000 | [76] |
| 212 | [D-Asp3]MC-WR | 95 ± 10 | [216] |
| 222 | MC-HtyR | 160–300 | [147] |
| 242 | MC-WR | 150–200 | [76] |
| 251 | [D-Asp3, ADMAdda5,Dhb7]MC-HtyR | 100 | [150] |
| 272 | MC-LR Cys conjugate | 1000 | [80] |
Most of the differences in in vivo toxicity between MC congeners can be attributed to a combination of differences in their uptake into the liver via a carrier-mediated transport system (organic anion transporting polypeptides (OATPs)) and/or their inhibitory potency toward hepatic PPs that are widely recognized as the principal targets of MCs [95,204,217,218,219,220,221,222,223]. MC toxicity requires active transport by OATP1B1 and OATP1B3, members of the OATP family of transporters, for uptake into cells and the high expression of these transporters in the liver accounts for their selective hepatotoxicity [95,224]. While some studies suggest that differences in in vivo and in vitro toxicity are related to PP inhibition rather than transport [216,225], other reports show that in vivo toxicity depends mainly on OATP transport kinetics rather than on differences in PP inhibition [215,224]. Therefore, future study is required to determine the relative importance/contribution of OATP versus PP inhibition to overall toxicity of MCs.
6.2. In Vitro Toxicity: Inhibition of Serine/Threonine PPs
The nature of the interaction of MCs with serine/threonine PPs has been intensively studied [118,217,221,228,229,230,231]. The concentrations required to inhibit 50% of the enzyme activity (IC50) of serine/threonine phosphatases (PP1–6) by MC congeners are shown in Table 3. MC-LR, the most toxic congener, has similar potency in the inhibition of PP1, PP2, and PP4–6, presenting an IC50 of around 0.1–1 nM [232,233,234,235]. However, it is 1000-fold less potent towards PP3 (IC50 ca. 1 µM) and seems to have no effect on the activity of PP7 [235,236].
Table 3.
The half inhibitory concentration (IC50) values (nM) for inhibition of serine/threonine protein phosphatases (PPs) by MC congeners.
| Entry a | Microcystin | Type of PP | Origin of PP | IC50 | Reference |
|---|---|---|---|---|---|
| 7 | MC-LA | PP1 | Rabbit muscle | 2.3 | [237] |
| PP2A | Human hepatocytes | 0.56 | [238] | ||
| PP2A | Rabbit muscle | 0.05 | [237] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.161 ± 0.002 | [239] | ||
| 14 | MC-LV | PP1 | Rabbit skeletal muscle | 0.06–0.45 | [133] |
| 17 | MC-LL | PP1 | Rabbit skeletal muscle | 0.06–0.45 | [133] |
| 25 | [D-Asp3,Dha7] MC-LR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.254 ± 0.004 | [239] |
| 34 | MC-LM | PP1 | Rabbit skeletal muscle | 0.06–0.45 | [133] |
| 39 | [D-Asp3]MC-LR | PP2A | Rabbit skeletal muscle | 0.09 | [215] |
| 40 | [D-Asp3,(E)-Dhb7]MC-LR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.201 ± 0.003 | [239] |
| 41 | [D-Asp3,(Z)-Dhb7]MC-LR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.16 ± 0.01 | [239] |
| 42 | [Dha7]MC-LR | PP1 | Rabbit skeletal muscle | 0.54–5 | [240,241] |
| PP2A | Bovine kidney | 0.11 ± 0.04 | [241] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.167 ± 0.003 | [239] | ||
| 43 | [DMAdda5]MC-LR | rPP1c | Recombinant rabbit skeletal muscle PP1 | 1.5 | [240] |
| 52 | MC-LF | rPP1c | Recombinant rabbit skeletal muscle PP1 | 1.8 | [224] |
| PP1 | Rabbit skeletal muscle | 0.06–0.45 | [133] | ||
| PP2A | Human hepatocytes | 0.57 | [238] | ||
| PP2A | Human red blood cells | 1.1 | [224] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.10 ± 0.02 | [239] | ||
| 66 | MC-LR | PP1 | Rabbit muscle | 0.1–1.9 | [215,217,237,240,241,242,243,244,245] |
| PP1 | Chicken gizzard myosin B | 6 | [246] | ||
| PP1 | Liver of grass carp | 0.90 | [247] | ||
| rPP1c | Recombinant rabbit skeletal muscle PP1 | 1.2 | [224] | ||
| PP2A | Rabbit skeletal muscle | 0.04–0.5 | [215,217,237,243,248] | ||
| PP2A | Human hepatocytes | 0.46 | [238] | ||
| PP2A | Human erythrocytes | 0.03–2.2 | [241,242,249,250] | ||
| PP2A | Bovine heart | 0.05-2 | [241,246,251] | ||
| PP2A | Bovine kidney | 0.2 ± 0.1 | [241] | ||
| PP2A | Mouse brain | 0.28–3.15 | [218,247,252,253] | ||
| PP2A | Liver of grass carp | 0.28 | [247] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.032 ± 0.004 | [239] | ||
| 68 | [(6Z)-Adda5]MC-LR | PP2A | Mouse brain | 80 | [252] |
| rPP1c | Recombinant rabbit skeletal muscle PP1 | >100 | [240] | ||
| 83 | MC-LY | PP2A | Human hepatocytes | 0.34 | [238] |
| 89 | [D-Asp3,ADMAdda5]MC-LR | PP2Ab | 4 | [178] | |
| 94 | [D-Glu(OMe)6]MC-LR | rPP1c | Recombinant rabbit skeletal muscle PP1 | >100 | [240] |
| 97 | [D-Asp3,Dha7]MC-RR | rPP1c | Recombinant rabbit skeletal muscle PP1 | 0.22 ± 0.01 | [239] |
| 129 | [D-Asp3]MC-RR | PP2A rPP2Ac |
Human red blood cells Recombinant human PP2A catalytic subunit |
0.45–11.5 0.30 ± 0.01 |
[254] [239] |
| 130 | [Dha7]MC-RR | PP1 | Rabbit muscle | 8.3 ± 0.8 | [241] |
| PP2A | Bovine kidney | 4 ± 1 | [241] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.29 ± 0.01 | [239] | ||
| PP1 | Rabbit skeletal muscle | 2.6–5.7 | [254] | ||
| 131 | [D-Asp3,(E)-Dhb7]MC-RR | PP1 | Rabbit skeletal muscle | 1.8–56.4 | [215,254] |
| PP2A | Rabbit skeletal muscle | 2.4 | [215] | ||
| PP2A | Human red blood cells | 17.9–49.4 | [254] | ||
| 135 | MC-LW | rPP1c | Recombinant rabbit skeletal muscle PP1 | 1.9 | [224] |
| PP2A | Human hepatocytes | 0.29 | [238] | ||
| PP2A | Human red blood cells | 1.1 | [224] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.114 ± 0.003 | [239] | ||
| 142 | MC-FR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.069 ± 0.003 | [239] |
| 149 | [Dha7]MC-YR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.379 ± 0.003 | [239] |
| 164 | [D-Leu1]MC-LR | PP1 | Recombinant rabbit skeletal muscle PP1 | 0.5–4.43 | [167,168] |
| 168 | MC-RR | PP1 | Rabbit skeletal muscle | 0.68 | [215] |
| PP1 | Chicken gizzard myosin B | 3 | [246] | ||
| PP1 | Liver of grass carp | 3.60 | [247] | ||
| rPP1c | Recombinant rabbit skeletal muscle PP1 | 1.5 | [224] | ||
| PP2A | Human red blood cells | 0.241–175 | [241,249,250] | ||
| PP2A | Human hepatocytes | 0.60 | [238] | ||
| PP2A | Mouse brain | 0.72–1.4 | [218,224,247,252] | ||
| PP2A | Bovine cardiac muscle | 1 | [246] | ||
| PP2A | Bovine Kidney | 10 ± 2 | [241] | ||
| PP2A | Rabbit skeletal muscle | 0.1 | [215] | ||
| PP2A | Liver of grass carp | 0.64 | [247] | ||
| rPP2Ac | Recombinant human PP2A catalytic subunit | 0.056 ± 0.002 | [239] | ||
| 169 | [(6Z)-Adda5]MC-RR | rPP2Ac | Recombinant human PP2A catalytic subunit | 10.1 ± 0.3 | [239] |
| PP2A | Mouse brain | 80 | [252] | ||
| 183 | [D-Asp3]MC-HtyR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.098 ± 0.006 | [239] |
| 185 | [D-Asp3,(E)-Dhb7]MC-HtyR | rPP2Ac | Recombinant human PP2A catalytic subunit |
0.122 ± 0.005 | [239] |
| 186 | [D-Asp3,(Z)-Dhb7]MC-HtyR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.110 ± 0.008 | [239] |
| 188 | MC-YR | PP1 PP1 PP2A PP2A PP2A PP2A PP2A PP2A rPP2Ac |
Rabbit skeletal muscle Liver of grass carp Human red blood cells Human hepatocytes Bovine kidney Rabbit skeletal muscle Mouse brain Liver of grass carp Recombinant human PP2A catalytic subunit |
1.0 0.90 0.26–9.0 0.84 0.09 ± 0.02 0.26 0.39–1.3 0.40 0.125 ± 0.005 |
[215] [247] [241,249,250] [238] [241] [215] [218,247] [247] [239] |
| 242 | MC-WR | rPP2Ac | Recombinant human PP2A catalytic subunit | 0.18 ± 0.01 | [239] |
| 251 | [D-Asp3,ADMAdda5,Dhb7]MC-HtyR | PP1 | Rabbit skeletal muscle | 0.15–0.24 | [254] |
| PP2A | Human red blood cells | 0.06 | [254] | ||
| PP2A | Bovine heart | 0.06 | [251] | ||
| PP4 | Porcine testis | 0.04 | [251] | ||
| PP5 | Recombinant human PP5 expressed in E. coli | 0.5 | [251] |
The inhibition of the PP activity by MCs results primarily from non-covalent interactions largely mediated by the hydrophobic Adda side-chain in position 5, and the Glu6 moiety [229,255,256]. The Adda side-chain is closely accommodated in the hydrophobic groove of the catalytic subunit of the enzyme (Figure 3) and accounts for a significant portion of the toxin’s binding potential. Xing et al. [119] reported that in the catalytic subunit of PP2A, four amino acids Gln122, Ile123, His191, and Trp200 form a hydrophobic cage that accommodates the long hydrophobic Adda side-chain in MC-LR (Figure 3). The second important interaction consist of those of the carboxylic acid of the Glu6 residue and its adjacent carbonyl group (Adda-amide), which make hydrogen bonds to metal-bound water molecules in the active site of the catalytic subunits of PPs (Figure 3). These obsevations from X-ray crystallography agree well with in vitro and in vivo toxicity data that found Glu and Adda residues to be essential for the toxicity of MCs, and that modifications to the configuration of the Adda side-chain and esterification of the free carboxylic acid of Glu6 both lead to loss of toxic potential [76,255,256].
Figure 3.
X-ray crystallographic structure showing the interaction of microcystin-LR (MC-LR) with the PP2A catalytic subunit and its adjacent amino acid side chains (based on data provided in [257]). Important interactions are described in the text (Section 6.2). The Mdha7, D-Glu6 and Adda5 moities, and selected atoms in these amino acids, are labelled in magenta, red, and pale blue, respectively. The large blue spheres are the catalytic metal ions, and the dashed yellow line shows the location of the covalent bond formed over time between the thiol of Cys269 and the Mdha7 moiety of MC-LR.
A third interaction also occurs between MCs and the catalytic sub-units of PPs via covalent linkage of the electrophilic α,β-unsaturated carbonyl of the Mdha residue in position 7 to the thiol of the cysteine residue in the catalytic subunits of six serine⁄threonine-specific phosphatases (PP1, PP2, PP3, PP4, PP5, and PP6) by Michael addition (reviewed in [256]). Interactions in the binding pocket between PP1 [118,228] and PP2A [119] and MC-LR are strengthened by a covalent linkage between the Sγ atom of Cys-273 and Cys-269, respectively and the terminal carbon atom (C-3) of the Mdha side chain. However, formation of the covalent bond occurs slowly, and is not essential for the inactivation of the PPs by MCs [100,228,229,251]. For example, Miles et al. [108,109] have shown that Dhb7-containing MCs react with thiols about 200 times more slowly than Mdha7 and Dha7-containing MCs [108]. Nodularin-R, which is structurally related to MC-LR but is a pentapeptide containing N-methyldehydrobutyrine (Mdhb) at position-5, did not react at a detectable rate with mercaptoethanol [108]. However, nodularin-R exhibits an IC50 (1.8 nM), like that of MC-LR (2.2 nM) for human red blood cells PP2A [250] and nodularins do not form a covalent linkage to thiol group of Cys-273 in PP1 [120,258]. Similarly, MC congeners containing Dha7 (e.g., [D-Asp3,Dha7]MC-LR) and its analogue containing Dhb7 ([D-Asp3,Dhb7]MC-LR) have similar IC50 values for PP2A of 0.254 ± 0.004 and 0.201 ± 0.003 nM, respectively [239].
Two other interactions that also play an important role in the inactivation of PPs have been recently described [256]. The first consists of a hydrogen-bonding interaction between the carboxyl group of the Masp3 residue to the conserved Arg and Tyr residues in PP’s sub-catalytic units (Arg96 and Tyr134 of PP1; and Arg89 and Tyr127 of PP2). The second consists of a hydrophobic interaction between the aromatic ring of a conserved tyrosine (Tyr272 of PP1; and Tyr265 of PP2) in the PP’s β12–β13 loop to the L-Leu2 residue of MC-LZ congeners [256]. The presence of L-leucine in position 2 can, therefore, explain the higher PP-inhibition potential of MC-LR and its analogues with hydrophobic amino acids in the same position (Table 3).
7. Conclusions and Future Perspectives
Many MCs have been isolated and fully characterized over the last three decades using a combination of analytical techniques and advances in LC-MS/MS instrumentation have recently provided a rapid influx of new MC congeners by tentative identification of trace variants in complex samples. As described in this review, multiple combinations of the variable L-amino acids at positions 2 and 4 and modifications to the other amino acids lead to the structural diversity of 279 MC variants characterized to date. These are formed because of genetic diversity in the mcy gene cluster as well as subsequent chemical and biochemical transformations that can occur after biosynthesis. Although the discovery of myc gene clusters has led to new avenues for investigating MC regulation at the molecular level, gaps remain in the understanding of how environmental factors can modulate the relative abundance of MC congeners in a bloom. Consequently, because the toxicity of a cyanobacterial bloom depends not only on the total MC accumulation, but also on the structure and the abundance of each congener, further research focusing on the effects of environmental factors on modulating the MC profile during bloom episodes is therefore recommended. This is particularly important in the context of new public health guidelines emerging around the world based on ‘total microcystins’, which remains a poorly defined concept in the absence of comprehensive data on the strucutural diversity and toxic potential of the broader class.
Variations in in vivo toxicity between MC congeners can be attributed to the relative importance/contribution of OATP transport versus PP inhibition to overall toxicity of MCs. However, the large range of values reported in the literature for the LD50 by the same route of exposure within each congener can in part be explained by a combination of biological variability as well as uncertainty in toxin purity and quantitation. It is, therefore, very important for researchers to carefully assess and report on the purity and amount of toxins used for toxicity assessments to ensure comparability of results between different studies and different MC variants.
Acknowledgments
We thank Alistair L. Wilkins and Patricia LeBlanc for helpful discussions.
Abbreviations
| Aha | aminoheptanoic acid |
| Abu(Aba) | 2-aminobutyric acid (L-2-aminobutanoic acid |
| Apa | aminopropionic acid |
| Aib | aminoisobutyric acid (aminoisobutyrate) |
| Bu | 2-aminobutyric acid (L-butyrine) |
| Cit | citrulline |
| ADMAdda | O-acetyl-9-O-desmethylAdda |
| Dha | dehydroalanine |
| Dhb | dehydrobutyrine |
| DMAdda | 9-O-desmethylAdda |
| dmAdda | Adda demethylated at C-2, C-6, or C-8 |
| E(OMe) | glutamic acid methylester |
| H(2)Y | dihydrotyrosine |
| (H4)Y | 4,5,6,7-tetrahydrotyrosine |
| Har | homoarginine |
| Hil | homoisoleucine |
| Hph | homophenylalanine |
| Hty | homotyrosine |
| Kyn | kynurenine |
| Mdhb | N-methyldehydrobutyrine |
| M(O) | methionine-S-oxide |
| M(O2) | methionine S-sulfone |
| Mser | N-methylserine |
| Nfk | N-formylkynurenine |
| Oia | oxindolyalanine |
| Y(OMe) | methoxytyrosine |
Author Contributions
N.B. provided conceptualization of the need for a review on this topic, led writing of this article, and additional editing. C.O.M. analysis of data including extensive literature investigation and additional input on literature sources for microcystin congeners, Figure 3, drafting and editing of manuscript. D.G.B. analysis of data including extensive literature investigation on microcystin congeners, Figure 2, drafting and editing of manuscript. Z.L. recompilation of data and input on literature sources for microcystin congeners, Table 1. A.D. recompilation of data and input on literature sources of toxicity of microcystin congeners, Table 2 and Table 3. N.Y.B. recompilation of data, Figure 1 and analysis and verification of bibliographic references. T.N.-Q. provided conceptualization and additional editing of manuscript.
Funding
This work received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
An up to date review surrounding the diversity, biosynthesis, characterization, and toxicology of MC congeners reported in the literature as identified from cyanobacterial cultures and field samples. At present, 279 MCs have been characterized using a wide range of combinations of analytical techniques. Modifications of each amino acid residue in their structure, especially in the positions 5 and 6, affect both their in vivo and in vitro toxicities.
References
- 1.Mooij W.M., Hülsmann S., De Senerpont Domis L.N., Nolet B.A., Bodelier P.L.E., Boers P.C.M., Pires L.M.D., Gons H.J., Ibelings B.W., Noordhuis R., et al. The impact of climate change on lakes in the Netherlands: A review. Aquat. Ecol. 2005;39:381–400. doi: 10.1007/s10452-005-9008-0. [DOI] [Google Scholar]
- 2.Rigosi A., Carey C.C., Ibelings B.W., Brookes J.D. The interaction between climate warming and eutrophication to promote cyanobacteria is dependent on trophic state and varies among taxa. Limnol. Oceanogr. 2014;59:99–114. doi: 10.4319/lo.2014.59.1.0099. [DOI] [Google Scholar]
- 3.Botana L.M. Toxicological perspective on climate change: Aquatic toxins. Chem. Res. Toxicol. 2016;29:619–625. doi: 10.1021/acs.chemrestox.6b00020. [DOI] [PubMed] [Google Scholar]
- 4.Visser P.M., Verspagen J.M.H., Sandrini G., Stal L.J., Matthijs H.C.P., Davis T.W., Paerl H.W., Huisman J. How rising CO2 and global warming may stimulate harmful cyanobacterial blooms. Harmful Algae. 2016;54:145–159. doi: 10.1016/j.hal.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Miller A., Russell C. Food crops irrigated with cyanobacteria-contaminated water: An emerging public health issue in Canada. Environ. Health Rev. 2017;60:58–63. doi: 10.5864/d2017-021. [DOI] [Google Scholar]
- 6.Sivonen K., Jones G. Cyanobacterial toxins. In: Chorus I., Bartram J., editors. Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management. E&FN Spon; London, UK: 1999. pp. 41–111. [Google Scholar]
- 7.Corbel S., Mougin C., Bouaïcha N. Cyanobacterial toxins: Modes of actions, fate in aquatic and soil ecosystems, phytotoxicity and bioaccumulation in agricultural crops. Chemosphere. 2014;96:1–15. doi: 10.1016/j.chemosphere.2013.07.056. [DOI] [PubMed] [Google Scholar]
- 8.Testai E., Buratti F.M., Funari E., Manganelli M., Vichi S., Arnich N., Biré R., Fessard V., Sialehaamoa A. Review and Analysis of Occurrence, Exposure and Toxicity of Cyanobacteria Toxins in Food. European Food Safety Authority; Parma, Italy: 2016. p. 309. EFSA Supporting Publication EN-998. [Google Scholar]
- 9.Svirčev Z., Drobac D., Tokodi N., Mijović B., Codd G.A., Meriluoto J. Toxicology of microcystins with reference to cases of human intoxications and epidemiolocal investigations of exposures to cyanobacteria and cyanotoxins. Arch. Toxicol. 2017;91:621–650. doi: 10.1007/s00204-016-1921-6. [DOI] [PubMed] [Google Scholar]
- 10.Pham T.L., Utsumi M. An overview of the accumulation of microcystins in aquatic ecosystems. J. Environ. Manag. 2018;213:520–529. doi: 10.1016/j.jenvman.2018.01.077. [DOI] [PubMed] [Google Scholar]
- 11.Ettoumi A., El Khalloufi F., El Ghazali I., Oudra B., Amrani A., Nasri H., Bouaïcha N. Bioaccumulation of cyanobacterial toxins in aquatic organisms and its consequences for public health. In: Kattel G., editor. Zooplankton and Phytoplankton. Nova Science Publishers, Inc.; Hauppauge, NY, USA: 2011. pp. 1–34. [Google Scholar]
- 12.Puddick J., Prinsep M.R., Wood S.A., Kaufononga S.A.F., Cary S.C., Hamilton D.P. High levels of structural diversity observed in microcystins from Microcystis CAWBG11 and characterization of six new microcystin congeners. Mar. Drugs. 2014;12:5372–5395. doi: 10.3390/md12115372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louw P.G.J., Smit J.D. The active constituent of the poisonous algae, Microcystis toxica Stephens. With a note on experimental cases of algae poisoning in small animals. S. Afr. Ind. Chem. 1950;4:62–66. [Google Scholar]
- 14.Bishop C.T., Anet E.F.L.J., Gorham P.R. Isolation and identification of the fast death factor in Microcystis aeruginosa NRC-1. Can. J. Biochem. Physiol. 1959;37:453–471. doi: 10.1139/y59-047. [DOI] [PubMed] [Google Scholar]
- 15.Botes D.P., Tuinman A.A., Wessels P.L., Viljoen C.C., Kruger H., Williams D.H., Santikarn S., Smith R.J., Hammond S.J. The structure of cyanoginosin-LA, a cyclic heptapeptide toxin from the cyanobacterium Microcystis aeruginosa. J. Chem. Soc. Perkin Trans. 1. 1984:2311–2318. doi: 10.1039/p19840002311. [DOI] [Google Scholar]
- 16.Botes D.P., Wessels P.L., Kruger H., Runnegar M.T.C., Santikarn S., Smith R.J., Barna J.C.J., Williams D.H. Structural studies on cyanoginosins-LR, -YR, -YA, and -YM, peptide toxins from Microcystis aeruginosa. J. Chem. Soc. Perkin Trans. 1. 1985:2747. doi: 10.1039/p19850002747. [DOI] [Google Scholar]
- 17.Botes D.P., Kruger H., Viljoen C.C. Isolation and characterization of four toxins from the blue-green alga, Microcystis aeruginosa. Toxicon. 1982;20:945–954. doi: 10.1016/0041-0101(82)90097-6. [DOI] [PubMed] [Google Scholar]
- 18.Botes D.P., Vilioen C.C., Kruger H., Wessels P.L., Williams D.H. Configuration assignments of the amino acid residues and the presence of N-methyldehydroalanine in toxins of the blue-green alga Microcystis aeruginosa. Toxicon. 1982;20:1037–1042. doi: 10.1016/0041-0101(82)90105-2. [DOI] [PubMed] [Google Scholar]
- 19.Botes D.P. Cyanoginosins-isolation and structure. In: Steyn P.S., Vleggaar R., editors. Mycotoxins and Phycotoxins, Bioactive Molecules. Volume 1. Elsevier; Amsterdam, The Netherlands: 1986. p. 167. [Google Scholar]
- 20.Santikarn S., Williams D.H., Smith R.J., Hammond S.J., Botes D.P., Tuinman A., Wessels P.L., Viljoen C.C., Kruger H. A partial structure for the toxin BE-4 from the blue-green algae, Microcystis aeruginosa. J. Chem. Soc. Chem. Commun. 1983;12:652. doi: 10.1039/c39830000652. [DOI] [Google Scholar]
- 21.Nishizawa T., Asayama M., Fujii K., Harada K.-I., Shirai M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 1999;126:520–529. doi: 10.1093/oxfordjournals.jbchem.a022481. [DOI] [PubMed] [Google Scholar]
- 22.Nishizawa T., Ueda A., Asayama M., Fujii K., Harada K.-I., Ochi K., Shirai M. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 2000;127:779–789. doi: 10.1093/oxfordjournals.jbchem.a022670. [DOI] [PubMed] [Google Scholar]
- 23.Tillett D., Dittmann E., Erhard M., von Döhren H., Börner T., Neilan B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide–polyketide synthetase system. Chem. Biol. 2000;7:753–764. doi: 10.1016/S1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 24.Dittmann E., Börner T. Genetic contributions to the risk assessment of microcystin in the environment. Toxicol. Appl. Pharmacol. 2005;203:192–200. doi: 10.1016/j.taap.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Song L., Sano T., Li R., Watanabe M.M., Liu Y., Kaya K. Microcystin production of Microcystis viridis (cyanobacteria) under different culture conditions. Physiol. Res. 1998;46:19–23. doi: 10.1046/j.1440-1835.1998.00120.x. [DOI] [Google Scholar]
- 26.Kameyama K., Sugiura N., Inamori Y., Maekawa T. Characteristics of microcystin production in the cell cycle of Microcystis viridis. Environ. Toxicol. 2004;19:20–25. doi: 10.1002/tox.10147. [DOI] [PubMed] [Google Scholar]
- 27.Amé M.V., Wunderlin D.A. Effects of iron, ammonium and temperature on microcystin content by a natural concentrated Microcystis aeruginosa population. Water Air Soil Pollut. 2005;168:235–248. doi: 10.1007/s11270-005-1774-8. [DOI] [Google Scholar]
- 28.Tonk L., Visser P.M., Christiansen G., Dittmann E., Snelder E.O.F.M., Wiedner C., Mur L.R., Huisman J. The microcystin composition of the cyanobacterium Planktothrix agardhii changes toward a more toxic variant with increasing light intensity. Appl. Environ. Microbiol. 2005;71:5177–5181. doi: 10.1128/AEM.71.9.5177-5181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonk L., Van De Waal D.B., Slot P., Huisman J., Matthijs H.C.P., Visser P.M. Amino acid availability determines the ratio of microcystin variants in the cyanobacterium Planktothrix agardhii: Amino acid availability determines microcystin variants. FEMS Microbiol. Ecol. 2008;65:383–390. doi: 10.1111/j.1574-6941.2008.00541.x. [DOI] [PubMed] [Google Scholar]
- 30.Van de Waal D.B., Verspagen J.M.H., Lürling M., Van Donk E., Visser P.M., Huisman J. The ecological stoichiometry of toxins produced by harmful cyanobacteria: An experimental test of the carbon-nutrient balance hypothesis: Ecological stoichiometry of toxin production. Ecol. Lett. 2009;12:1326–1335. doi: 10.1111/j.1461-0248.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 31.Dziallas C., Grossart H.-P. Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS ONE. 2011;6:e25569. doi: 10.1371/journal.pone.0025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puddick J., Prinsep M.R., Wood S.A., Cary S.C., Hamilton D.P. Modulation of microcystin congener abundance following nitrogen depletion of a Microcystis batch culture. Aquat. Ecol. 2016;50:235–246. doi: 10.1007/s10452-016-9571-6. [DOI] [Google Scholar]
- 33.Fewer D.P., Rouhiainen L., Jokela J., Wahlsten M., Laakso K., Wang H., Sivonen K. Recurrent adenylation domain replacement in the microcystin synthetase gene cluster. BMC Evol. Biol. 2007;7:183. doi: 10.1186/1471-2148-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fewer D.P., Tooming-Klunderud A., Jokela J., Wahlsten M., Rouhiainen L., Kristensen T., Rohrlack T., Jakobsen K.S., Sivonen K. Natural occurrence of microcystin synthetase deletion mutants capable of producing microcystins in strains of the genus Anabaena (Cyanobacteria) Microbiology. 2008;154:1007–1014. doi: 10.1099/mic.0.2007/016097-0. [DOI] [PubMed] [Google Scholar]
- 35.Fewer D.P., Halinen K., Sipari H., Bernardová K., Mänttäri M., Eronen E., Sivonen K. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea: Inactivation of mcy genes in Baltic Sea Anabaena. Environ. Microbiol. Rep. 2011;3:189–194. doi: 10.1111/j.1758-2229.2010.00207.x. [DOI] [PubMed] [Google Scholar]
- 36.Kurmayer R., Christiansen G., Fastner J., Borner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 2004;6:831–841. doi: 10.1111/j.1462-2920.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- 37.Tooming-Klunderud A., Mikalsen B., Kristensen T., Jakobsen K.S. The mosaic structure of the mcyABC operon in Microcystis. Microbiology. 2008;154:1886–1899. doi: 10.1099/mic.0.2007/015875-0. [DOI] [PubMed] [Google Scholar]
- 38.del Campo F.F., Ouahid Y. Identification of microcystins from three collection strains of Microcystis aeruginosa. Environ. Pollut. 2010;158:2906–2914. doi: 10.1016/j.envpol.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 39.Niedermeyer T. Microcystin congeners described in the literature. 2013 doi: 10.6084/m9.figshare.880756. [DOI] [Google Scholar]
- 40.Bouhaddada R., Nélieu S., Nasri H., Delarue G., Bouaïcha N. High diversity of microcystins in a Microcystis bloom from an Algerian lake. Environ. Pollut. 2016;216:836–844. doi: 10.1016/j.envpol.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 41.Miller T.R., Beversdorf L.J., Weirich C.A., Bartlett S.L. Cyanobacterial toxins of the Laurentian Great Lakes, their toxicological effects, and numerical limits in drinking water. Mar. Drugs. 2017;15:160. doi: 10.3390/md15060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spoof L., Catherine A. Appendix 3: Tables of microcystins and nodularins. In: Meriluoto J., Spoof L., Codd G.A., editors. Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis. John Wiley & Sons, Ltd.; Chichester, UK: 2017. pp. 526–537. [Google Scholar]
- 43.Hughes E.O., Gorham P.R., Zehnder A. Toxicity of a unialgal culture of Microcystis aeruginosa. Can. J. Microbiol. 1958;4:225–236. doi: 10.1139/m58-024. [DOI] [PubMed] [Google Scholar]
- 44.Konst H., McKercher P.D., Gorham P.R., Robertson A., Howell J. Symptoms and pathology produced by Toxic Microcystis aeruginosa NRC-1 in laboratory and domestic animals. Can. J. Comp. Med. Vet. Sci. 1965;29:221–228. [PMC free article] [PubMed] [Google Scholar]
- 45.Murthy J.R., Capindale J.B. A new isolation and structure for the endotoxin from Microcystis aeruginosa NRC-1. Can. J. Biochem. 1970;48:508–510. doi: 10.1139/o70-081. [DOI] [PubMed] [Google Scholar]
- 46.Rabin P., Darbre A. An improved extraction procedure for the endotoxin from Microcystis aeruginosa NRC-1. Biochem. Soc. Trans. 1975;3:428–430. doi: 10.1042/bst0030428. [DOI] [PubMed] [Google Scholar]
- 47.Painuly P., Perez R., Fukai T., Shimizu Y. The structure of a cyclic peptide toxin, cyanogenosin-RR from Microcystis aeruginosa. Tetrahedron Lett. 1988;29:11–14. doi: 10.1016/0040-4039(88)80002-9. [DOI] [Google Scholar]
- 48.Kusumi T., Ooi T., Watanabe M.M., Takahashi H., Kakisawa H. Cyanoviridin RR, a toxin from the cyanobacterium (blue-green alga) Microcystis viridis. Tetrahedron Lett. 1987;28:4695–4698. doi: 10.1016/S0040-4039(00)96600-0. [DOI] [Google Scholar]
- 49.Carmichael W.W., Eschedor J.T., Patterson G.M.L., Moore R.E. Toxicity and partial structure of a hepatotoxic peptide produced by the cyanobacterium Nodularia spumigena Mertens emend. L575 from New Zealand. Appl. Environ. Microbiol. 1988;54:2257–2263. doi: 10.1128/aem.54.9.2257-2263.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmichael W.W., Beasley V., Bunner D.L., Eloff J.N., Falconer I., Gorham P., Harada K., Krishnamurthy T., Min-Juan Y., Moore R.E., et al. Naming of cyclic heptapeptide toxins of cyanobacteria (blue-green algae) Toxicon. 1988;26:971–973. doi: 10.1016/0041-0101(88)90195-X. [DOI] [PubMed] [Google Scholar]
- 51.Du X., Liu H., Yuan L., Wang Y., Ma Y., Wang R., Chen X., Losiewicz M.D., Guo H., Zhang H. The diversity of cyanobacterial toxins on structural characterization, distribution and identification: A systematic review. Toxins. 2019;11:530. doi: 10.3390/toxins11090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon H.B.F., Cornish-Bowden A., Liebecq C., Loening K.L., Moss G.P., Reedijk J., Velick S.F., Vliegenthart J.F.G., Bielka H., Sharon N., et al. International Union of pure and applied chemistry and international union of biochemistry, join commission on biochemical nomenclature. Nomenclature and symbolism for amino acids and peptides. Pure Appl. Chem. 1984;56:595–624. [Google Scholar]
- 53.Miles C.O., Stirling D. Toxin Mass List, Version 16. [(accessed on 24 October 2019)]; doi: 10.13140/RG.2.2.12580.22402. Available online: https://www.researchgate.net/publication/316605326_Toxin_mass_list_version_16. [DOI]
- 54.Dittmann E., Fewer D.P., Neilan B.A. Cyanobacterial toxins: Biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 2013;37:23–43. doi: 10.1111/j.1574-6976.2012.12000.x. [DOI] [PubMed] [Google Scholar]
- 55.Christiansen G., Fastner J., Erhard M., Borner T., Dittmann E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouhiainen L., Vakkilainen T., Siemer B.L., Buikema W., Haselkorn R., Sivonen K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena Strain 90. Appl. Environ. Microbiol. 2004;70:686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittmann E., Neilan B.A., Erhard M., von Döhren H., Börner T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 1997;26:779–787. doi: 10.1046/j.1365-2958.1997.6131982.x. [DOI] [PubMed] [Google Scholar]
- 58.Pearson L.A., Dittmann E., Mazmouz R., Ongley S.E., D’Agostino P.M., Neilan B.A. The genetics, biosynthesis and regulation of toxic specialized metabolites of cyanobacteria. Harmful Algae. 2016;54:98–111. doi: 10.1016/j.hal.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Christiansen G., Kurmayer R., Liu Q., Borner T. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 2006;72:117–123. doi: 10.1128/AEM.72.1.117-123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikalsen B., Boison G., Skulberg O.M., Fastner J., Davies W., Gabrielsen T.M., Rudi K., Jakobsen K.S. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003;185:2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shishido T.K., Jokela J., Humisto A., Suurnäkki S., Wahlsten M., Alvarenga D.O., Sivonen K., Fewer D.P. The biosynthesis of rare homo-amino acid containing variants of microcystin by a benthic cynabacterium. Mar. Drugs. 2019;17:271. doi: 10.3390/md17050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaebernick M., Neilan B.A., Borner T., Dittmann E. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 2000;66:3387–3392. doi: 10.1128/AEM.66.8.3387-3392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh H.-M., Lee S.J., Jang M.-H., Yoon B.-D. Microcystin production by Microcystis aeruginosa in a phosphorus-limited chemostat. Appl. Environ. Microbiol. 2000;66:176–179. doi: 10.1128/AEM.66.1.176-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sevilla E., Martin-Luna B., Vela L., Bes M.T., Fillat M.F., Peleato M.L. Iron availability affects mcyD expression and microcystin-LR synthesis in Microcystis aeruginosa PCC7806: Iron starvation triggers microcystin synthesis. Environ. Microbiol. 2008;10:2476–2483. doi: 10.1111/j.1462-2920.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- 65.Jähnichen S., Long B.M., Petzoldt T. Microcystin production by Microcystis aeruginosa: Direct regulation by multiple environmental factors. Harmful Algae. 2011;12:95–104. doi: 10.1016/j.hal.2011.09.002. [DOI] [Google Scholar]
- 66.Tao M., Xie P., Chen J., Qin B., Zhang D., Niu Y., Zhang M., Wang Q., Wu L. Use of a generalized additive model to investigate key abiotic factors affecting microcystin cellular quotas in heavy bloom areas of Lake Taihu. PLoS ONE. 2012;7:e32020. doi: 10.1371/journal.pone.0032020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner S., Fastner J., Dittmann E. Microcystin production revisited: Conjugate formation makes a major contribution: Production of cyanobacterial toxins strongly underestimated. Environ. Microbiol. 2013;15:1810–1820. doi: 10.1111/1462-2920.12072. [DOI] [PubMed] [Google Scholar]
- 68.Pimentel J.S.M., Giani A. Microcystin production and regulation under nutrient stress conditions in toxic Microcystis strains. Appl. Environ. Microbiol. 2014;80:5836–5843. doi: 10.1128/AEM.01009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Z., Kong F. UV-B Exposure affects the biosynthesis of microcystin in toxic Microcystis aeruginosa cells and its degradation in the extracellular space. Toxins. 2015;7:4238–4252. doi: 10.3390/toxins7104238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horst G.P., Sarnelle O., White J.D., Hamilton S.K., Kaul R.B., Bressie J.D. Nitrogen availability increases the toxin quota of a harmful cyanobacterium, Microcystis aeruginosa. Water Res. 2014;54:188–198. doi: 10.1016/j.watres.2014.01.063. [DOI] [PubMed] [Google Scholar]
- 71.Dittmann E., Gugger M., Sivonen K., Fewer D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015;23:642–652. doi: 10.1016/j.tim.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 72.Van de Waal D.B., Smith V.H., Declerck S.A.J., Stam E.C.M., Elser J.J. Stoichiometric regulation of phytoplankton toxins. Ecol. Lett. 2014;17:736–742. doi: 10.1111/ele.12280. [DOI] [PubMed] [Google Scholar]
- 73.Monchamp M.E., Pick F.R., Beisner B.E., Maranger R. Nitrogen forms influence microcystin concentration and composition via changes in cyanobacterial community structure. PLoS ONE. 2014;9:e85573. doi: 10.1371/journal.pone.0085573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walsh C.T., Chen H., Keating T.A., Hubbard B.K., Losey H.C., Luo L., Marshall C.G., Miller D.A., Patel H.M. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 2001;5:525–534. doi: 10.1016/S1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 75.Shishido T., Kaasalainen U., Fewer D.P., Rouhiainen L., Jokela J., Wahlsten M., Fiore M., Yunes J., Rikkinen J., Sivonen K. Convergent evolution of [D-Leucine1]microcystin-LR in taxonomically disparate cyanobacteria. BMC Evol. Biol. 2013;13:1–15. doi: 10.1186/1471-2148-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Namikoshi M., Rinehart K.L., Sakai R., Stotts R.R., Dahlem A.M., Beasley V.R., Carmichael W.W., Evans W.R. Identification of 12 hepatotoxins from a Homer lake bloom of the cyanobacteria Microcystis aeruginosa, Microcystis viridis, Microcystis wesenbergii; nine new microcystins. J. Org. Chem. 1992;57:866–872. doi: 10.1021/jo00029a016. [DOI] [Google Scholar]
- 77.Miles C.O., Sandvik M., Nonga H.E., Rundberget T., Wilkins A.L., Rise F., Ballot A. Identification of microcystins in a Lake Victoria cyanobacterial bloom using LC–MS with thiol derivatization. Toxicon. 2013;70:21–31. doi: 10.1016/j.toxicon.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 78.Namikoshi M., Rinehart K.L., Sakai R., Sivonen K., Carmichael W.W. Structures of three new cyclic heptapeptide hepatotoxins produced by the cyanobacterium (blue-green alga) Nostoc sp. strain 152. J. Org. Chem. 1990;55:6135–6139. doi: 10.1021/jo00312a019. [DOI] [Google Scholar]
- 79.Sivonen K., Carmichael W.W., Namikoshi M., Rinehart K.L., Dahlem A.M. Isolation and characterization of hepatotoxic microcystin homologs from the filamentous freshwater cyanobacterium Nostoc sp. strain 152. Appl. Environ. Microbiol. 1990;56:2650–2657. doi: 10.1128/aem.56.9.2650-2657.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Namikoshi M., Sun F., Choi B.W., Rinehart K.L., Carmichael W.W., Evans W.R., Beasley V.R. Seven more microcystins from Homer lake cells: Application of the general method for structure assignment of peptides containing dehydroamino acid unit(s) J. Org. Chem. 1995;60:3671–3679. doi: 10.1021/jo00117a017. [DOI] [Google Scholar]
- 81.Foss A.J., Miles C.O., Samdal I.A., Løvberg K.E., Wilkins A.L., Rise F., Jaabæk J.A.H., McGowan P.C., Aubel M.T. Analysis of free and metabolized microcystins in samples following a bird mortality event. Harmful Algae. 2018;80:117–129. doi: 10.1016/j.hal.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 82.Yilmaz M., Foss A.J., Miles C.O., Özen M., Demir N., Balci M., Beach D.G. Comprehensive multi-technique approach reveals the high diversity of microcystins in field collections and an associated isolate of Microcystis aeruginosa from a Turkish lake. Toxicon. 2019;167:87–100. doi: 10.1016/j.toxicon.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 83.Harada K., Ogawa K., Matsuura K., Nagai H., Murata H., Suzuki M., Itezono Y., Nakayama N., Shirai M., Nakano M. Isolation of two toxic heptapeptide microcystins from an axenic strain of Microcystis aeruginosa, K-139. Toxicon. 1991;29:479–489. doi: 10.1016/0041-0101(91)90022-J. [DOI] [PubMed] [Google Scholar]
- 84.Namikoshi M., Sivonen K., Evans W.R., Carmichael W.W., Sun F., Rouhiainen L., Luukkainen R., Rinehart K.L. Two new L-serine variants of microcystins-LR and -RR from Anabaena sp. strains 202 A1 and 202 A2. Toxicon. 1992;30:1457–1464. doi: 10.1016/0041-0101(92)90521-6. [DOI] [PubMed] [Google Scholar]
- 85.Sivonen K., Skulberg O.M., Namikoshi M., Evans W.R., Carmichael W.W., Rinehart K.L. Two methyl ester derivatives of microcystins, cyclic heptapeptide hepatotoxins, isolated from Anabaena flos-aquae strain CYA 83/1. Toxicon. 1992;30:1465–1471. doi: 10.1016/0041-0101(92)90522-7. [DOI] [PubMed] [Google Scholar]
- 86.Sivonen K., Namikoshi M., Evans W.R., Fardig M., Carmichael W.W., Rinehart K.L. Three new microcystins, cyclic heptapeptide hepatotoxins, from Nostoc sp. strain 152. Chem. Res. Toxicol. 1992;5:464–469. doi: 10.1021/tx00028a003. [DOI] [PubMed] [Google Scholar]
- 87.Lee T.H., Chou H.N. Isolation and identification of seven microcystins from a cultured M.TN-2 strain of Microcystis aeruginosa. Bot. Bull. Acad. Sin. 2000;41:197–202. [Google Scholar]
- 88.Welker M., Christiansen G., von Döhren H. Diversity of coexisting Planktothrix (Cyanobacteria) chemotypes deduced by mass spectral analysis of microcystins and other oligopeptides. Arch. Microbiol. 2004;182:288–298. doi: 10.1007/s00203-004-0711-3. [DOI] [PubMed] [Google Scholar]
- 89.Vasas G., Szydlowska D., Gaspar A., Welker M., Trojanowicz M., Borbély G. Determination of microcystins in environmental samples using capillary electrophoresis. J. Biochem. Biophys. Methods. 2006;66:87–97. doi: 10.1016/j.jbbm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 90.Puddick J., Prinsep M.R., Wood S.A., Miles C.O., Rise F., Cary S.C., Hamilton D.P., Wilkins A.L. Structural characterization of new microcystins containing tryptophan and oxidized tryptophan residues. Mar. Drugs. 2013;11:3025–3045. doi: 10.3390/md11083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Puddick J., Prinsep M.R., Wood S.A., Cary S.C., Hamilton D.P., Holland P.T. Further characterization of glycine-containing microcystins from the McMurdo dry valleys of Antarctica. Toxins. 2015;7:493–515. doi: 10.3390/toxins7020493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wood S.A., Mountfort D., Selwood A.I., Holland P.T., Puddick J., Cary S.C. Widespread distribution and identification of eight novel microcystins in Antarctic cyanobacterial mats. Appl. Environ. Microbiol. 2008;74:7243–7251. doi: 10.1128/AEM.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miles C.O., Melanson J.E., Ballot A. Sulfide oxidations for LC-MS Analysis of methionine-containing microcystins in Dolichospermum flos-aquae NIVA-CYA 656. Environ. Sci. Technol. 2014;48:13307–13315. doi: 10.1021/es5029102. [DOI] [PubMed] [Google Scholar]
- 94.Diehnelt C.W., Dugan N.R., Peterman S.M., Budde W.L. Identification of microcystin toxins from a strain of Microcystis aeruginosa by liquid chromatography introduction into a hybrid linear ion trap-fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 2006;78:501–512. doi: 10.1021/ac051556d. [DOI] [PubMed] [Google Scholar]
- 95.Niedermeyer T.H.J., Daily A., Swiatecka-Hagenbruch M., Moscow J.A. Selectivity and potency of microcystin congeners against OATP1B1 and OATP1B3 expressing cancer cells. PLoS ONE. 2014;9:e91476. doi: 10.1371/journal.pone.0091476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teta R., Della Sala G., Glukhov E., Gerwick L., Gerwick W.H., Mangoni A., Costantino V. Combined LC-MS/MS and molecular networking approach reveals new cyanotoxins from the 2014 cyanobacterial bloom in Green Lake, Seattle (WA, USA) Environ. Sci. Technol. 2015;49:14301–14310. doi: 10.1021/acs.est.5b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ortiz X., Korenkova E., Jobst K.J., MacPherson K.A., Reiner E.J. A high throughput targeted and non-targeted method for the analysis of microcystins and anatoxin-a using on-line solid phase extraction coupled to liquid chromatography-quadrupole time-of-flight high resolution mass spectrometry. Anal. Bioanal. Chem. 2017;409:4959–4969. doi: 10.1007/s00216-017-0437-0. [DOI] [PubMed] [Google Scholar]
- 98.Meriluoto J. Chromatography of microcystins. Anal. Chim. Acta. 1997;352:277–298. doi: 10.1016/S0003-2670(97)00131-1. [DOI] [Google Scholar]
- 99.Mallia V., Uhlig S., Rafuse C., Meija J., Miles Christopher O. Novel microcystins from Planktothrix prolifica NIVA-CYA 544 identified by LC-MS/MS, functional group derivatization and 15N-labeling. Mar. Drugs. 2019;17:643. doi: 10.3390/md17110643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kondo F., Ikai Y., Oka H., Okumura M., Ishikawa N., Harada K., Matsuura K., Murata H., Suzuki M. Formation, characterization, and toxicity of the glutathione and cysteine conjugates of toxic heptapeptide microcystins. Chem. Res. Toxicol. 1992;5:591–596. doi: 10.1021/tx00029a002. [DOI] [PubMed] [Google Scholar]
- 101.Miles C.O., Sandvik M., Nonga H.E., Ballot A., Wilkins A.L., Rise F., Jaabaek A.H., Loader J.I. Conjugation of microcystins with thiols is reversible: Base-catalyzed deconjugation for chemical analysis. Chem. Res. Toxicol. 2016;29:860–870. doi: 10.1021/acs.chemrestox.6b00028. [DOI] [PubMed] [Google Scholar]
- 102.Mayumi T., Kato H., Imanishi S., Kawasaki Y., Hasegawa M., Harada K. Structural characterization of microcystins by LC/MS/MS under ion trap conditions. J. Antibiot. 2006;59:710–719. doi: 10.1038/ja.2006.95. [DOI] [PubMed] [Google Scholar]
- 103.Stewart A.K., Strangman W.K., Percy A., Wright J.L.C. The biosynthesis of 15N-labeled microcystins and the comparative MS/MS fragmentation of natural abundance and their 15N-labeled congeners using LC-MS/MS. Toxicon. 2018;144:91–102. doi: 10.1016/j.toxicon.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 104.Ferranti P., Fabbrocino S., Nasi A., Caira S., Bruno M., Serpe L., Gallo P. Liquid chromatography coupled to quadrupole time of flight tandem mass spectrometry for microcystin analysis in freshwaters: Method performances and characterisation of a novel variant of microcystin-RR. Rapid Commun. Mass Spectrom. 2009;23:1328–1336. doi: 10.1002/rcm.4006. [DOI] [PubMed] [Google Scholar]
- 105.Dörr F.A., Oliveira-Silva D., Lopes N.P., Iglesias J., Volmer D.A., Pinto E. Dissociation of deprotonated microcystin variants by collision-induced dissociation following electrospray ionization. Rapid Commun. Mass Spectrom. 2011;25:1981–1992. doi: 10.1002/rcm.5083. [DOI] [PubMed] [Google Scholar]
- 106.Flores C., Caixach J. An integrated strategy for rapid and accurate determination of free and cell-bound microcystins and related peptides in natural blooms by liquid chromatography–electrospray-high resolution mass spectrometry and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry using both positive and negative ionization modes. J. Chromatogr. A. 2015;1407:76–89. doi: 10.1016/j.chroma.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 107.Ngoka L.C.M., Gross M.L. Multistep tandem mass spectrometry for sequencing cyclic peptides in an ion-trap mass spectrometer. J. Am. Soc. Mass Spectrom. 1999;10:732–746. doi: 10.1016/S1044-0305(99)00049-5. [DOI] [PubMed] [Google Scholar]
- 108.Miles C.O., Sandvik M., Nonga H.E., Rundberget T., Wilkins A.L., Rise F., Ballot A. Thiol derivatization for LC-MS identification of microcystins in complex matrices. Environ. Sci. Technol. 2012;46:8937–8944. doi: 10.1021/es301808h. [DOI] [PubMed] [Google Scholar]
- 109.Miles C.O., Sandvik M., Haande S., Nonaga H., Ballot A. LC-MS analysis with thiol derivatization to differentiate [Dhb7] from [Mdha7]microcystins: Analysis of cyanobacterial blooms, Planktothrix cultures and European crayfish from Lake Steinsfjorden, Norway. Environ. Sci. Technol. 2013;47:4080–4087. doi: 10.1021/es305202p. [DOI] [PubMed] [Google Scholar]
- 110.Zemskov I., Kropp H.M., Wittmann V. Regioselective cleavage of thioether linkages in microcystin conjugates. Chem. Eur. J. 2016;22:10990–10997. doi: 10.1002/chem.201601660. [DOI] [PubMed] [Google Scholar]
- 111.Miles C.O. Rapid and convenient oxidative release of thiol-conjugated forms of microcystins for chemical analysis. Chem. Res. Toxicol. 2017;30:1599–1608. doi: 10.1021/acs.chemrestox.7b00121. [DOI] [PubMed] [Google Scholar]
- 112.Harada K., Murata H., Qiang Z., Suzuki M., Kondo F. Mass spectrometric screening method for microcystins in cyanobacteria. Toxicon. 1996;34:701–710. doi: 10.1016/0041-0101(95)00163-8. [DOI] [PubMed] [Google Scholar]
- 113.Davidson I. Hydrolysis of samples for amino acid analysis. In: Smith B.J., editor. Protein Sequencing Protocols. Methods in Molecular Biology. Volume 64. Humana Press; Totowa, NJ, USA: 1997. [DOI] [PubMed] [Google Scholar]
- 114.Luukkainen R., Sivonen K., Namikoshi M., Fardig M., Rinehart K.L., Niemelä S.I. Isolation and identification of eight microcystins from thirteen Oscillatoria agardhii strains and structure of a new microcystin. Appl. Environ. Microbiol. 1993;59:2204–2209. doi: 10.1128/aem.59.7.2204-2209.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harada K., Ogawa K., Matsuura K., Murata H., Suzuki M., Watanabe M.F., Itezono Y., Nakayama N. Structural determination of geometrical isomers of microcystins LR and RR from cyanobacteria by two-dimensional NMR spectroscopic techniques. Chem. Res. Toxicol. 1990;3:473–481. doi: 10.1021/tx00017a014. [DOI] [PubMed] [Google Scholar]
- 116.Burton I.W., Quilliam M.A., Walter J.A. Quantitative 1H NMR with external standards: Use in preparation of calibration solutions for algal toxins and other natural products. Anal. Chem. 2005;77:3123–3131. doi: 10.1021/ac048385h. [DOI] [PubMed] [Google Scholar]
- 117.National Research Council Canada List of CRM Products. [(accessed on 25 September 2019)]; Available online: www.nrc-cnrc.gc.ca/eng/solutions/advisory/crm/list_product.html.
- 118.Goldberg J., Huang H., Kwon Y., Greengard P., Nairn A.C., Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 119.Xing Y., Xu Y., Chen Y., Jeffrey P.D., Chao Y., Lin Z., Li Z., Strack S., Stock J.B., Shi Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell. 2006;127:341–353. doi: 10.1016/j.cell.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 120.Maynes J.T., Luu H.A., Cherney M.M., Andersen R.J., Williams D., Holmes C.F.B., James M.N.G. Crystal structures of protein phosphatase-1 bound to motuporin and dihydromicrocystin-LA: Elucidation of the mechanism of enzyme inhibition by cyanobacterial toxins. J. Mol. Biol. 2006;356:111–120. doi: 10.1016/j.jmb.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 121.Cho U.S., Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 122.Xu Y., Chen Y., Zhang P., Jeffrey P.D., Shi Y. Structure of a protein phosphatase 2A holoenzyme: Insights into B55-mediated tau dephosphorylation. Mol. Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Z., Cetin B., Anger M., Cho U.S., Helmhart W., Nasmyth K., Xu W. Structure and Function of the PP2A-Shugoshin Interaction. Mol. Cell. 2009;35:426–441. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wlodarchak N., Guo F., Satyshur K.A., Jiang L., Jeffrey P.D., Sun T., Stanevich V., Mumby M.C., Xing Y. Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 2013;23:931. doi: 10.1038/cr.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen E., Choy M.S., Petrényi K., Kónya Z., Erdődi F., Dombrádi V., Peti W., Page R. Molecular insights into the fungus-specific serine/threonine protein phosphatase Z1 in Candida albicans. mBio. 2016;7:e00872-16. doi: 10.1128/mBio.00872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kumar G.S., Choy M.S., Koveal D.M., Lorinsky M.K., Lyons S.P., Kettenbach A.N., Page R., Peti W. Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme. Sci. Adv. 2018;4:eaau6044. doi: 10.1126/sciadv.aau6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rudolph-Böhner S., Mierke D.F., Moroder L. Molecular structure of the cyanobacterial tumor-promoting microcystins. FEBS Lett. 1994;349:319–323. doi: 10.1016/0014-5793(94)00680-6. [DOI] [PubMed] [Google Scholar]
- 128.Bagu J.R., Sönnichsen F.D., Williams D., Andersen R.J., Sykes B.D., Holmes C.F.B. Comparison of the solution structures of microcystin-LR and motuporin. Nat. Struct. Mol. Biol. 1995;2:114–116. doi: 10.1038/nsb0295-114. [DOI] [PubMed] [Google Scholar]
- 129.Mierke D.F., Rudolph-Böhner S., Müller G., Moroder L. Structure of two microcystins: Refinement with nuclear Overhauser effects and ensemble calculations. Biopolymers. 1995;36:811–828. doi: 10.1002/bip.360360613. [DOI] [Google Scholar]
- 130.Trogen G.B., Edlund U., Larsson G., Sethson I. The solution NMR structure of a blue-green algae hepatotoxin, microcystin-RR. A comparison with the structure of microcystin-LR. Eur. J. Biochem. 1998;258:301–312. doi: 10.1046/j.1432-1327.1998.2580301.x. [DOI] [PubMed] [Google Scholar]
- 131.Ballot A., Sandvik M., Rundberget T., Botha C.J., Miles C.O. Diversity of cyanobacteria and cyanotoxins in Hartbeespoort Dam, South Africa. Mar. Freshw. Res. 2014;65:175–189. doi: 10.1071/MF13153. [DOI] [Google Scholar]
- 132.Mahakhant A., Sano T., Ratanachot P., Tong-a-ram T., Srivastava V.C., Watanabe M.M., Kaya K. Detection of microcystins from cyanobacterial water blooms in Thailand fresh water. Phycol. Res. 1998;42:25–29. doi: 10.1046/j.1440-1835.1998.00119.x. [DOI] [Google Scholar]
- 133.Craig M., McCready T.L., Luu H.A., Smillie M.A., Dubord P., Holmes C.F.B. Identification and characterization of hydrophobic microcystins in Canadian freshwater cyanobacteria. Toxicon. 1993;31:1541–1549. doi: 10.1016/0041-0101(93)90338-J. [DOI] [PubMed] [Google Scholar]
- 134.Puddick J., Prinsep M.R., Wood S.A., Cary S.C., Hamilton D.P., Wilkins A.L. Isolation and structure determination of two new hydrophobic microcystins from Microcystis sp. (CAWBG11) Phytochem. Lett. 2013;6:575–581. doi: 10.1016/j.phytol.2013.07.011. [DOI] [Google Scholar]
- 135.Lee T.H., Chen Y.M., Chou H.N. First report of microcystins in Taiwan. Toxicon. 1998;36:247–255. doi: 10.1016/S0041-0101(97)00128-1. [DOI] [PubMed] [Google Scholar]
- 136.Prakash S., Lawton L.A., Edwards C. Stability of toxigenic Microcystis blooms. Harmful Algae. 2009;8:377–384. doi: 10.1016/j.hal.2008.08.014. [DOI] [Google Scholar]
- 137.Kaasalainen U., Jokela J., Fewer D.P., Sivonen K., Rikkinen J. Microcystin production in the tripartite cyanolichen Peltigera leucophlebia. Mol. Plant Microbe Interact. 2009;22:695–702. doi: 10.1094/MPMI-22-6-0695. [DOI] [PubMed] [Google Scholar]
- 138.Namikoshi M., Yuan M., Sivonen K., Carmichael W.W., Rinehart K.L., Rouhiainen L., Sun F., Brittain S., Otsuki A. Seven new microcystins possessing two L-glutamic acid units, isolated from Anabaena sp. Strain 186. Chem. Res. Toxicol. 1998;11:143–149. doi: 10.1021/tx970120t. [DOI] [PubMed] [Google Scholar]
- 139.Robillot C., Vinh J., Puiseux-Dao S., Hennion M.-C. Hepatotoxin production kinetics of the cyanobacterium Microcystis aeruginosa PCC 7820, as determined by HPLC−mass spectrometry and protein phosphatase bioassay. Environ. Sci. Technol. 2000;34:3372–3378. doi: 10.1021/es991294v. [DOI] [Google Scholar]
- 140.Bateman K.P., Thibault P., Douglas D.J., White R.L. Mass spectral analyses of microcystins from toxic cyanobacteria using on-line chromatographic and electrophoretic separations. J. Chromatogr. A. 1995;712:253–268. doi: 10.1016/0021-9673(95)00438-S. [DOI] [PubMed] [Google Scholar]
- 141.Krishnamurthy T., Szafraniec L., Hunt D.F., Shabanowitz J., Yates J.R., Hauer C.R., Carmichael W.W., Skulberg O., Codd G.A., Missler S. Structural characterization of toxic cyclic peptides from blue-green algae by tandem mass spectrometry. Proc. Nat. Acad. Sci. USA. 1989;86:770–774. doi: 10.1073/pnas.86.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sano T., Kaya K. Two new (E)-2-amino-2-butenoic acid (Dhb)-containing microcystins isolated from Oscillatoria agardhii. Tetrahedron. 1998;54:463–470. doi: 10.1016/S0040-4020(97)10291-5. [DOI] [Google Scholar]
- 143.Mazur-Marzec H., Browarczyk-Matusiak G., Forycka K., Kobos J., Plinski M. Morphological, genetic, chemical and ecophysiological characterisation of two Microcystis aeruginosa isolates from the Vistula Lagoon, southern Baltic. Oceanologia. 2010;52:127–146. doi: 10.5697/oc.52-1.127. [DOI] [Google Scholar]
- 144.Sano T., Takagi H., Kaya K.A. Dhb-microcystin from the filamentous cyanobacterium Planktothrix rubescens. Phytochemistry. 2004;65:2159–2162. doi: 10.1016/j.phytochem.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 145.Sano T., Takagi H., Nishikawa M., Kaya K. NIES certified reference material for microcystins, hepatotoxic cyclic peptide toxins from cyanobacterial blooms in eutrophic water bodies. Anal. Bioanal. Chem. 2008;391:2005–2010. doi: 10.1007/s00216-008-2040-x. [DOI] [PubMed] [Google Scholar]
- 146.Krishnamurthy T., Carmichael W.W., Sarver E.W. Toxic peptides from freshwater cyanobacteria (blue-green algae). I. Isolation, purification and characterization of peptides from Microcystis aeruginosa and Anabaena flos-aquae. Toxicon. 1986;24:865–873. doi: 10.1016/0041-0101(86)90087-5. [DOI] [PubMed] [Google Scholar]
- 147.Harada K., Matsuura K., Suzuki M., Watanabe M.F., Oishi S., Dahlem A.M., Beasley V.R., Carmichael W.W. Isolation and characterization of the minor components associated with microcystins LR and RR in the cyanobacterium (blue-green algae) Toxicon. 1990;28:55–64. doi: 10.1016/0041-0101(90)90006-S. [DOI] [PubMed] [Google Scholar]
- 148.Rudolph-Böhner S., Wu J.T., Moroder L. Identification and characterization of microcystin-LY from Microcystis aeruginosa (strain 298) Biol. Chem. 1993;374:635–640. doi: 10.1515/bchm3.1993.374.7-12.635. [DOI] [PubMed] [Google Scholar]
- 149.Oksanen I., Jokela J., Fewer D.P., Wahlsten M., Rikkinen J., Sivonen K. Discovery of rare and highly toxic microcystins from lichen-associated cyanobacterium Nostoc sp. strain IO-102-I. Appl. Environ. Microbiol. 2004;70:5756–5763. doi: 10.1128/AEM.70.10.5756-5763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Beattie K.A., Kaya K., Sano T., Codd G.A. Three dehydrobutyrine (Dhb)-containing microcystins from the cyanobacterium Nostoc sp. Phytochemistry. 1998;47:1289–1292. doi: 10.1016/S0031-9422(97)00769-3. [DOI] [Google Scholar]
- 151.Brittain S., Mohamed Z.A., Wang J., Lehmann V.K.B., Carmichael W.W., Rinehart K.L. Isolation and characterization of microcystins from a River Nile strain of Oscillatoria tenuis Agardh ex Gomont. Toxicon. 2000;38:1759–1771. doi: 10.1016/S0041-0101(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 152.Qi Y., Rosso L., Sedan D., Giannuzzi L., Andrinolo D., Volmer D.A. Seven new microcystin variants discovered from a native Microcystis aeruginosa strain—Unambiguous assignment of product ions by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2015;29:1–5. doi: 10.1002/rcm.7075. [DOI] [PubMed] [Google Scholar]
- 153.Kaasalainen U., Fewer D.P., Jokela J., Wahlsten M., Sivonen K., Rikkinen J. Cyanobacteria produce a high variety of hepatotoxic peptides in lichen symbiosis. Proc. Natl. Acad. Sci. USA. 2012;109:5886–5891. doi: 10.1073/pnas.1200279109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Choi B.W., Namikoshi M., Sun F., Rinehart K.L., Carmichael W.W., Kaup A.M., Evans W.R., Beasley V.R. Isolation of linear peptides related to the hepatotoxins nodularin and microcystins. Tetrahedron Lett. 1993;34:7881–7884. doi: 10.1016/S0040-4039(00)61500-9. [DOI] [Google Scholar]
- 155.Luukkainen R., Namikoshi M., Sivonen K., Rinehart K.L., Niemela S.I. Isolation and identification of 12 microcystins from four strains and two bloom samples of Microcystis spp.: Structure of a new hepatotoxin. Toxicon. 1994;32:133–139. doi: 10.1016/0041-0101(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 156.Meriluoto J.A.O., Sandström A., Eriksson J.E., Remaud G., Grey Graig A., Chattopadhyaya J. Structure and toxicity of a peptide hepatotoxin from the cyanobacterium Oscillatoria agardhii. Toxicon. 1989;27:1021–1034. doi: 10.1016/0041-0101(89)90153-0. [DOI] [PubMed] [Google Scholar]
- 157.Kiviranta J., Namikoshi M., Sivonen K., Evans W.R., Carmichael W.W., Rinehart K.L. Structure determination and toxicity of a new microcystin from Microcystis aeruginosa strain 205. Toxicon. 1992;30:1093–1098. doi: 10.1016/0041-0101(92)90054-9. [DOI] [PubMed] [Google Scholar]
- 158.Sano T., Kaya K. A 2-amino-2-butenoic acid (Dhb)-containing microcystin isolated from Oscillatoria agardhii. Tetrahedron Lett. 1995;36:8603–8606. doi: 10.1016/0040-4039(95)01824-2. [DOI] [Google Scholar]
- 159.Kleinteich J., Puddick J., Wood S., Hildebrand F., Laughinghouse IV H., Pearce D., Dietrich D., Wilmotte A. Toxic cyanobacteria in Svalbard: Chemical diversity of microcystins detected using a liquid chromatography mass spectrometry precursor ion screening method. Toxins. 2018;10:147. doi: 10.3390/toxins10040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gathercole P.S., Thiel P.G. Liquid chromatographic determination of the cyanoginosins, toxins produced by the cyanobacterium Microcystis aeruginosa. J. Chromatogr. 1987;408:435–440. doi: 10.1016/S0021-9673(01)81837-9. [DOI] [PubMed] [Google Scholar]
- 161.Namikoshi M., Sivonen K., Evans W.R., Carmichael W.W., Rouhiainen L., Luukkainen R., Rinehart K.L. Structures of three new homotyrosine-containing microcystins and a new homophenylalanine variant from Anabaena sp. strain 66. Chem. Res. Toxicol. 1992;5:661–666. doi: 10.1021/tx00029a011. [DOI] [PubMed] [Google Scholar]
- 162.Sivonen K., Namikoshi M., Evans W.R., Gromov B.V., Carmichael W.W., Rinehart K.L. Isolation and structures of five microcystins from a Russian Microcystis aeruginosa strain CALU 972. Toxicon. 1992;30:1481–1485. doi: 10.1016/0041-0101(92)90524-9. [DOI] [PubMed] [Google Scholar]
- 163.Okello W., Ostermaier V., Portmann C., Gademann K., Kurmayer R. Spatial isolation favours the divergence in microcystin net production by Microcystis in Ugandan freshwater lakes. Water Res. 2010;44:2803–2814. doi: 10.1016/j.watres.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Okello W., Portmann C., Erhard M., Gademann K., Kurmayer R. Occurrence of microcystin-producing cyanobacteria in Ugandan fresh waters. Environ. Toxicol. 2010;25:367–380. doi: 10.1002/tox.20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Namikoshi M., Sivonen K., Evans W.R., Sun F., Carmichael W.W., Rinehart K.L. Isolation and structures of microcystins from a cyanobacterial water bloom (Finland) Toxicon. 1992;30:1473–1479. doi: 10.1016/0041-0101(92)90523-8. [DOI] [PubMed] [Google Scholar]
- 166.Elleman T., Falconer I., Jackson A., Runnegar M. Isolation, characterization and pathology of the toxin from a Microcystis aeruginosa (= Anacystis cyanea) bloom. Aust. J. Biol. Sci. 1978;31:209. doi: 10.1071/BI9780209. [DOI] [PubMed] [Google Scholar]
- 167.Matthiensen A., Beattie K.A., Yunes J.S., Kaya K., Codd G.A. [D-Leu1]Microcystin-LR, from the cyanobacterium Microcystis RST 9501 and from a Microcystis bloom in the Patos Lagoon estuary, Brazil. Phytochemistry. 2000;55:383–387. doi: 10.1016/S0031-9422(00)00335-6. [DOI] [PubMed] [Google Scholar]
- 168.Park H., Namikoshi M., Brittain S., Carmichael W., Murphy T. [D-Leu1]Microcystin-LR, a new microcystin isolated from waterbloom in a Canadian prairie lake. Toxicon. 2001;39:855–862. doi: 10.1016/S0041-0101(00)00224-5. [DOI] [PubMed] [Google Scholar]
- 169.Grach-Pogrebinsky O., Sedmak B., Carmeli S. Seco[D-Asp3]microcystin-RR and [D-Asp3,D-Glu(OMe)6]microcystin-RR, two new microcystins from a toxic water bloom of the cyanobacterium Planktothrix rubescens. J. Nat. Prod. 2004;67:337–342. doi: 10.1021/np034036r. [DOI] [PubMed] [Google Scholar]
- 170.Roy-Lachapelle A., Solliec M., Sauvé S., Gagnon C. A data-independent methodology for the structural characterization of microcystins and anabaenopeptins leading to the identification of four new congeners. Toxins. 2019;11:619. doi: 10.3390/toxins11110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Welker M., Erhard M. Consistency between chemotyping of single filaments of Planktothrix rubescens (cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J. Mass Spectrom. 2007;42:1062–1068. doi: 10.1002/jms.1237. [DOI] [PubMed] [Google Scholar]
- 172.Hollingdale C., Thomas K., Lewis N., Bekri K., McCarron P., Quillam M.A. Feasibility study on production of a matrix reference material for cyanobacterial toxins. Anal. Bioanal. Chem. 2015;407:5353–5363. doi: 10.1007/s00216-015-8695-1. [DOI] [PubMed] [Google Scholar]
- 173.Harada K., Ogawa K., Kimura Y., Murata H., Suzuki M., Thorn P.M., Evans W.R., Carmichael W.W. Microcystins from Anabaena flos-aquae NRC 525-17. Chem. Res. Toxicol. 1991;4:535–540. doi: 10.1021/tx00023a008. [DOI] [PubMed] [Google Scholar]
- 174.Birbeck J.A., Peraino N.J., O’Neill G.M., Coady J., Westrick J.A. Dhb microcystins discovered in USA using an online concentration LC–MS/MS platform. Toxins. 2019;11:653. doi: 10.3390/toxins11110653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Christiansen G., Yoshida W.Y., Blom J.F., Portmann C., Gademann K., Hemscheidt T., Kurmayer R. Isolation and structure determination of two microcystins and sequence comparison of the McyABC adenylation domains in Planktothrix species. J. Nat. Prod. 2008;71:1881–1886. doi: 10.1021/np800397u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kleinteich J., Wood S.A., Puddick J., Schleheck D., Kupper F.C., Dietrich D. Potent toxins in Arctic environments—Presence of saxitoxins and an unusual microcystin variant in Arctic freshwater ecosystem. Chem. Biol. Interact. 2013;206:423–431. doi: 10.1016/j.cbi.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 177.Frias H.V., Mendes M.A., Cardozo K.H., Carvalho V.M., Tomazela D., Colepicolo P., Pinto E. Use of electrospray tandem mass spectrometry for identification of microcystins during a cyanobacterial bloom event. Biochem. Biophys. Res. Commun. 2006;344:741–746. doi: 10.1016/j.bbrc.2006.03.199. [DOI] [PubMed] [Google Scholar]
- 178.Laub J., Henriksen P., Brittain S.M., Wang J., Carmichael W.W., Rinehart K.L., Moestrup Ø. [ADMAdda5]-microcystins in Planktothrix agardhii strain PH-123 (cyanobacteria)? Importance for monitoring of microcystins in the environment. Environ. Toxicol. 2002;17:351–357. doi: 10.1002/tox.10042. [DOI] [PubMed] [Google Scholar]
- 179.Rapala J., Sivonen K., Lyra C., Niemela S.I. Variation of microcystins, cyanobacterial hepatotoxins, in Anabaena spp. as a function of growth stimuli. Appl. Environ. Microbiol. 1997;63:2206–2212. doi: 10.1128/aem.63.6.2206-2212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ndlela L.L., Oberholster P.J., Van Wyk J.H., Cheng P.H. An overview of cyanobacterial bloom occurrences and research in Africa over the last decade. Harmful Algae. 2016;60:11–26. doi: 10.1016/j.hal.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 181.Pearson L.A., Barrow K.D., Neilan B.A. Characterization of the 2-hydroxy-acid dehydrogenase McyI, encoded within the microcystin biosynthesis gene cluster of Microcystis aeruginosa PCC7806. J. Biol. Chem. 2007;282:4681–4692. doi: 10.1074/jbc.M606986200. [DOI] [PubMed] [Google Scholar]
- 182.Sielaff H., Dittmann E., Tandeau De Marsac N., Bouchier C., von Döhren H., Börner T., Schwecke T. The mcyF gene of the microcystin biosynthetic gene cluster from Microcystis aeruginosa encodes an aspartate racemase. Biochem. J. 2003;373:909–916. doi: 10.1042/bj20030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fewer D.P., Wahlsten M., Österholm J., Jokela J., Rouhiainen L., Kaasalainen U., Rikkinen J., Sivonen K. The genetic basis for O-acetylation of the microcystin toxin in cyanobacteria. Chem. Biol. 2013;20:861–869. doi: 10.1016/j.chembiol.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 184.Sieber S.A., Marahiel M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 185.Kurmayer R., Christiansen G., Gumpenberger M., Fastner J. Genetic identification of microcystin ecotypes in toxic cyanobacteria of the genus Planktothrix. Microbiology. 2005;151:1525–1533. doi: 10.1099/mic.0.27779-0. [DOI] [PubMed] [Google Scholar]
- 186.Marahiel M.A., Stachelhaus T., Mootz H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 187.Moffitt M.C., Neilan B.A. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Environ. Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Namikoshi M., Choi B.W., Sakai R., Sun F., Rinehart K.L., Carmichael W.W., Evans W.R., Cruz P., Munro M.H.G., Blunt J.W. New nodularins: A general method for structure assignment. J. Org. Chem. 1994;59:2349–2357. doi: 10.1021/jo00088a014. [DOI] [Google Scholar]
- 189.Rinehart K.L., Namikoshi M., Choi B.W. Structure and biosynthesis of toxins from blue-green algae (cyanobacteria) J. Appl. Phycol. 1994;6:159–176. doi: 10.1007/BF02186070. [DOI] [Google Scholar]
- 190.Tsuji K., Watanuki T., Kondo F., Watanabe M.F., Suzuki S., Nakazawa H., Suzuki M., Uchida H., Harada K.I. Stability of microcystins from cyanobacteria-II. Effect of UV light on decomposition and isomerization. Toxicon. 1995;33:1619–1631. doi: 10.1016/0041-0101(95)00101-8. [DOI] [PubMed] [Google Scholar]
- 191.Kaya K., Sano T.A. Photodetoxification mechanism of the cyanobacterial hepatotoxin microcystin-lr by ultraviolet irradiation. Chem. Res. Toxicol. 1998;11:159–163. doi: 10.1021/tx970132e. [DOI] [PubMed] [Google Scholar]
- 192.Song W., Bardowell S., O’Shea K.E. Mechanistic study and the influence of oxygen on the photosensitized transformations of microcystins (cyanotoxins) Environ. Sci. Technol. 2007;41:5336–5341. doi: 10.1021/es063066o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fastner J., Erhard M., von Döhren H. Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2001;67:5069–5076. doi: 10.1128/AEM.67.11.5069-5076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Buratti F.M., Scardala S., Funari E., Testai E. The conjugation of microcystin-RR by human recombinant GSTs and hepatic cytosol. Toxicol. Lett. 2013;219:231–238. doi: 10.1016/j.toxlet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 195.Dai M., Xie P., Liang G., Chen J., Lei H. Simultaneous determination of microcystin-LR and its glutathione conjugate in fish tissues by liquid chromatography–tandem mass spectrometry. J. Chromatogr. B. 2008;862:43–50. doi: 10.1016/j.jchromb.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 196.Díez-Quijada L., Puerto M., Gutiérrez-Praena D., Llana-Ruiz-Cabello M., Jos A., Cameán A.M. Microcystin-RR: Occurrence, content in water and food and toxicological studies. A review. Environ. Res. 2019;168:467–489. doi: 10.1016/j.envres.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 197.Díez-Quijada L., Prieto A.I., Guzmán-Guillén R., Jos A., Cameán A.M. Occurrence and toxicity of microcystin congeners other than MC-LR and MC-RR: A review. Food Chem. Toxicol. 2019;125:106–132. doi: 10.1016/j.fct.2018.12.042. [DOI] [PubMed] [Google Scholar]
- 198.Zechmann B., Tomašić A., Horvat L., Fulgosi H. Subcellular distribution of glutathione and cysteine in cyanobacteria. Protoplasma. 2010;246:65–72. doi: 10.1007/s00709-010-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meister A. Glutathione metabolism and its selective modification. J. Biol. Chem. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 200.Li J., Li R., Li J. Current research scenario for microcystins biodegradation—A review on fundamental knowledge, application prospects and challenges. Sci. Total Environ. 2017;595:615–632. doi: 10.1016/j.scitotenv.2017.03.285. [DOI] [PubMed] [Google Scholar]
- 201.Ashworth C.T., Mason M.F. Observations on the pathological changes produced by a toxic substance present in blue-green algae (Microcystis aeruginosa) Am. J. Pathol. 1946;XXII:369–383. [PubMed] [Google Scholar]
- 202.McBarron E.J., May V. Poisoning of sheep in New South Wales by the blue-green alga Anacystis cyanea (Kuetz.) Dr. And dail. Aust. Vet. J. 1966;42:449–453. doi: 10.1111/j.1751-0813.1966.tb14471.x. [DOI] [PubMed] [Google Scholar]
- 203.Fawell J.K., James C.P., James H.A. Toxins from Blue-Green Algae: Toxicological Assessment of Microcystin-LR and a Method for Its Determination in Water. Foundation for Water Research; Denver, CO, USA: 1994. Report No: FR 0359/2/DoE 3358/2. [Google Scholar]
- 204.Kuiper-Goodman T., Falconer I.R., Fitzgerald J. Human health aspects. In: Chorus I., Bartram J., editors. Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring and Management. E & FN Spon; New York, NY, USA: 1999. pp. 113–153. [Google Scholar]
- 205.Chorus I., Bartram J. Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management. E & FN Spon; New York, NY, USA: 1999. [Google Scholar]
- 206.Stoner R.D., Adams W.H., Slatkin D.N., Siegelman H.W. The effects of single L-amino acid substitutions on the lethal potencies of the microcystins. Toxicon. 1989;27:825–828. doi: 10.1016/0041-0101(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 207.Creasia D.A. Acute inhalation toxicity of microcystin-LR with mice. Toxicon. 1990;28:605. doi: 10.1016/0041-0101(90)90186-b. [DOI] [Google Scholar]
- 208.Miura G.A., Robinson N.A., Lawrence W.B., Pace J.G. Hepatotoxicity of microcystin-LR in fed and fasted rats. Toxicon. 1991;29:337–346. doi: 10.1016/0041-0101(91)90287-2. [DOI] [PubMed] [Google Scholar]
- 209.Ito E., Kondo F., Harada K.-I. Intratracheal administration of microcystin-LR, and its distribution. Toxicon. 2001;39:265–271. doi: 10.1016/S0041-0101(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 210.Fawell J.K., Mitchell R.E., Everett D.J., Hill R.E. The toxicity of cyanobacterial toxins in the mouse: I Microcystin-LR. Hum. Exp. Toxicol. 1999;18:162–167. doi: 10.1177/096032719901800305. [DOI] [PubMed] [Google Scholar]
- 211.Smith J.L., Schulz K.L., Zimba P.V., Boyer G.L. Possible mechanism for the foodweb transfer of covalently bound microcystins. Ecotoxicol. Environ. Saf. 2010;73:757–761. doi: 10.1016/j.ecoenv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 212.Freitas M., Azevedo J., Carvalho A.P., Campos A., Vasconcelos V. Effects of storage, processing and proteolytic digestion on microcystin-LR concentration in edible clams. Food Chem. Toxicol. 2014;66:217–223. doi: 10.1016/j.fct.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 213.Gulledge B.M., Aggen J.B., Eng H., Sweimeh K., Chamberlin A.R. Microcystin analogues comprised only of Adda and a single additional amino acid retain moderate activity as PP1/PP2A inhibitors. Bioorg. Med. Chem. Lett. 2003;13:2907–2911. doi: 10.1016/S0960-894X(03)00588-2. [DOI] [PubMed] [Google Scholar]
- 214.Gulledge B.M., Aggen J.B., Chamberlin A.R. Linearized and truncated microcystin analogues as inhibitors of protein phosphatases 1 and 2A. Bioorg. Med. Chem. Lett. 2003;13:2903–2906. doi: 10.1016/S0960-894X(03)00589-4. [DOI] [PubMed] [Google Scholar]
- 215.Blom J.F., Jüttner F. High crustacean toxicity of microcystin congeners does not correlate with high protein phosphatase inhibitory activity. Toxicon. 2005;46:465–470. doi: 10.1016/j.toxicon.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 216.Chen Y.-M., Lee T.-H., Lee S.-J., Huang H.-B., Huang R., Chou H.-N. Comparison of protein phosphatase inhibition activities and mouse toxicities of microcystins. Toxicon. 2006;47:742–746. doi: 10.1016/j.toxicon.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 217.MacKintosh C., Beattie K.A., Klumpp S., Cohen P., Codd G.A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 1990;264:187–192. doi: 10.1016/0014-5793(90)80245-E. [DOI] [PubMed] [Google Scholar]
- 218.Matsushima R., Yoshizawa S., Watanabe M.F., Harada K., Furusawa M., Carmichael W.W., Fujiki H. In vitro and in vivo effects of protein phosphatase inhibitors, microcystins and nodularin, on mouse skin and fibroblasts. Biochem. Bioph. Res. Commun. 1990;171:867–874. doi: 10.1016/0006-291X(90)91226-I. [DOI] [PubMed] [Google Scholar]
- 219.Yoshizawa S., Matsushima R., Watanabe M.F., Harada K., Ichihara A., Carmichael W.W., Fujiki H. Inhibition of protein phosphatases by microcystis and nodularin associated with hepatotoxicity. J. Cancer Res. Clin. 1990;116:609–614. doi: 10.1007/BF01637082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Honkanen R.E., Zwiller J., Daily S., Khatra B.S., Dukelow M., Boynton A.L. Identification, purification, and characterization of novel serine/threonine protein phosphatase from bovine brain. J. Biol. Chem. 1991;266:6614–6619. [PubMed] [Google Scholar]
- 221.Runnegar M., Berndt N., Kong S.M., Lee E.Y.C., Zhang L.F. In vivo and in vitro binding of microcystin to protein phosphatase 1 and 2A. Biochem. Biophys. Res. Commun. 1995;216:162–169. doi: 10.1006/bbrc.1995.2605. [DOI] [PubMed] [Google Scholar]
- 222.Dawson R. The toxicology of microcystins. Toxicon. 1998;36:953–962. doi: 10.1016/S0041-0101(97)00102-5. [DOI] [PubMed] [Google Scholar]
- 223.Guzman R.E., Solter P.F., Runnegar M.T. Inhibition of nuclear protein phosphatase activity in mouse hepatocytes by the cyanobacterial toxin microcystin-LR. Toxicon. 2003;41:773–781. doi: 10.1016/S0041-0101(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 224.Fischer A., Hoeger S.J., Stemmer K., Feurstein D.J., Knobeloch D., Nussler A., Dietrich D.R. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: A comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol. Appl. Pharmacol. 2010;245:9–20. doi: 10.1016/j.taap.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 225.Monks N.R., Liu S., Xu Y., Yu H., Bendelow A.S., Moscow J.A. Potent cytotoxicity of the phosphatase inhibitor microcystin-LR and microcystin analogues in OATP1B1- and OATP1B3-expressing HeLa cells. Mol. Cancer Ther. 2007;6:587–598. doi: 10.1158/1535-7163.MCT-06-0500. [DOI] [PubMed] [Google Scholar]
- 226.Kaya K., Watanabe M.M. Microcystin composition of an axenic clonal strain of Microcystis viridis and Microcystis viridis-containing waterblooms in Japanese freshwaters. J. Appl. Phycol. 1990;2:173–178. doi: 10.1007/BF00023379. [DOI] [Google Scholar]
- 227.Zurawell R.W., Chen H., Burke J.M., Prepas E.E. Hepatotoxic cyanobacteria: A review of the biological importance of microcystins in freshwater environments. J. Toxicol. Environ. Health Part B. 2005;8:1–37. doi: 10.1080/10937400590889412. [DOI] [PubMed] [Google Scholar]
- 228.MacKintosh R.W., Dalby K.N., Campbell D.G., Cohen P.T.W., Cohen P., MacKintosh C. The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1. FEBS Lett. 1995;371:236–240. doi: 10.1016/0014-5793(95)00888-g. [DOI] [PubMed] [Google Scholar]
- 229.Craig M., Luu H.A., McCready T.L., Holmes C.F.B., Williams D., Andersen R.J. Molecular mechanisms underlying the interaction of motuporin and microcystins with type-1 and type-2A protein phosphatases. Biochem. Cell Biol. 1996;74:569–578. doi: 10.1139/o96-061. [DOI] [PubMed] [Google Scholar]
- 230.Zhang L., Zhang Z., Long F., Lee E.Y.C. Tyrosine-272 is involved in the inhibition of protein phosphatase-1 by multiple toxins. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 231.Dawson J.F., Holmes C.F.B. Molecular mechanisms underlying inhibition of protein phosphatases by marine toxins. Front. Biosci. 1999;4:646–658. doi: 10.2741/A461. [DOI] [PubMed] [Google Scholar]
- 232.Chen M.X., McPartlin A.E., Brown L., Chen Y.H., Barker H.M., Cohen P.T. A novel human protein serine/threonine phosphatase, which possesses four tetratricopeptide repeat motifs and localizes to the nucleus. EMBO J. 1994;13:4278–4290. doi: 10.1002/j.1460-2075.1994.tb06748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hastie C.J., Cohen P.T.W. Purification of protein phosphatase 4 catalytic subunit: Inhibition by the antitumour drug fostriecin and other tumour suppressors and promoters. FEBS Lett. 1998;431:357–361. doi: 10.1016/S0014-5793(98)00775-3. [DOI] [PubMed] [Google Scholar]
- 234.Prickett T.D., Brautigan D.L. The α4 Regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J. Biol. Chem. 2006;281:30503–30511. doi: 10.1074/jbc.M601054200. [DOI] [PubMed] [Google Scholar]
- 235.Swingle M., Ni L., Honkanen R.E. Small-molecule inhibitors of Ser/Thr protein phosphatases: Specificity, use and common forms of abuse. In: Ludlow J.W., editor. Methods in Molecular Biology: Protein Phosphatase Protocols. Volume 365. Humana Press; Totowa, NJ, USA: 2006. pp. 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang X., Honkanen R.E. Molecular cloning, expression, and characterization of a novel human Serine/Threonine protein phosphatase, PP7, that is homologous to Drosophila retinal degeneration C gene product (rdgC) J. Biol. Chem. 1998;273:1462–1468. doi: 10.1074/jbc.273.3.1462. [DOI] [PubMed] [Google Scholar]
- 237.Honkanen R.E., Codispoti B.A., Tse K., Boynton A.L. Characterization of natural toxins with inhibitory activity against serine/threonine protein phosphatases. Toxicon. 1994;32:339–350. doi: 10.1016/0041-0101(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 238.Robillot C., Hennion M.-C. Issues arising when interpreting the results of the protein phosphatase 2A inhibition assay for the monitoring of microcystins. Anal. Chim. Acta. 2004;512:339–346. doi: 10.1016/j.aca.2004.03.004. [DOI] [Google Scholar]
- 239.Ikehara T., Imamura S., Sano T., Nakashima J., Kuniyoshi K., Oshiro N., Yoshimoto M., Yasumoto T. The effect of structural variation in 21 microcystins on their inhibition of PP2A and the effect of replacing Cys269 with glycine. Toxicon. 2009;54:539–544. doi: 10.1016/j.toxicon.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 240.An J., Carmichael W.W. Use of a colorimetric protein phosphatase inhibition assay and enzyme-linked immunosorbent assay for the study of microcystins and nodularins. Toxicon. 1994;32:1495–1507. doi: 10.1016/0041-0101(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 241.Ufelmann H., Krüger T., Luckas B., Schrenk D. Human and rat hepatocyte toxicity and protein phosphatase 1 and 2A inhibitory activity of naturally occurring desmethyl-microcystins and nodularins. Toxicology. 2012;293:59–67. doi: 10.1016/j.tox.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 242.Suganuma M., Fujiki H., Okabe S., Nishiwaki S., Brautigan D., Ingebritsen T.S., Rosner M.R. Structurally different members of the okadaic acid class selectively inhibit protein serine/threonine but not tyrosine phosphatase activity. Toxicon. 1992;30:873–878. doi: 10.1016/0041-0101(92)90385-I. [DOI] [PubMed] [Google Scholar]
- 243.Holmes C.F.B., Boland M.P. Inhibitors of protein phosphatase-1 and -2A; two of the major serine/threonine protein phosphatases involved in cellular regulation. Curr. Opin. Struct. Biol. 1993;3:934–943. doi: 10.1016/0959-440X(93)90159-I. [DOI] [Google Scholar]
- 244.Wirsing B., Flury T., Wiedner C., Neumann U., Weckesser J. Estimation of the microcystin content in cyanobacterial field samples from German lakes using the colorimetric protein-phosphatase inhibition assay and RP-HPLC. Environ. Toxicol. 1999;14:23–29. doi: 10.1002/(SICI)1522-7278(199902)14:1<23::AID-TOX5>3.0.CO;2-9. [DOI] [Google Scholar]
- 245.Catanante G., Espin L., Marty J.-L. Sensitive biosensor based on recombinant PP1α for microcystin detection. Biosens. Bioelectron. 2015;67:700–707. doi: 10.1016/j.bios.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 246.Eriksson J.E., Toivola D., Meriluoto J.A.O., Karaki H., Han Y.-G., Hartshorne D. Hepatocyte deformation induced by cyanobacterial toxins reflects inhibition of protein phosphatases. Biochem. Biophys. Res. Commun. 1990;173:1347–1353. doi: 10.1016/S0006-291X(05)80936-2. [DOI] [PubMed] [Google Scholar]
- 247.Xu L., Lam P.K.S., Chen J., Zhang Y., Harada K. Comparative study on in vitro inhibition of grass carp (Ctenopharyngodon idellus) and mouse protein phosphatases by microcystins. Environ. Toxicol. 2000;15:71–75. doi: 10.1002/(SICI)1522-7278(2000)15:2<71::AID-TOX1>3.0.CO;2-E. [DOI] [Google Scholar]
- 248.Heresztyn T., Nicholson B.C. Determination of cyanobacterial hepatotoxins directly in water using a protein phosphatase inhibition assay. Water Res. 2001;35:3049–3056. doi: 10.1016/S0043-1354(01)00018-5. [DOI] [PubMed] [Google Scholar]
- 249.Rivasseau C., Racaud P., Deguin A., Hennion M.-C. Development of a bioanalytical phosphatase inhibition test for the monitoring of microcystins in environmental water samples. Anal. Chim. Acta. 1999;394:243–257. doi: 10.1016/S0003-2670(99)00301-3. [DOI] [Google Scholar]
- 250.Mountfort D.O., Holland P., Sprosen J. Method for detecting classes of microcystins by combination of protein phosphatase inhibition assay and ELISA: Comparison with LC-MS. Toxicon. 2005;45:199–206. doi: 10.1016/j.toxicon.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 251.Hastie C., Borthwick E., Morrison L., Codd G., Cohen P. Inhibition of several protein phosphatases by a non-covalently interacting microcystin and a novel cyanobacterial peptide, nostocyclin. BBA Gen. Subj. 2005;1726:187–193. doi: 10.1016/j.bbagen.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 252.Nishiwaki-Matsushima R., Nishiwaki S., Ohta T., Yoshizawa S., Suganuma M., Harada K.-I., Watanabe M.F., Fujiki H. Structure-function relationships of microcystins, liver tumor promoters, in interaction with protein phosphatase. Jpn. J. Cancer Res. 1991;82:993–996. doi: 10.1111/j.1349-7006.1991.tb01933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wong B.S.F., Lam P.K.S., Xu L., Zhang Y., Richardson B.J. A colorimetric assay for screening microcystin class compounds in aquatic systems. Chemosphere. 1999;38:1113–1122. doi: 10.1016/S0045-6535(98)00354-3. [DOI] [Google Scholar]
- 254.Hoeger S.J., Schmid D., Blom J.F., Ernst B., Dietrich D.R. Analytical and functional characterization of microcystins [Asp3]MC-RR and [Asp3,Dhb7]MC-RR: Consequences for risk assessment? Environ. Sci. Technol. 2007;41:2609–2616. doi: 10.1021/es062681p. [DOI] [PubMed] [Google Scholar]
- 255.Bagu J.R., Sykes B.D., Craig M.M., Holmes C.F.B. A molecular basis for different interactions of marine toxins with protein phosphatase-1: Molecular models for bound motuporin, microcystins, okadaic acid, and calyculin A. J. Biol. Chem. 1997;272:5087–5097. doi: 10.1074/jbc.272.8.5087. [DOI] [PubMed] [Google Scholar]
- 256.Pereira S.R., Vasconcelos V.M., Antunes A. The phosphoprotein phosphatase family of Ser/Thr phosphatases as principal targets of naturally occurring toxins. Crit. Rev. Toxicol. 2011;41:83–110. doi: 10.3109/10408444.2010.515564. [DOI] [PubMed] [Google Scholar]
- 257.Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., Yu J.W., Strack S., Jeffrey P.D., Shi1 Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 258.Kelker M.S., Page R., Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: A novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J. Mol. Biol. 2009;385:11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]