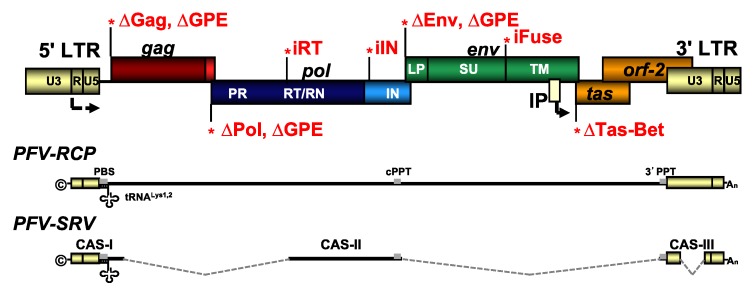
Figure A1.
PFV genome organization and viral expression systems used. Schematic outline of the wild type (wt) PFV proviral genome structure with gag, pol, and env ORFs. The locations of the point mutations for the individual proviral mutants resulting in variants with enzymatically inactive integrase (iIN) or reverse transcriptase (iRT); fusion-deficient Env (iFuse); or deficiencies in the translation of, Tas (∆Tas-Bet); Gag (∆Gag); Pol (∆Pol); Env (∆Env); or Gag, Pol, and Env (∆GPE), due to inactivation of the respective translation initiation sites. PFV-RCP supernatants contain full-length viral genomic RNA (vgRNA) whereas PFV-SRV supernatants harbor vgRNA comprising the minimal essential cis-acting viral sequences (CAS). LTR: long terminal repeat; U3: unique 3′ LTR region; U5: unique 5′ LTR region; R: repeat LTR region; PR: protease; RT/RN: reverse transcriptase—RNase H; IN: integrase; LP leader peptide; SU: surface subunit; TM: transmembrane subunit; IP: internal promoter; ©: cap; An: poly-A tail; PBS: primer binding site; 3′ PPT: 3′ poly purine tract; cPPT: central PPT.