Abstract
Introduction
The study evaluated the patterns of local innate immune response in bronchoalveolar lavage fluid (BALF) cells of pigs infected with porcine reproductive and respiratory syndrome virus (PRRSV) alone or co-infected with swine influenza virus (SIV).
Material and Methods
The study was performed on 26 seven-week-old pigs in three groups: PRRSV-infected (n = 11), PRRSV and SIV-infected (n = 11), and control (n = 4). BALF was collected post euthanasia at 2 and 4 dpi (three piglets per inoculated group) and at 21 dpi (all remaining pigs). Expression of IFN-α, IFN-γ, IL-1β, IL-6, IL-8, and IL-10 mRNA was quantified in BALF cells. PRRSV RNA was quantified in BALF samples using a commercial real-time RT-PCR kit.
Results
The three cytokines IFN-α, IFN-γ, and IL-1β presented significant expression changes in all experimental pigs. In PRRSV-infected animals IL-8 also did, but in co-infected subjects IL-6 and IL-10 were the additional upregulated cytokines. The highest number of differentially expressed genes was observed at 4 dpi, and significant differences in cytokine gene expression did not occur between the experimental groups at any other time point. The mean PRRSV load in the BALF of PRRSV-infected pigs was higher than that of co-infected pigs at each time point, having statistical significance only at 4 dpi.
Conclusion
The results of the study indicate that infection with PRRSV alone as well as with SIV interferes with innate and adaptive immune response in the infected host. They also showed that co-infection demonstrates additive effects on IL-6 and IL-10 mRNA expression levels.
Keywords: pigs, porcine reproductive and respiratory syndrome virus; swine influenza virus; local immunity; cytokines
Introduction
Porcine respiratory disease complex (PRDC) caused by mixed viral and bacterial infections is a common problem in the modern swine industry worldwide. Among viruses, porcine reproductive and respiratory syndrome virus (PRRSV) and swine influenza virus (SIV), alone or in combination, are the top two most significant contributors to PRDC and are frequently detected in the field (2, 8). Porcine reproductive and respiratory syndrome virus, an enveloped, positive-sense single-strand RNA virus within the Arteriviridae family, is a causative agent of PRRS responsible for late-term abortions in pregnant gilts and sows and respiratory distress in piglets and growing pigs (26). Some of the hallmarks of PRRSV infection in pigs which are pivotal in diagnosis are suppression of type I interferon (IFN-α/β) production, modulation
of cytokine expression, apoptotic responses, and adaptive immunity. These mechanisms may lead to a failure of virus clearance and to viral persistence in the host (14). The additive effect of modulation of the host immune response by PRRSV is increased predisposition to secondary infections of the respiratory tract (27, 28). SIV is the causative agent of swine influenza (SI), a highly contagious acute respiratory viral disease of swine. It is an enveloped, segmented, single-stranded RNA virus belonging to the Orthomyxoviridae family. In SIV-infected pigs mortality is usually low, while morbidity may reach 100% (18). During the acute phase of SI, production of inflammatory cytokines such as interferon alpha (IFN-α), tumour necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β), interleukin-6 (IL-6), interleukin-12 (IL-12), and interferon gamma (IFN-γ) has been well documented as an important factor determining disease severity (1, 15). In acutely SIV-infected pigs, a significant correlation was found between lung lesions and lung concentrations of IL-1β, interleukin-8 (IL-8), and TNF-α (24).
So far, experimental studies dealing with PRRSV and SIV infections conducted in conventional pigs have been focused on clinical manifestation and production performance. Nevertheless, little is known about the effect of concurrent infection with PRRSV and SIV on local innate immune response at the molecular level in conventional pigs. Only one study generated valuable insight on the impact of concomitant PRRSV and SIV infections on the development of the innate immune response. However, the study was performed in vitro and ex vivo on porcine alveolar macrophages (PAMs) and precision-cut lung slices (PCLS), respectively (5). Therefore, our study took up an in vivo objective: to explore the patterns of local innate immune response in BALF cells of pigs singly infected with PRRSV or co-infected with PRRSV and SIV using a model of in vivo experimental challenge.
Material and Methods
Viruses. PL15-33 strain of PRRSV 1 was isolated from lung tissue of a Polish-farmed pig with respiratory clinical signs by three passages in PAMs. The avian-like H1N1 A/Poland/Swine/14131/2014 (hereafter referred to as SwH1N1) of SIV used in this study had been isolated from lung tissue of a pig with severe swine influenza clinical manifestations. The viral inoculum was prepared from third-passage SPF embryonated chicken eggs, and its concentration was evaluated in Madin-Darby canine kidney (MDCK) cells.
Animals and infection studies. A total of 26 seven-week-old pigs obtained from a conventional healthy herd were used. The pigs were randomly divided into three groups – two experimental and one control. Before the start of the study, all pigs were tested for being negative both for PRRSV and influenza A virus specific antibodies with an IDEXX PRRS X3 ELISA kit (USA) and haemagglutination inhibition assays using SwH1N1, A/swine/England/96 (H1N2), A/swine/Flanders/1/98 (H3N2), and pdm-like H1N1 (A/swine/Poland/031951/12) strains. The pigs were also tested for infection with PRRSV on nasal swabs and serum and SIV on nasal swabs in real-time PCRs.
During the experiment, the animals were housed at the BSL3 animal facility in three independent units: one for the control and others for the infected pigs. On day 0, 11 pigs were inoculated intranasally (IN) with 105 TCID50 of PRRSV in 3 ml of phosphate-buffered saline (PBS) (1.5 ml for each nostril). Another 11 pigs were co-inoculated IN with 105 TCID50 of PRRSV and 107 TCID50 SwH1N1. Four pigs were mock-inoculated IN with 3 ml of PBS and served as a negative control group.
Clinical examination of pigs, including measurement of rectal temperature and observation for respiratory and general signs, was performed daily, from day 7 before inoculation until euthanasia. Fever was recorded when the rectal temperature was equal to or higher than 40°C. At 2 and 4 dpi, three pigs per infected group were euthanised. The remaining inoculated and control pigs were euthanised at 21 dpi. A complete necropsy was performed on each animal, with special emphasis on the respiratory tract. Lung lesions were evaluated, using the method and procedure described previously (13, 24).
BALF collection and total cell counts. Bronchoalveolar lavage fluid was obtained from each animal post mortem. A tracheal tube was inserted into an incision made in the trachea, and each lung was lavaged with 10 ml of PBS, yielding approximately 5 mL of recovery. Cells were collected from BALF by centrifugation at 500 × g for 10 min at 4°C. The concentration of nucleated cells in BALF was determined by counting in Türk’s solution, and approximately 3 × 106 BALF cells were prepared for cytokine gene expression evaluation. The supernatant was frozen at − 80°C for further detection of viral RNA in real-time PCR experiments.
Cytokine gene expressions. RNA was extracted from BALF cells using an RNeasyMini kit (Qiagen, Germany), according to the manufacturer’s instructions. RNA integrity numbers (RIN) determined using an Agilent 2100 Bioanalyzer (Agilent, USA) ranged from 7.5 to 9.9 (mean RIN was 8.9). The purity and quantity of RNA were measured on a NanoPhotometer (Implen, Germany). A range of 78 to 577 ng/μL described the RNA concentrations, the mean RNA concentration being 270.6 ng/μL. Synthesis of cDNA from total RNA was carried out using M-MLV reverse transcriptase (Invitrogen, USA), PCR-dNTP mix (Invitrogen), Recombinant RNasin Ribonuclease Inhibitor (Promega, USA), and the Random Primer (Invitrogen). The protocol for cDNA synthesis was as follows: 10 min at 25°C, 60 min at 42°C, and 15 min at 70°C.
A quantitative real-time PCR was conducted to detect relative mRNA expression levels using β-actin as the internal control gene and a QuantiTect SYBR Green PCR Kit (Qiagen), according to the manufacturer’s instructions. Primer sequences for β-actin were those designed by Duvigneau et al. (6), primer sequences for IL-1β, IL-6, and IFN-γ were as in Meurens et al. (20), and those for IL-8 and IFN-α were first specified by Rola-Łuszczak et al. (25). Reaction mixtures included 22 μL of total volume of reaction mix, and 3 μL cDNA. The PCR was performed with an initial denaturation step at 95°C for 15 min, followed by 40 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, using a Stratagene Mx3005P thermocycler (Agilent). After amplification, a melting curve analysis of the real-time PCR products was performed to confirm the presence of the expected amplicons.
The relative expression (R) of target genes was calculated using the following equation: R = (Etarget)ΔCt target (control−sample)/(Eref)ΔCt ref (control−sample) which expresses mRNA from the cells of infected pigs relative to the cells from control pigs after normalising to β-actin (23).
PRRSV quantification with real-time PCR in BALF. Viral RNA was extracted from BALF supernatant with a QIAamp viral RNA mini kit (Qiagen), according to the manufacturer’s instructions. For the detection of PRRSV in BALF samples, a real-time PCR was performed with EZ-PRRSV MPX 4.0 master mix and reagents (Tetracore Inc., USA). For consistency, each plate contained Tetracore quantification standards and control sets for use with the Tetracore reagents. Viral RNA concentration was expressed as the number of RNA copies/ml of sample. The virus concentration was calculated by linear extrapolation of the cycle threshold values against a standard curve generated from serial dilutions of quantified standards (1 × 102 to 1 × 105 copies per μL).
Statistical analysis. The obtained data were subjected to the Shapiro–Wilk test for normality and Levene’s test for equality of variances. After rejection of normality and variance homogeneity, differences between viral loads were compared between groups by the nonparametric Mann–Whitney U test. For the mean mRNA levels of cytokines in the BALF cells, the nonparametric Kruskal–Wallis test with post hoc Dunn’s test were used for analysing the specific sample pairs. Differences were considered significant at p < 0.05. All calculations were performed with Statistica 8.0 (Tibco, USA).
Results
Clinical signs, gross pathology. Seven pigs infected with PRRSV alone had short-term fever (40.1°C–41.5°C). In the group inoculated with both viruses, long-term fever (over three days) was observed in 8 out of 11 animals. The most severe course of the disease was observed in the co-infected piglets of which 10 out of 11 demonstrated clinical signs of infection. In PRRSV-infected pigs, clinical signs were recorded in 8 out of 11 animals. No symptoms were observed in control pigs. All animals from the PRRSV-infected and co-infected groups had lung gross lesions typical for PRRSV and/or SIV infection, exemplified most severely by the co-infected group at 21 dpi.
PRRSV RNA quantification. Mean viral loads, measured as viral genomic copies (copies of RNA/mL of BALF), were quantified at each time point, with the peaks of viral load coming at 4 dpi and 21 dpi in the PRRSV-infected and co-infected groups, respectively (Fig. 1). The mean concentration of the PRRSV load in BALF of PRRSV-infected pigs was higher than that of pigs infected with PRRSV and SIV over the entire study; however, statistical significance was observed only at 4 dpi (p < 0.05). No viral RNA was detected in control animals (data not shown).
Fig. 1.
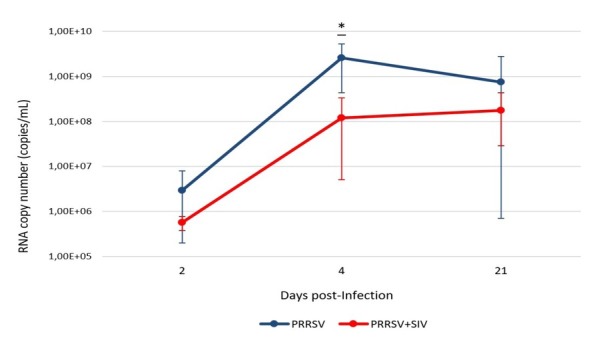
Mean, minimum and maximum value of PRRSV copy number (copies/mL) in BALF of pigs infected with PRRSV or co-infected with PRRSV and SIV over the observation period. Asterisks indicate statistically significant differences between groups at the same number of days post infection (p < 0.05)
Cytokine gene expression in BALF cells. Detailed data on the mean (± SEM) mRNA levels of cytokine in the BALF cells of experimental pigs are summarised in Fig. 2. In general, statistically significant expression changes were observed in four gene transcripts in pigs infected with PRRSV alone and in five gene transcripts in pigs infected with both viruses along the time course analysed. Overall, the highest number of differentially expressed genes was observed at 4 dpi, and only at this time point were significant differences in cytokine gene expression observed between pigs solely infected with PRRSV and those which were co-infected.
Fig. 2.
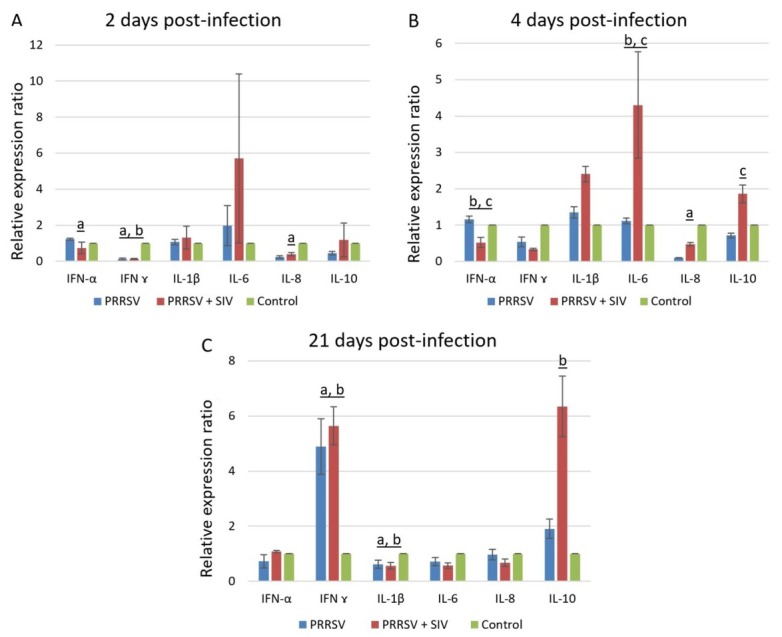
Mean (± SEM) cytokines mRNA expression profile in BALF cells following infection with PRRSV or co-infection with PRRSV and SIV at 2 dpi (A), 4 dpi (B), and 21 dpi (C). a – p < 0.05 between PRRSV-infected group and control group; b – p < 0.05 between co-infected group and control group; c – p < 0.05 between PRRSV-infected group and co-infected group
On the 2nd dpi in pigs challenged with PRRSV alone, significant mRNA upregulation of IFN-α and downregulation of IL-8 and IFN-γ mRNA was observed (p < 0.05) as compared to the controls. Also, in co-infected pigs significant downregulation of IFN-γ mRNA was recorded when evaluated against the controls.
On the 4th dpi in PRRSV-infected pigs, only IL-8 mRNA expression was significantly changed compared to the controls, whereas in co-infected pigs significant mRNA downregulation of IFN-α and upregulation of IL-6 was recorded when contrasted with the control pigs. Moreover, for the co-infected pigs at this time point, the transcript level of IFN-α was significantly downregulated, and IL-6 and IL-10 were significantly upregulated in BALF cells as compared to singly infected animals (p < 0.05).
On day 21, only the transcript levels of IFN-γ, IL-1β, and in some cases IL-10 were still significantly changed as compared to uninfected pigs. With respect to IFN-γ mRNA expression, its relative level was significantly higher in the PRRSV-infected and co-infected groups as compared to the controls. In the co-infected group, the transcript level of IL-10 was significantly greater than that of uninfected animals. By contrast, in the PRRSV-infected and co-infected groups significant mRNA down-regulation of IL-1β relative to the control pigs was observed.
Discussion
In this investigation, we studied the expression of local inflammatory mediators in BALF cells of pigs following infection with PRRSV or co-infection with PRRSV and SIV under in vivo experimental challenge. The innate immune response plays a pivotal role in controlling pathogen spread and in orchestrating effective adaptive immune responses. Type I IFNs (IFN-α/β) are central elements of antiviral immunity, and their production is critical to activation of the antiviral innate immune response and regulation of the induction of the adaptive immune response (29). It has been documented previously that PRRSV is a poor inducer of IFN-α, and its level remains low throughout the course of infection (9, 14). Here, IFN-α transcript levels were slightly increased in BALF cells of pigs infected with PRRSV alone, however, only early post infection (2 dpi). Our findings are in agreement with earlier experiments and support the theory that PRRSV has developed efficient strategies to circumvent the IFN response (7, 9). Also in pigs infected with both viruses there was a dampened expression profile of IFN-α transcript level observed, most noticeably at 4 dpi. Similar results were found in PCLS infected with PRRSV and SIV or superinfected with both viruses (5). Interestingly, in the same study, a significant increase in IFN-α transcript expression in PAMs under superinfection conditions was recorded (5). According to Dobrescu et al. (5), the higher expression of IFN-α transcripts in superinfection conditions could be a result of both additive effects and viral kinetics. Regarding IFN-γ, downregulation at 2 dpi and upregulation of its expression at 21 dpi was observed in the PRRSV-infected group. The delayed and dampened onset of cellular immune response in PRRSV-infected pigs was also documented in previous studies (4, 9, 19, 31). In co-infected pigs, the same trend of IFN-γ expression profile was found. SIV infection is known to induce several inflammatory cytokines including IFN-γ (3, 15, 16, 24). Therefore, it can be hypothesised that PRRSV could affect SIV-induced IFN-γ production in the BALF cells of pigs. Furthermore, increased mRNA levels of IFN-γ at 21 dpi coincided with a decreased amount of PRRSV RNA in BALF in pigs infected with PRRSV. Similarly in a previous study, the last detection of viraemia in PRRSV-infected pigs corresponded to the appearance of interferon-γ-secreting cells (IFN-γ-SC) (4). This finding indicates that IFN-γ could play an important role in controlling PRRSV infection.
IL-1β, an endogenous pyrogen, is one of the main cytokines produced by alveolar macrophages and has a variety of immune regulation functions, including participation in the body’s defence and promoting systemic inflammatory reaction. In this study, a dampened expression profile of IL-1β transcript levels in BALF cells of PRRSV-infected pigs was detected at each time point. Our results are in line with previous in vivo studies, in which no significant differences of IL-1β mRNA expression in peripheral blood mononuclear cells (PBMCs), tracheobronchial lymph nodes (TBLNs), or lungs of pigs infected with PRRSV were found (21, 30). Interestingly, in co-infected pigs upregulation of IL-1β expression at 4 dpi was detected, however, when compared with the control group the difference was not significant. Similar results were reported by Tu et al. (30), who investigated the mRNA expression profiles of Toll-like receptors (TLRs) and related inflammatory cytokines including IL-1β in PBMCs from pigs infected with PRRSV, porcine circovirus type 2 (PCV2), or both. Their findings implicated that the exacerbated clinical signs and lesion scores in co-infected pigs were related to the increased secretion of IL-1β (30).
Regarding IL-8, its mRNA expression level was downregulated at 2 and 4 dpi and remained unchanged at 21 dpi in PRRSV-infected pigs. IL-8 is a neutrophil chaemotactic factor and plays a central role in the inflammation process. According to the majority of studies, PRRSV induces IL-8 production in PAMs, BALF, serum, and TBLNs (12, 17). Contradictory findings between studies may be related to the use of different PRRSV isolates for inoculation. One of the latest pieces of research demonstrated that expression of IL-8 is strain-dependent. Rola-Łuszczak et al. (25) found that PRRSV type 1 strains including a subtype 1 strain isolated in Denmark and a Russian subtype 2 isolate either did not alter or downregulated the IL-8 signalling pathway, whereas upregulation was observed after infection with a subtype 2 Belarusian isolate. In our study in co-infected pigs, no significant differences of IL-8 expression were observed at any of the time points. Previously, swine influenza virus co-infection with Bordetella bronchiseptica resulted in a considerable upregulation of IL-8 expression compared to its expression in single viral or bacterial infection, and pneumonia induced by SIV was associated with production of IL-8 (16, 18). It can be hypothesised that PRRSV impairs SIV-induced proinflammatory cytokine production.
In the present study, among the innate proinflammatory cytokines evaluated, only the IL-6 mRNA level was markedly upregulated at 4 dpi, the difference being observed only in co-infected pigs. Similarly, Dobrescu et al. (5) found that in PCLS, IL-6 mRNA expression level was upregulated in response to SIV infection but not to PRRSV infection, whereas in PAMs, the upregulation was more significant in co-infection and superinfection conditions than in single viral infection. In contrast, Gómez-Laguna et al. (11) showed a significant correlation between expression of IL-6 and lung lesions in PRRSV-infected pigs. The differences between studies may be related to the diversity of PRRSV isolates used for inoculation, as different isolates may induce different expression patterns of genes involved in the IL-6 signalling pathway (25). It is worth emphasising that the increased IL-6 mRNA level at 4 dpi in co-infected pigs coincided with a significantly lower PRRSV load in BALF. It may suggest that the IL-6 contributes virus clearance during PRRSV and SIV infection.
IL-10 is an immunomodulatory cytokine that is able to inhibit the synthesis and release of other cytokines (22). In the present study, no significant changes in IL-10 transcript level were detected in PRRSV-infected pigs at any time-point post infection. Previous reports suggested that different PRRSV isolates were able to induce different patterns of IL-10 expression (9, 10). However, in pigs infected with both viruses, the IL-10 transcript level was significantly upregulated at 4 dpi compared to PRRSV-infected pigs and at 21 dpi compared to controls. In a previous study in which the same swine influenza virus subtype (H1N1) was used for inoculation, no changes in mRNA expression of IL-10 in SIV-infected pigs were demonstrated (16). These results imply that there is a positive synergistic interaction between the viruses in respect to IL-10 mRNA expression in concomitant infections.
In conclusion, our results indicate that infection with PRRSV alone and with SIV affected the expression of IFN-α and delayed the onset of IFN-γ expression. Together, these findings highlight the interference of PRRSV with the innate and cell-mediated immune response in the infected host. In addition, the results of our study show that co-infection with PRRSV and SIV demonstrates additive effects on the mRNA expression of IL-6 and IL-10. However, the impact of such synergy on the viral load and severity of clinical disease are not clear and require further investigation.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This work was supported by the National Science Centre (DEC-2014/13/B/NZ6/02566) and by the KNOW (Leading National Research Centre) Scientific Consortium “Healthy Animal - Safe Food”, under Ministry of Science and Higher Education resolution no. 05-1/KNOW2/2015.
Animal Rights Statement: The experiment was approved by the Local Ethics Committee at the University of Life Sciences in Lublin.
References
- 1.Barbé F., Atanasova K., Van Reeth K.. Cytokines and acute phase proteins associated with acute swine influenza infection in pigs. Vet J. 2011;187:48–53. doi: 10.1016/j.tvjl.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi Y.K., Goyal S.M., Joo H.S.. Retrospective analysis of etiologic agents associated with respiratory diseases in pigs. Can Vet J. 2003;44:735–737. [PMC free article] [PubMed] [Google Scholar]
- 3.Czyżewska-Dors E., Dors A., Kwit K., Stasiak E., Pomorska-Mól M.. Pig lung immune cytokine response to the swine influenza virus and the Actinobacillus pleuropneumoniae infection. J Vet Res. 2017;61:259–265. doi: 10.1515/jvetres-2017-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Díaz I., Darwich L., Pappaterra G., Pujols J., Mateu E.. Immune responses of pigs after experimental infection with a European strain of porcine reproductive and respiratory syndrome virus. J Gen Virol. 2005;86:1943–1951. doi: 10.1099/vir.0.80959-0. [DOI] [PubMed] [Google Scholar]
- 5.Dobrescu I., Levast B., Lai K., Delgado-Ortega M., Walker S., Banman S., Townsend H., Simon G., Zhou Y., Gerdts V., Meurens F.. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Vet Microbiol. 2014;169:18–32. doi: 10.1016/j.vetmic.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duvigneau J.C., Hartl R.T., Groiss S., Gemeiner M.. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J Immunol Methods. 2005;306:16–27. doi: 10.1016/j.jim.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 7.Dwivedi V., Manickam C., Binjawadagi B., Linhares D., Murtaugh M.P., Renukaradhya G.J.. Evaluation of immune responses to porcine reproductive and respiratory syndrome virus in pigs during early stage of infection under farm conditions. Virol J. 2012;9:45. doi: 10.1186/1743-422X-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fablet C., Marois-Crehan C., Simon G., Grasland B., Jestin A., Kobisch M., Madec F., Rose N.. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: a cross-sectional study. Vet Microbiol. 2012;157:152–163. doi: 10.1016/j.vetmic.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 9.García-Nicolás O., Rosales R.S., Pallarés F.J., Risco D., Quereda J.J., Graham S.P., Frossard J.P., Morgan S.B., Steinbach F., Drew T.W., Strickland T.S., Salguero F.J.. Comparative analysis of cytokine transcript profiles within mediastinal lymph node compartments of pigs after infection with porcine reproductive and respiratory syndrome genotype 1 strains differing in pathogenicity. Vet Res. 2015;46:34. doi: 10.1186/s13567-015-0161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimeno M., Darwich L., Diaz I., de la Torre E., Pujols J., Martin M., Inumaru S., Cano E., Domingo M., Montoya M., Mateu E.. Cytokine profiles and phenotype regulation of antigen presenting cells by genotype-I porcine reproductive and respiratory syndrome virus isolates. Vet Res. 2011;42:9. doi: 10.1186/1297-9716-42-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-Laguna J., Salguero F.J., Barranco I., Pallarés F.J., Rodríguez-Gómez I.M., Bernabé A., Carrasco L.. Cytokine expression by macrophages in the lung of pigs infected with the porcine reproductive and respiratory syndrome virus. J Comp Pathol. 2010;142:51–60. doi: 10.1016/j.jcpa.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo B., Lager K.M., Henningson J.N., Miller L.C., Schlink S.N., Kappes M.A., Kehrli M.E. Jr, Brockmeier S.L., Nicholson T.L., Yang H.C., Faaberg K.S.. Experimental infection of United States swine with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. Virology. 2013;435:372–384. doi: 10.1016/j.virol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbur P.G., Paul P.S., Frey M.L., Landgraf J., Eernisse K., Meng X.J., Lum M.A., Andrews J.J., Rathje J.A.. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 14.Huang C., Zhang Q., Feng W.H.. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015;202:101–111. doi: 10.1016/j.virusres.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatri M., Dwivedi V., Krakowka S., Manickam C., Ali A., Wang L.. Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: a potential animal model for human H1N1 influenza virus. J Virol. 2010;84:11210–11218. doi: 10.1128/JVI.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalczyk A., Pomorska-Mól M., Kwit K., Pejsak Z., Rachubik J., Markowska-Daniel I.. Cytokine and chemokine mRNA expression profiles in BALF cells isolated from pigs single infected or co-infected with swine influenza virus and Bordetella bronchiseptica. Vet Microbiol. 2014;170:206–212. doi: 10.1016/j.vetmic.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Du Y., Wang H., Du L., Feng W.H.. Porcine reproductive and respiratory syndrome virus (PRRSV) up-regulates IL-8 expression through TAK-1/JNK/AP-1 pathways. Virology. 2017;506:64–72. doi: 10.1016/j.virol.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loving C.L., Brockmeier S.L., Vincent A.L., Palmer M.V., Sacco R.E., Nicholson T.L.. Influenza virus coinfection with Bordetella bronchiseptica enhances bacterial colonization and host responses exacerbating pulmonary lesions. Microb Pathog. 2010;49:237–245. doi: 10.1016/j.micpath.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Meier W.A., Galeota J., Osorio F.A., Husmann R.J., Schnitzlein W.M., Zuckermann F.A.. Gradual development of the interferon-γ response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology. 2003;309:18–31. doi: 10.1016/s0042-6822(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 20.Meurens F., Berri M., Auray G., Melo S., Levast B., Virlogeux-Payant I., Chevaleyre C., Gerdts V., Salmon H.. Early immune response following Salmonella enterica subspecies enterica serovar Typhimurium infection in porcine jejunal gut loops. Vet Res. 2009;40:5. doi: 10.1051/vetres:2008043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miguel J.C., Chen J., Van Alstine W.G., Johnson R.W.. Expression of inflammatory cytokines and Toll-like receptors in the brain and respiratory tract of pigs infected with porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2010;135:314–319. doi: 10.1016/j.vetimm.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Moore K.W., de Waal Malefyt R., Coffman R.L., O’Garra A.. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl M.W.. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pomorska-Mól M., Markowska-Daniel I., Kwit K., Czyżewska E., Dors A., Rachubik J., Pejsak Z.. Immune and inflammatory response in pigs during acute influenza caused by H1N1 swine influenza virus. Arch Virol. 2014;159:2605–2614. doi: 10.1007/s00705-014-2116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rola-Łuszczak M., Materniak-Kornas M., Pluta A., Podgórska K., Nielsen J., Stadejek T., Kuźmak J.. Transcriptional profiles of PBMCs from pigs infected with three genetically diverse porcine reproductive and respiratory syndrome virus strains. Mol Biol Rep. 2018;45:675–688. doi: 10.1007/s11033-018-4204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossow K.D.. Porcine reproductive and respiratory syndrome. Vet Pathol. 1998;35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- 27.Solano G.I., Segalés J., Collins J.E., Molitor T.W., Pijoan C.. Porcine reproductive and respiratory syndrome virus (PRRSV) interaction with Haemophilus parasuis. Vet Microbiol. 1997;55:247–257. doi: 10.1016/S0378-1135(96)01325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanawongnuwech R., Brown G.B., Halbur P.G., Roth J.A., Royer R.L., Thacker B.J.. Pathogenesis of porcine reproductive and respiratory syndrome virus-induced increase in susceptibility to Streptococcus suis infection. Vet Pathol. 2000;37:143–152. doi: 10.1354/vp.37-2-143. [DOI] [PubMed] [Google Scholar]
- 29.Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H.. Type I interferons (α/β) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 30.Tu P.Y., Tsai P.C., Lin Y.H., Liu P.C., Chang H.L., Kuo T.Y., Chung W.B.. Expression profile of Toll-like receptor mRNA in pigs co-infected with porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Res Vet Sci. 2015;98:134–141. doi: 10.1016/j.rvsc.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Zuckermann F.A., Garcia E.A., Luque I.D., Christopher-Hennings J., Doster A., Brito M., Osorio F.. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet Microbiol. 2007;123:69–85. doi: 10.1016/j.vetmic.2007.02.009. [DOI] [PubMed] [Google Scholar]


