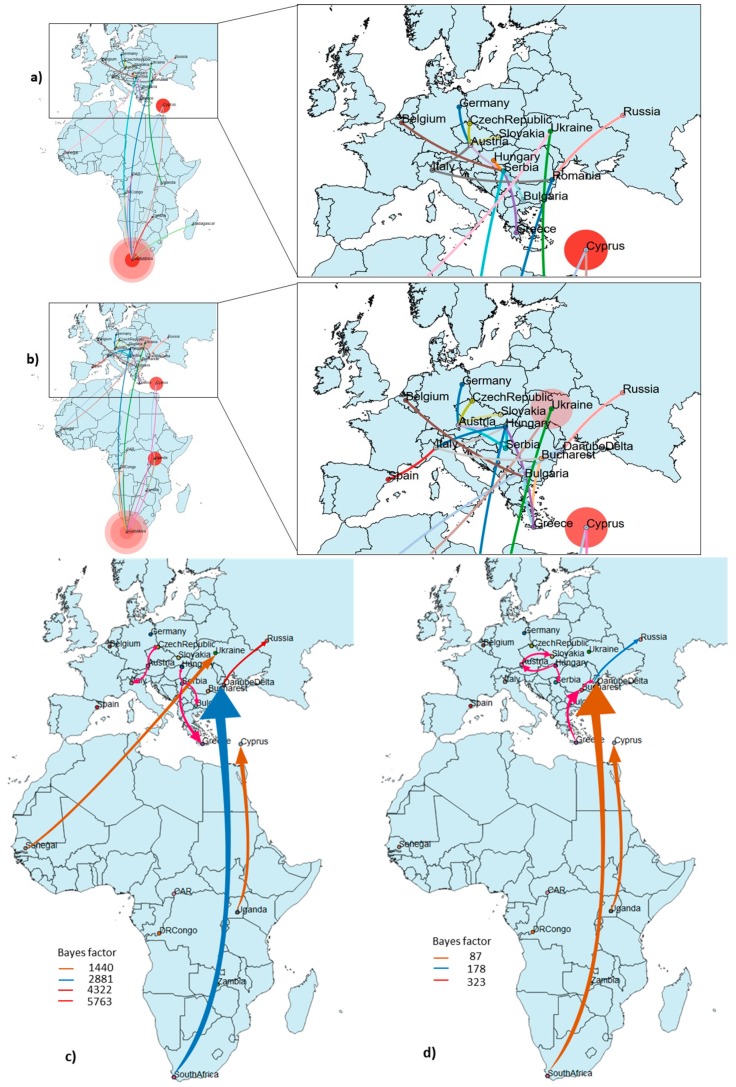
Figure 3.
Spatial dynamics of the WNV lineage 2 reconstructed from the (a) complete genome and (b) partial NS5 based on MCC tree, a flexible demographic prior (coalescent Gaussian Markov Random field Bayesian Skyride model, GMRF) with location states and a Bayesian Stochastic Search Variable Selection (BSSVS) with location states. The directed lines between locations connect the sources and target countries (color coded) of viral strains and represent branches in the MCC tree along which the relevant location transition occurs. Location circle diameters are proportional to the square root of the number of MCC branches maintaining a particular location state at each time-point. Migration pattern of WNV between Africa and Europe and within Europe based on Bayes factor (BF) test for significant non-zero rates using complete genome (c) and partial NS5 dataset (d). Viral migration patterns are indicated between the different regions and countries and are proportional to the strength of the transmission rate (Bayes factor [BF]). The color of the connections indicates the origin and the direction of migration and are proportional with the strength of connections. Only well supported paths between locations are shown.