Abstract
RfaH, a two-domain protein from a universally conserved NusG/Spt5 family of regulators, is required for the transcription and translation of long virulence and conjugation operons in many Gram-negative bacterial pathogens. Escherichia coli RfaH action is controlled by a unique large-scale structural rearrangement triggered by recruitment to transcription elongation complexes through a specific DNA element. Upon recruitment, the C-terminal domain of RfaH refolds from an α-hairpin, which is bound to RNA polymerase binding site within the N-terminal domain, into an unbound β-barrel that interacts with the ribosome. Although structures of the autoinhibited (α-hairpin) and active (β-barrel) states and plausible refolding pathways have been reported, how this reversible switch is encoded within RfaH sequence and structure is poorly understood. Here, we combined hydrogen-deuterium exchange measurements by mass spectrometry and nuclear magnetic resonance with molecular dynamics to evaluate the differential local stability between both RfaH folds. Deuteron incorporation reveals that the tip of the C-terminal hairpin (residues 125–145) is stably folded in the autoinhibited state (∼20% deuteron incorporation), whereas the rest of this domain is highly flexible (>40% deuteron incorporation), and its flexibility only decreases in the β-folded state. Computationally predicted ΔG agree with these results by displaying similar anisotropic stability within the tip of the α-hairpin and on neighboring N-terminal domain residues. Remarkably, the β-folded state shows comparable structural flexibility than nonmetamorphic homologs. Our findings provide information critical for understanding the metamorphic behavior of RfaH and other chameleon proteins and for devising targeted strategies to combat bacterial infections.
Significance
Infections caused by Gram-negative bacteria are a worldwide health threat because of rapid acquisition of antibiotic resistance. RfaH, a protein essential for virulence in several Gram-negative pathogens, undergoes a large-scale structural rearrangement in which one RfaH domain completely refolds. Refolding transforms RfaH from an inactive state that restricts RfaH recruitment to a few target genes into an active state that binds to, and couples, transcription and translation machineries to elicit dramatic activation of gene expression. However, the molecular basis of this unique conformational change is poorly understood. Here, we combine molecular dynamics and structural biology to unveil the hotspots that differentially stabilize both states of RfaH. Our findings provide novel insights that will guide the design of inhibitors blocking RfaH action.
Introduction
Metamorphic proteins can access multiple structurally different and yet energetically stable states in solution (1), directly challenging the uniqueness of the native state considered in the thermodynamic hypothesis proposed by Anfinsen (2), typically interpreted as one sequence-one fold. This process takes place by major architectural rearrangements and is commonly related to changes in protein function and dynamics (3).
Escherichia coli RfaH is a metamorphic protein branching from the universally conserved NusG family of transcription elongation factors (4,5), which enable processive RNA synthesis by RNA polymerase (RNAP) while simultaneously coupling it to concurrent processes (6). In NusG proteins, this coupling is achieved by two domains connected via a flexible linker. The N-terminal domain (NTD) is a structurally conserved α/β sandwich that freely binds the transcription elongation complex (TEC) by contacting the two largest RNAP subunits to form a processivity clamp around the DNA (7, 8, 9); the C-terminal domain (CTD) is commonly folded as a small five-stranded, antiparallel β-barrel able to interact with diverse cellular targets (6,10).
Despite sharing 41% sequence similarity, the structure of free E. coli RfaH displays striking differences from its paralog NusG. Instead of the canonical β-barrel, RfaH CTD is folded as an α-helical hairpin (αRfaH), which is tightly bound to the NTD, occluding the RNAP-binding site (Fig. 1; (11,12)). This autoinhibition is relieved upon domain dissociation, which is elicited during RfaH recruitment to the TEC or when the interdomain linker is cleaved, and the released CTD spontaneously refolds into the canonical β-barrel structure (βRfaH) observed in most NusG proteins (Fig. 1 A; (12, 13, 14, 15)). This unique structural transformation is required to restrict RfaH action to just a few genes. Autoinhibited RfaH is specifically recruited to a paused TEC in which an ops sequence in the nontemplate DNA strand forms a surface-exposed hairpin (16,17); subsequently, domain dissociation leads to RfaH activation by CTD fold switching to attain a NusG-like structure (12,13) and by binding of the NTD to its high-affinity binding site on RNAP (7,13). Remarkably, RfaH transformation is fully reversible as the autoinhibited state is restored upon RfaH dissociation from the RNAP (14).
Figure 1.
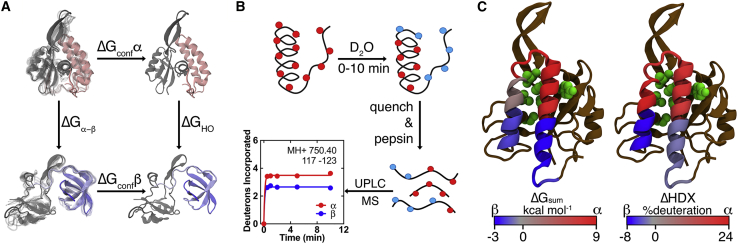
Computational and experimental assessment of local stability in the metamorphic protein RfaH. (A) Shown is the thermodynamic cycle of the confinement MD approach (25) used to estimate the per-residue ΔG between both RfaH folds. The autoinhibited form with the CTD in the α-state (pink, PDB: 5OND) and the active form with the CTD in the refolded β-state (light blue, PDB: 6C6S) are confined toward a deeply minimized state through a harmonic constraint (ΔGconf), allowing calculation of the difference in free energy between these structures (ΔGHO). (B) Shown is a scheme of HDXMS experiments (52). Both full-length RfaH and the isolated CTD were incubated in deuterated buffer for different reaction times, quenched, and pepsin digested for analyzing the local extent of deuteron incorporation. (C) Shown are cartoon representations of the full-length αRfaH, in which the CTD covers the RNAP-binding residues from the NTD (green), summarizing our findings from simulations (left) and experiments (right) on the differential local stability toward the α- (red) and β-state (blue) of the CTD. To see this figure in color, go online.
Because the trigger for RfaH metamorphosis is the complete ops-paused TEC (14,17), it is challenging to study this process experimentally. Instead, most of the thermodynamic and kinetic studies have used computational approaches to directly explore this fold switch by simulating either the isolated CTD (18, 19, 20) or the entire RfaH protein (21,22). Although the RfaH CTD is composed of only 51 residues (residues 112–162), its α-to-β transition has not been observed through conventional MD (19) but through the use of enhanced sampling techniques (18,20,22) or reduced system granularity (21,23). A way to circumvent such computing barriers is the use of confinement simulations, which rely on a discontinuous thermodynamic integration to estimate the absolute free energy of a clearly defined energy well (24). By evaluating two alternative states within the same system, one can calculate the energy required for the structural interconversion without explicitly observing such a transition (25).
In this work, we employed hydrogen-deuterium exchange mass spectrometry (HDXMS), [1H,15N] heteronuclear single quantum coherence (HSQC)-based NMR spectroscopy, and confinement molecular dynamics (MD) to assess the differences in local stability between the autoinhibited and active folds of RfaH. By using deuterium as a probe to experimentally assess the solvent accessibility of peptides and individual backbone amides along RfaH in combination with simulations to estimate per-residue free-energy changes upon refolding (Fig. 1), we aimed to trace back localized regions, preferentially favoring the α- or β-fold, and determine how this reversible switch is encoded within RfaH sequence.
Materials and Methods
Confinement simulations
Structures of RfaH were built using the crystal structure of αRfaH (Protein Data Bank, PDB: 5OND) (17) and cryo-electron microscopy (cryoEM) composition of βRfaH (PDB: 6C6S) (13). Rosetta3 suite was used to model and relax the flexible interdomain linker in both structures (26). To calculate the free energy between both structures, implicit solvent MD simulations were performed on Amber16 along with CUDA as previously reported (27,28). Although water molecules are not being directly computed, the per-residue root mean-square fluctuations of RfaH in both structures is comparable to explicit solvent models previously reported (29, 30, 31). Furthermore, this method requires an initial step of deep energy minimization of the system, suggesting that the phase change the solvent would undergo during this process would result in more artifactual dynamics than those arising from a steady solvation potential. In total, 26 independent simulations were performed per system. For each, a harmonic position-restraining potential was used to drive the atoms toward a deeply energy-minimized configuration (see Supporting Materials and Methods) for either the α- or β-state of RfaH. The stiffness of this potential was increased exponentially from mostly free (2.5∙10−5 kcal mol−1 Å−2) to highly restrained (419.2 kcal mol−1 Å−2). Fluctuations and free energy were calculated for each basin as previously reported (Fig. S1; (27)). Briefly, the estimated free energy is the result of a thermodynamic cycle (Fig. 1 A) comprising two energy terms: confinement and convert. The former corresponds to the work applied by the external potential, exerted through all 26 simulations. Once confined, the magnitude of this work depends solely on the stiffness of the harmonic potential and not the configuration of the system (Fig. S1). Thus, beyond this point, the confinement free-energy difference between two distinct basins converges to a single value. The convert term corresponds to the free-energy difference between the already confined states, represented as the deeply energy-minimized configuration for each basin. This is calculated by using a harmonic oscillator approximation (27), in which the absolute free energy of each state is calculated from the canonical partition function for a system of vibrating particles, whose frequencies are obtained from normal-mode analysis for both deeply minimized configurations (25). A per-residue free-energy decomposition scheme was also used as indicated previously (28). Formula derivations are described in the Supporting Materials and Methods.
Gene expression and protein purification
All protein sequences were encoded in plasmids harboring either a tobacco etch virus protease cleaving site (TEV) or thrombin cleaving site. Full-length E. coli RfaH was encoded in pIA777, a derivative of pET36b(+) containing its NTD-TEV-CTD-[His6] (32). The isolated CTD (i.e., RfaH residues 101–162) was harbored in a pETGB1A vector, containing [His6]-GB1-TEV-CTD (12). E. coli NusG was encoded in pIA244, a derivate of pET33 (33), encoding [His6]-thrombin cleaving site-NusG. For protein production, the E. coli BL21 (DE3) strain was used. Bacteria were grown at 37°C until reaching an optical density at 600 nm (OD600) = 0.6–0.7, and protein expression was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (US Biological, Salem, MA) at 30°C overnight in the case of RfaH and its isolated CTD or 30°C for 3 h for NusG. The cells were harvested by centrifugation at 4°C.
Cells were disrupted by sonication at a high intensity in buffer A containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 10 mM imidazole (pH 7.5). The supernatant was obtained by centrifugation at 12,000 × g for 30 min, loaded onto a His-Trap HP column (GE Healthcare, Chicago, IL), washed with buffer A, and then eluted using a linear gradient of 10–250 mM imidazole in the same buffer. For isolated CTD, this eluate was incubated in buffer A with a noncleavable His-tagged TEV protease at 4°C overnight in a ratio of 20:1 mg of CTD/TEV protease. This mixture was then separated using another His-Trap HP column, collecting its flow-through enriched in isolated RfaH CTD. Purity of protein samples was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Finally, all proteins were subjected to size exclusion chromatography before all experiments. This was performed on a Sephadex S75 column (GE Healthcare) connected onto an ÄKTA fast protein liquid chromatography (GE Healthcare), using 20 mM Tris-HCl (pH 7.9), 40 mM KCl, 5.0 mM MgCl2, 1.0 mM β-mercaptoethanol, and 6.0% (v/v) glycerol as the mobile phase.
HDXMS
HDXMS was performed on each protein using a Synapt G2Si system with hydrogen-deuterium exchange (HDX) technology (Waters, Milford, MA) as in previous works (34). In these experiments, 5 μL of protein solution at an initial concentration of 11 μM were allowed to exchange at 25°C for 0–10 min in 55 μL of deuterated buffer containing 20 mM Tris-HCl (pH 7.9), 40 mM KCl, 5.0 mM MgCl2, 1.0 mM β-mercaptoethanol, and 6.0% (v/v) glycerol. Then, reactions were quenched for 2 min at 1°C using an equal volume of a solution containing 2 M GndHCl and 1% formic acid (pH 2.66). The quenched samples were injected onto a custom-built pepsin-agarose column (Thermo Fisher Scientific, Waltham, MA), and the resulting peptic peptides were separated by analytical chromatography at 1°C. The analytes were electrosprayed into a Synapt G2-Si quadrupole time-of-flight mass spectrometer (Waters) set to MSE-ESI + mode for initial peptide identification and to Mobility-time-of-flight-ESI + mode to collect HDX data. Deuterium uptake was determined by calculating the shift in the centroids of the mass envelopes for each peptide compared with the undeuterated controls, using the DynamX 3.0 software (Waters; Tables S1 and S2). The difference in deuteron incorporation of overlapping peptides was used for calculating the incorporation of overhanging regions when the difference in mass exceeded five times its uncertainty (see Supporting Materials and Methods and Tables S3–S6). Incorporation was fitted to a single negative exponential per region (Fig. S3) to obtain the maximal deuteron incorporation per peptide, which was expressed as a percentage over the total number of amides. For the formulation, see Supporting Materials and Methods, and for the raw and processed data, see Tables S3–S5.
Hydrogen-deuterium exchange HSQC spectroscopy
15N-labeled full-length RfaH and RfaH CTD were produced as described (12). In brief, expression was carried out by growing E. coli in M9 minimal medium (35,36) supplemented with (15NH4)2SO4 (Campro Scientific, Berlin, Germany) as the only nitrogen source. For the HDX experiments, the proteins were in 25 mM HEPES (pH 7.5), 60 mM NaCl, 5% (v/v) glycerol, and 1 mM dithiothreitol, and spectra were recorded at 288 K for full-length RfaH and 298 K for the isolated CTD on Bruker Avance 700 MHz and Avance 800 MHz spectrometers using cryo-cooled triple-resonance bearing pulse field-gradient capabilities. After lyophilization proteins were dissolved in 500 μL D2O (99.98%), the decay of signal intensities was observed in a series of [1H,15N]-HSQC spectra over 24 h. After the experiment, the pD was measured using a pH meter. Resonance assignment of backbone amide protons of RfaH were taken from a previous study (12). Exchange rates were determined by fitting the signal decay to a monoexponential curve (Table S6) using only signals whose intensity had not completely decayed within the first 90 min, so that at least five data points were used for fitting. The pD was corrected by adding 0.4 units to the experimentally determined value. The protection factors were calculated by dividing the experimental exchange rates (kex) by the intrinsic exchange rates calculated from the amino acid sequence (krc) and experimental conditions with tabulated parameters and were finally converted to ΔG values (Table S6; (37,38)).
Data availability
All experimental and computational data are available from the corresponding authors upon request.
Results
Confinement MD show localized differential stability
To computationally ascertain the difference in local stability between both native states of RfaH, confinement MD simulations were used to estimate their global and local free-energy differences. Given that interdomain interactions are critical for the stability of the CTD in the autoinhibited state (12), both structural states were modeled on the full-length RfaH to perform the confine-convert-release (CCR) approach (27,28). These models were built in both states using the crystallographic αRfaH structure (corresponding to the autoinhibited state with NTD and CTD in the α-fold (17)) and the cryoEM βRfaH structure (corresponding to the activated, open state with NTD and CTD in the β-fold (13)), further refining the flexible loop connecting both domains using the knowledge-based Rosetta software (26). Then, these refined structures were used for confinement MD, thoroughly exploring the fluctuations from mostly free to highly restrained states, integrating and then decomposing the free-energy difference between αRfaH and βRfaH required for such process (Figs. 2 and S1).
Figure 2.
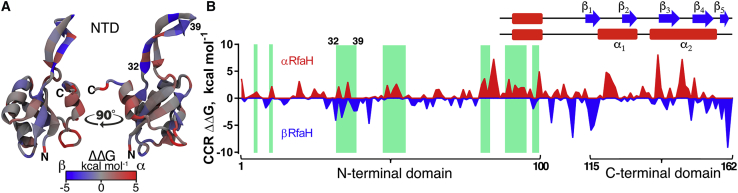
Confinement MD estimates anisotropic per-residue energetic contributions behind RfaH metamorphosis. Using the CCR approximation, the contribution toward stabilizing either αRfaH (red) or βRfaH (blue) state was calculated (27,28) and displayed on the NTD structure (A) and RfaH sequence (B). The green stripes highlight the NTD residues, forming close contacts with the CTD in the crystal structure of αRfaH (17), with the β-hairpin residues 32–39 (also indicated in A) being the only NTD interface residues showing both stabilizing and destabilizing energetic contributions toward αRfaH. A visual guide for the fold-dependent secondary structure is shown for the CTD. To see this figure in color, go online.
Free-energy decomposition at a per-residue level shows that localized groups of residues differentially stabilize either RfaH state. As shown experimentally (12,15), most interdomain contacts deeply stabilize the autoinhibited αRfaH, with the exception of one region comprising the first strand in the β-hairpin of the NTD (residues 32–39 in Fig. 2) in which some residues are destabilizing. The behavior observed for these residues when compared to other interdomain NTD regions can be attributed to interactions between the CTD and the NTD β-hairpin observed in the structures of αRfaH (17) and βRfaH (13) used as starting configurations for the CCR approach. As this method drives the atom positions toward highly restrained states starting from these initial structures, the CTD-NTD interactions are stably kept for βRfaH throughout the confinement MD, even though for the E. coli paralog NusG, they have been determined to be intermittent in solution (39). The persistence of these interactions could potentially lead to overestimations of the contribution of these regions toward the stability of RfaH in the β-fold in our simulations. Thus, we modeled βRfaH with a fully extended loop and lacking NTD-CTD interactions. Subsequent loop relaxation in Rosetta led to a structure similar to that determined by cryoEM for βRfaH, in which CTD-NTD interactions are reestablished even when forcing Rosetta to explore extended loop configurations (Fig. S2). Consequently, we consider the NTD-CTD interacting βRfaH seen in cryoEM as the functionally relevant structure for our studies. An alternative but energetically less favored structure obtained after Rosetta relaxation shows an even larger interaction surface between NTD and CTD domains in the β-fold but contributes to changes in differential stability of the CTD in only a few residues (Fig. S2).
Although most of the CTD (residues 115–162) interacts with the NTD in αRfaH, per-residue free-energy differences show localized heterogeneity in preferential stability toward each native CTD state within this region. The C-terminal region of the linker (residues 110–114) as well as initial (residues 115–119) and terminal (residues 151–162) regions of the CTD show clear preference toward forming the strands β1 and β4-β5, respectively, whereas remaining residues 120–150 are more stabilized when forming the tip of the α-hairpin rather than strands β2 and β3 (Figs. 1 C and 2). These results are consistent with previous simulations of the refolding pathway of RfaH in the context of the full-length protein using structure-based models (21). This is not the first example of heterogeneous and alternating ΔG along the primary structure using the CCR method; chameleonic proteins GA30 and GB30 provide per-residue free energies that strongly correlate to the sequence content from which they were engineered, effectively tracing back conformational space information from these simulations to their primary structure (28).
Hydrogen-deuterium exchange confirms the differential stability of metamorphic RfaH states
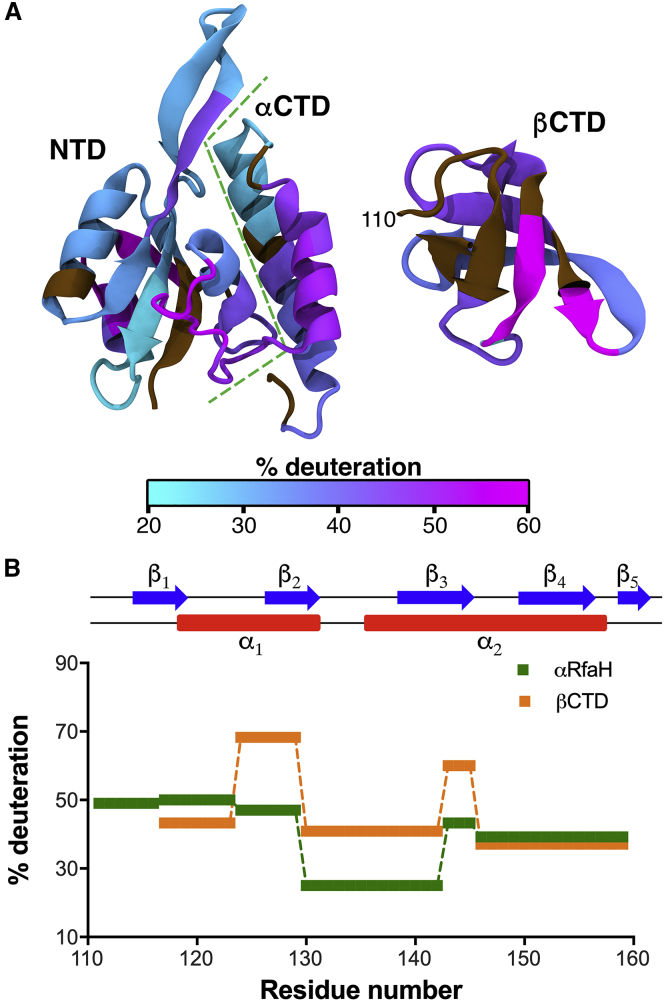
To determine how confinement calculations correlate with experimentally determined stabilities, HDXMS was performed with full-length RfaH (i.e., the CTD is in the α-fold) and isolated CTD, which is in the β-fold. This analysis identified a total of 43 different peptides derived from pepsin digestion of the 162 residue-long full-length RfaH (31 peptides for the NTD, 12 for the CTD), covering residues 7–159 (Fig. S3; Tables S1 and S3). A total of 27 peptides were identified from pepsin digestion of the isolated CTD (residues 110–162), covering positions 117–162 (Fig. S3; Tables S2 and S4). Given that many of the CTD peptides have varying lengths and overlapping regions between them, backbone amide deuteron incorporation was deconvoluted into six unique regions that were observable for RfaH CTD in both the α-fold and the β-fold (see Supporting Materials and Methods) and covered almost the entirety of the CTD (residues 117–159).
As measure for flexibility, we determined the relative deuterium uptake of full-length αRfaH and the isolated βCTD (Fig. 3), corresponding to the ratio between the maximal deuterium incorporation, calculated as the saturation value of an exponential fit to the deuterium uptake (Fig. S3), and the maximal theoretical incorporation, which depends on the peptic fragment sequence and length (40). In the NTD, buried regions show deuterium uptakes of ∼30%, whereas solvent-exposed regions exhibit increased deuteration of around 50% (Figs. 3 A and S3). Strikingly, in full-length RfaH, almost all of the peptides of the CTD display between 40 and 50% deuteron incorporation, with the exception of a single region covering residues 130–142, whose incorporation reaches a maximum of only 24% (Fig. 3 B). This is reminiscent of the temperature factors observed in the crystal structure of full-length RfaH, wherein residues 115–128 and 153–156 display B-factor values over 50, whereas residues covering the tip of the α-hairpin display values of around 30 (17). These results indicate that the ends of the α-hairpin are highly flexible, whereas the tip is relatively more rigid.
Figure 3.
Mapping the differences in local flexibility between RfaH states by HDXMS. (A) Shown is structural mapping of the relative deuterium uptake of different regions of NTD and CTD in the context of the full-length RfaH (αRfaH, PDB: 5OND) or the CTD in isolation (βCTD, PDB: 2LCL). Regions are colored with a gradient from cyan (solvent protected against deuterium exchange) to magenta (solvent accessible). Residues whose deuteron incorporation could not be determined by mass spectrometry are shown in brown. (B) Shown is the relative deuterium uptake for RfaH CTD in its α- and β-states (green and orange, respectively) determined by mass spectrometry. To see this figure in color, go online.
In contrast to αRfaH, most regions of the isolated CTD display around 40% relative deuterium uptake with the exception of a short loop (residues 143–145) and strand β2 (residues 124–129), which exhibit deuteration extents of around 60 and 68%, respectively. For comparison, the experiment was also carried out with the full-length NusG protein (Fig. S4; Table S5), whose CTD is always folded as a β-barrel and that does not stably interact with its NTD (10,39). Relative deuterium uptake between 50 and 60% is observed for almost all regions within NusG CTD, slightly higher than those observed for its metamorphic paralog, except strand β2 that exhibits 30% less exchange as compared to RfaH. Despite having superimposable structures and 41% sequence similarity, RfaH CTD residues have overall larger aliphatic and more hydrophobic side chains than those of NusG, physicochemical features that are compatible with the observed lower flexibility of RfaH CTD in the β-fold. This analysis shows that, nevertheless, local flexibility of RfaH CTD in the β-fold are not drastically different from that of its paralog NusG while highlighting the strongly reduced flexibility of the tip of the α-helical hairpin in αRfaH.
To further confirm the heterogeneity in local stability and flexibility observed for RfaH, we performed NMR-based HDX experiments on full-length RfaH and the isolated CTD. The lyophilized 1H,15N-labeled proteins were dissolved in D2O, and HDX was monitored via the decay of signal intensities in a series of two dimensional [1H,15N]-HSQC spectra over 24 h. In full-length RfaH, only 17 signals in the NTD and five in the α-folded CTD corresponding to individual amides were detectable and analyzed (Fig. S5; Table S6). All other amide protons in full-length RfaH and all amide protons in the isolated CTD exchanged too fast (for the isolated CTD, the exchange was completed within the experimental time for the first spectrum), thus suggesting that these amides are either solvent exposed or not stably involved in hydrogen bonds to observe them in NMR-based HDX.
The decay rates in the signal intensity of the observable amides were fitted onto a single exponential and converted into exchange protection factors. These equate to an equilibrium measurement of local stabilization in a folded conformation as compared to the unfolded state and can be further used to determine the free-energy change involved in exposing the protein amides to the solvent (Table S6; (37,38)). Remarkably, all of the 22 analyzable RfaH NTD and CTD amide protons are located in regions of preferential stability toward the α-fold, according to CCR (Fig. S5), with all the CTD signals located on the tip of the α-helical hairpin. Also, these single amides are encompassed in peptides showing low deuteration in HDXMS experiments using full-length RfaH (Fig. S5). Thus, these data confirm that the tip of the α-helical hairpin exhibits a stability comparable to that of the NTD in full-length RfaH.
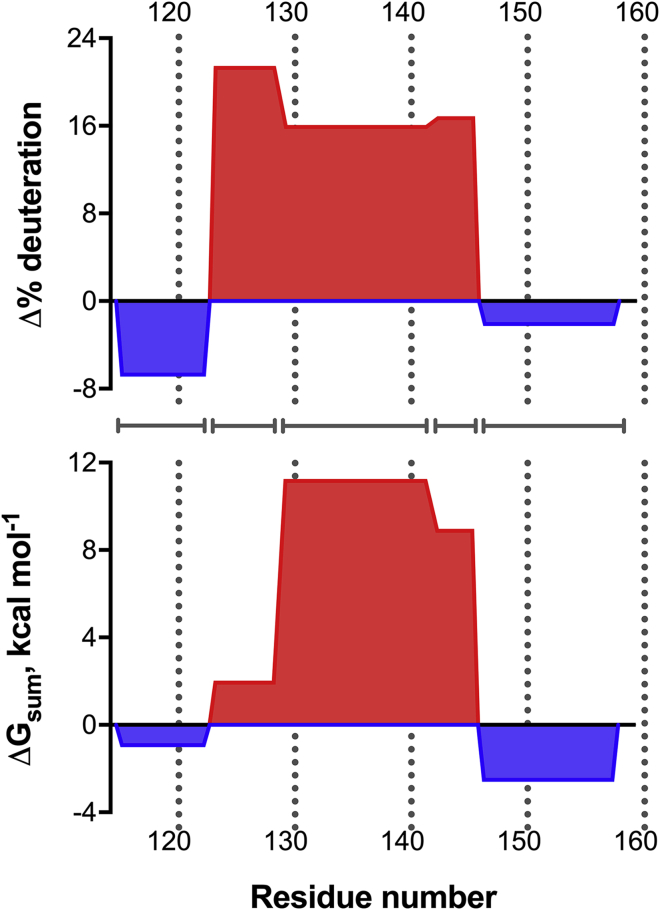
To provide greater detail into the similarities in the local stability of RfaH CTD in the α-fold and the β-fold observed through HDXMS and MD, the ΔG values were summed for residues matching five regions experimentally observed through HDXMS, excluding the region that contains the linker between domains. Comparison of the local differential stability patterns using both strategies revealed striking similarities in distribution as well as in magnitude (Fig. 4), with the exception of CTD residues 124–129 from helix α1 that constitute part of the tip of the α-helical hairpin (Fig. 1). This is partly explained by the high solvent accessibility of this region in the β-barrel fold as ascertained by HDXMS, exhibiting the highest extent of deuteration (Fig. 3). In contrast, the confinement procedure of the MD simulations might stabilize the interactions within this region, thus allowing their energetics to be comparable to those estimated for the CTD in the α-fold (Fig. 2).
Figure 4.
Comparison between computationally derived free energy (ΔGsum) and experimentally determined flexibility (Δ% deuteration) between five indicated regions of RfaH CTD (indicated by gray lines) in its α-fold and β-fold. Per-residue free-energy differences were added accordingly to match the length of the peptides analyzed via HDXMS. Regions in red and blue have a preferential stability toward the α-fold and β-fold, respectively. To see this figure in color, go online.
Discussion
The molecular mechanism by which the transformer protein RfaH completely refolds its CTD has been experimentally elusive. Computational and experimental approaches strongly support the importance of interdomain contacts in controlling RfaH metamorphosis (12,15,21,41). In this work, both HDX and confinement MD reveal that the interaction-rich upper region of the α-helical hairpin, comprising residues 125–145, provides the highest degree of stabilization toward the α-folded CTD (Figs. 1 C and 4). Moreover, confinement MD and [1H,15N]-HSQC-based HDX experiments show that interdomain contacts stabilize to a similar extent the tip of the α-helical CTD as well as the NTD (Fig. S5). They also suggest that the structural metamorphosis of RfaH from the autoinhibited to the active state is controlled not only by native contacts with the other domain but also by intrinsic CTD determinants within the aforementioned region. Thus, it comes as no surprise that the computational analysis of NusG and RfaH sequences identified seven residues that are highly conserved within the RfaH subfamily and significantly contribute to NTD-CTD binding, of which three NTD (E48, F51, and P52; E. coli numbering) and three CTD residues (F130, R138, and L142) are located in the vicinity of the tip of the α-helical hairpin (12,41).
Using structure-based models, we previously simulated RNAP binding to RfaH and concluded that contacts in the vicinity of NTD residue E48, adjacent to the tip of the hairpin, may suffice to favor CTD refolding into the β-state and thus the relief of autoinhibition (21). Our present results, along with the recent cryoEM structure of RfaH bound to the complete TEC (13), are consistent with this hypothesis. Binding of the autoinhibited αRfaH, which cannot contact its high-affinity site on the β′ subunit of RNAP, is thought to be mediated by its initial contacts to a hairpin that forms in the nontemplate DNA strand and to the β subunit gate loop (13,14,17,42), resulting in an encounter complex (14).
The preference toward the β-fold displayed by residues 110–119 and 151–162 (Fig. 2) strongly suggests that the refolding of the CTD toward the β-fold starts with the unfolding of the ends of the α-helical hairpin as they fluctuate toward a locally unfolded state even before dissociation (Fig. 3). Thus, the tip of the α-helical CTD seems to act as an anchor, preventing its spontaneous refolding into a β-barrel. This is supported by previous observations that the disruption of the E48/R138 salt bridge located in this region led RfaH to exist in equilibrium between the autoinhibited and active folds (12). However, this view contrasts with conclusions drawn from other computational approaches (18,22), which suggested that contacts involving RfaH CTD residues comprising strand β3 are particularly stable and nucleate the β-barrel, as solution dynamics do not display high stability in this region for RfaH or NusG CTD in the β-fold (Fig. S4).
Folding of RfaH into a stable, autoinhibited structure is essential for its function. Because RfaH has a higher affinity for the TEC than NusG (13), its binding to RNAP has to be tightly controlled to prevent the misregulation of NusG-dependent housekeeping genes. The emergence of the autoinhibited state, which is relieved only in the presence of a 12-bp ops DNA element with complex properties (13,17), presents an elegant solution to this problem. Our results suggest that establishing interdomain contacts at the tip of the hairpin, blocking most of the RNAP-binding residues (Fig. 1 C), is sufficient to enable autoinhibition. Moreover, the emergence of this novel fold causes only few changes in the local stability and dynamics of the canonical β-barrel of NusG CTDs (Figs. 3 and S4), supporting its ability to interact with the translational machinery (12,14). These interactions, established with the ribosomal protein S10, are formed through hydrophobic residues located in strands β2 (residues 122–126) and β4 (residues 145–148) (6,12,14), whose identities are mostly conserved between RfaH and NusG. Our results show that these residues are stably interfaced with the NTD, thus explaining why S10 is unable to elicit RfaH metamorphosis on its own (12).
The key differences between RfaH and NusG, metamorphosis and sequence divergence of the CTD, underpin their orthogonal cellular functions. Even though NusG and RfaH bind to the same site on the TEC and display similar effects on RNA synthesis (5,13), they paradoxically play opposite roles in the expression of horizontally acquired genes. NusG cooperates with Rho to silence foreign DNA, an activity that explains the essentiality of E. coli NusG (43) through direct contacts between NusG CTD and Rho (44). In contrast, RfaH does not interact with Rho and abolishes Rho-mediated termination in its target operons, all of which have foreign origin, in part by excluding NusG (42). Remarkably, grafting a five-residue NusG loop (S163IFGR) into RfaH (N144LINK) creates an even more potent activator of Rho (44). Thus, the loss of interactions with Rho, another pivotal step in the evolution of RfaH, also occurs around the tip of the helical hairpin.
Altogether, our results show that the tip of the α-helical hairpin is the main determinant for stabilizing the autoinhibited state of RfaH and that this localized stability arises from interdomain interactions and intrinsic sequence-encoded preferences. Both our present results and other available evidence suggest that targeted substitutions in this CTD region enabled both the acquisition of the autoinhibited state, in which this region forms the tip of the stabilizing α-hairpin, and the loss of termination-promoting contacts with Rho. These changes converted a nascent paralog of NusG, an essential xenogene silencer, into an activator of horizontally transferred virulence genes that encode capsules, toxins, and conjugation pili (45). We hypothesize that the molecular details about RfaH mechanism can be harnessed to design ligands that interfere specifically with RfaH activity and thus virulence (46). In addition to directly inhibiting the expression of virulence genes, these ligands may also limit the spread of plasmid-encoded antibiotic resistance determinants through conjugation and synergize with the existing drugs by compromising the cell wall integrity in Gram-negative pathogens.
Author Contributions
P.G.-D., E.A.K., I.A., and C.A.R.-S. designed the research. J.A.M., S.S., S.H.K., and C.A.R.-S. conducted the experiments. P.G.-D., E.A.K., S.H.K., and C.A.R.-S. analyzed the data. P.G.-D. and C.A.R.-S. conducted and analyzed the computational work. P.G.-D., E.A.K., S.H.K., I.A., and C.A.R.-S. wrote the manuscript.
Acknowledgments
We acknowledge the gracious help of Dr. Ken Dill’s group at Stony Brook University and Dr. Arijit Roy regarding the use and calculation of the CCR approach used in this work. We also acknowledge the help of Dr. Maira Rivera during the final stages of this work. We thank Prof. Paul Rösch and Dr. Kristian Schweimer for helpful discussions.
This research was funded by Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 11140601) and International Cooperation Grant (REDI170624) from Comisión Nacional de Investigación Científica y Tecnológica, the National Institutes of Health (National Institutes of Health 1S10OD016234), the NVIDIA GPU Grant Program, the Pontificia Universidad Católica de Chile Office of Research Grant (VRI Puente P1810), and the German Research Foundation (Ro 617/21-1). C.A.R.-S. was funded by American Society for Biochemistry and Molecular Biology, Pan-American Association for Biochemistry and Molecular Biology and International Union of Biochemistry and Molecular Biology through the PROLAB program. P.G.-D. and J.A.M. were funded by Comisión Nacional de Investigación Científica y Tecnológica Doctoral Scholarships (CONICYT-PFCHA 21181705 and 21181787, respectively).
Editor: Daniel Raleigh.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.11.014.
Contributor Information
Irina Artsimovitch, Email: artsimovitch.1@osu.edu.
César A. Ramírez-Sarmiento, Email: cesar.ramirez@uc.cl.
Supporting Citations
References (47, 48, 49, 50, 51) appear in the Supporting Material.
Supporting Material
References
- 1.Murzin A.G. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 2.Dishman A.F., Volkman B.F. Unfolding the mysteries of protein metamorphosis. ACS Chem. Biol. 2018;13:1438–1446. doi: 10.1021/acschembio.8b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lella M., Mahalakshmi R. Metamorphic proteins: emergence of dual protein folds from one primary sequence. Biochemistry. 2017;56:2971–2984. doi: 10.1021/acs.biochem.7b00375. [DOI] [PubMed] [Google Scholar]
- 4.Bailey M.J., Hughes C., Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 5.Belogurov G.A., Mooney R.A., Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmann B.M., Schweimer K., Rösch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 7.Belogurov G.A., Sevostyanova A., Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Rucobo F.W., Sainsbury S., Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirtreiter A., Damsma G.E., Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooney R.A., Schweimer K., Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belogurov G.A., Vassylyeva M.N., Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmann B.M., Knauer S.H., Rösch P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang J.Y., Mooney R.A., Darst S.A. Structural basis for transcript elongation control by NusG family universal regulators. Cell. 2018;173:1650–1662.e14. doi: 10.1016/j.cell.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuber P.K., Schweimer K., Knauer S.H. Reversible fold-switching controls the functional cycle of the antitermination factor RfaH. Nat. Commun. 2019;10:702. doi: 10.1038/s41467-019-08567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomar S.K., Knauer S.H., Artsimovitch I. Interdomain contacts control folding of transcription factor RfaH. Nucleic Acids Res. 2013;41:10077–10085. doi: 10.1093/nar/gkt779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artsimovitch I., Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 17.Zuber P.K., Artsimovitch I., Knauer S.H. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. eLife. 2018;7:e36349. doi: 10.7554/eLife.36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S., Xiong B., Jiang H. Mechanism of the all-α to all-β conformational transition of RfaH-CTD: molecular dynamics simulation and markov state model. J. Chem. Theory Comput. 2014;10:2255–2264. doi: 10.1021/ct5002279. [DOI] [PubMed] [Google Scholar]
- 19.Balasco N., Barone D., Vitagliano L. Structural conversion of the transformer protein RfaH: new insights derived from protein structure prediction and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2015;33:2173–2179. doi: 10.1080/07391102.2014.994188. [DOI] [PubMed] [Google Scholar]
- 20.Bernhardt N.A., Hansmann U.H.E. Multifunnel landscape of the fold-switching protein RfaH-CTD. J. Phys. Chem. B. 2018;122:1600–1607. doi: 10.1021/acs.jpcb.7b11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramírez-Sarmiento C.A., Noel J.K., Artsimovitch I. Interdomain contacts control native state switching of RfaH on a dual-funneled landscape. PLoS Comput. Biol. 2015;11:e1004379. doi: 10.1371/journal.pcbi.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gc J.B., Gerstman B.S., Chapagain P.P. The role of the interdomain interactions on RfaH dynamics and conformational transformation. J. Phys. Chem. B. 2015;119:12750–12759. doi: 10.1021/acs.jpcb.5b05681. [DOI] [PubMed] [Google Scholar]
- 23.Xiong L., Liu Z. Molecular dynamics study on folding and allostery in RfaH. Proteins. 2015;83:1582–1592. doi: 10.1002/prot.24839. [DOI] [PubMed] [Google Scholar]
- 24.Ovchinnikov V., Cecchini M., Karplus M. A simplified confinement method for calculating absolute free energies and free energy and entropy differences. J. Phys. Chem. B. 2013;117:750–762. doi: 10.1021/jp3080578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyka M.D., Clarke A.R., Sessions R.B. An efficient, path-independent method for free-energy calculations. J. Phys. Chem. B. 2006;110:17212–17220. doi: 10.1021/jp060734j. [DOI] [PubMed] [Google Scholar]
- 26.Qian B., Raman S., Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cecchini M., Krivov S.V., Karplus M. Calculation of free-energy differences by confinement simulations. Application to peptide conformers. J. Phys. Chem. B. 2009;113:9728–9740. doi: 10.1021/jp9020646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A., Perez A., Maccallum J.L. Computing the relative stabilities and the per-residue components in protein conformational changes. Structure. 2014;22:168–175. doi: 10.1016/j.str.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xun S., Jiang F., Wu Y.D. Intrinsically disordered regions stabilize the helical form of the C-terminal domain of RfaH: a molecular dynamics study. Bioorg. Med. Chem. 2016;24:4970–4977. doi: 10.1016/j.bmc.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Gc J.B., Bhandari Y.R., Chapagain P.P. Molecular dynamics investigations of the α-helix to β-barrel conformational transformation in the RfaH transcription factor. J. Phys. Chem. B. 2014;118:5101–5108. doi: 10.1021/jp502193v. [DOI] [PubMed] [Google Scholar]
- 31.Joseph J.A., Chakraborty D., Wales D.J. Energy landscape for fold-switching in regulatory protein RfaH. J. Chem. Theory Comput. 2019;15:731–742. doi: 10.1021/acs.jctc.8b00912. [DOI] [PubMed] [Google Scholar]
- 32.Svetlov V., Belogurov G.A., Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artsimovitch I., Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina E., Córdova C., Babul J. Three-dimensional domain swapping changes the folding mechanism of the forkhead domain of FoxP1. Biophys. J. 2016;110:2349–2360. doi: 10.1016/j.bpj.2016.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J., Russell D.W. Third Edition. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 36.Meyer O., Schlegel H.G. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 1983;37:277–310. doi: 10.1146/annurev.mi.37.100183.001425. [DOI] [PubMed] [Google Scholar]
- 37.Bai Y., Milne J.S., Englander S.W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Y., Milne J.S., Englander S.W. Protein stability parameters measured by hydrogen exchange. Proteins. 1994;20:4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- 39.Burmann B.M., Scheckenhofer U., Rösch P. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem. J. 2011;435:783–789. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z., Smith D.L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi D., Svetlov D., Artsimovitch I. Flipping states: a few key residues decide the winning conformation of the only universally conserved transcription factor. Nucleic Acids Res. 2017;45:8835–8843. doi: 10.1093/nar/gkx523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sevostyanova A., Belogurov G.A., Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitra P., Ghosh G., Sen R. Rho protein: roles and mechanisms. Annu. Rev. Microbiol. 2017;71:687–709. doi: 10.1146/annurev-micro-030117-020432. [DOI] [PubMed] [Google Scholar]
- 44.Lawson M.R., Ma W., Berger J.M. Mechanism for the regulated control of bacterial transcription termination by a universal adaptor protein. Mol. Cell. 2018;71:911–922.e4. doi: 10.1016/j.molcel.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagy G., Danino V., Hacker J. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 2006;74:5914–5925. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svetlov D., Shi D., Artsimovitch I. In silico discovery of small molecules that inhibit RfaH recruitment to RNA polymerase. Mol. Microbiol. 2018;110:128–142. doi: 10.1111/mmi.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leaver-Fay A., Tyka M., Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case D.A., Betz R.M., Kollman P.A. University of California, San Francisco; 2016. AMBER 2016. [Google Scholar]
- 50.Hawkins G.D., Cramer C.J., Truhlar D.G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 1996;100:19824–19839. [Google Scholar]
- 51.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 52.Ramirez-Sarmiento C.A., Komives E.A. Hydrogen-deuterium exchange mass spectrometry reveals folding and allostery in protein-protein interactions. Methods. 2018;144:43–52. doi: 10.1016/j.ymeth.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental and computational data are available from the corresponding authors upon request.