Abstract
Cytochrome c (cyt c) is known for its role in the electron transport chain but transitions to a peroxidase-active state upon exposure to oxidative species. The peroxidase activity ultimately results in the release of cyt c into the cytosol for the engagement of apoptosis. The accumulation of oxidative modifications that accompany the onset of the peroxidase function are well-characterized. However, the concurrent structural and conformational transitions of cyt c remain undercharacterized. Fast photochemical oxidation of proteins (FPOP) coupled with mass spectrometry is a protein footprinting technique used to structurally characterize proteins. FPOP coupled with native ion mobility separation shows that exposure to H2O2 results in the accumulation of a compact state of cyt c. Subsequent top-down fragmentation to localize FPOP modifications reveals changes in heme coordination between conformers. A time-resolved functional assay suggests that this compact conformer is peroxidase active. Altogether, combining FPOP, ion mobility separation, and top-down and bottom-up mass spectrometry allows us to discern individual conformations in solution and obtain a better understanding of the conformational ensemble and structural transitions of cyt c as it transitions from a respiratory role to a proapoptotic role.
Significance
Cytochrome c (cyt c) transitions from a respiratory to a proapoptotic role upon exposure to reactive oxidative species such as hydrogen peroxide. These species activate the peroxidase activity of cyt c, resulting in self-oxidation, the oxidation of cardiolipin, and ultimately, its release to the cytoplasm, where it engages in the formation of the apoptosome. Whereas the accumulation of oxidative modifications of cyt c has been extensively characterized and correlated with the onset of peroxidase activity, the structural and dynamic consequences of such modifications remained unclear. In this study, we used fast photochemical oxidation of proteins alone or in combination with ion mobility separation to describe the structural transition from a respiratory to a proapoptotic role and reveal an unexpected compact conformer.
Introduction
Functionally, cytochrome c (cyt c) participates in respiration as part of the electron transport chain. Located at the inner membrane of the mitochondria, it is known to interact and transfer electrons to cyt c oxidase. It is now established that the release of cyt c from the mitochondrial membrane into the cytosol is a crucial step for the onset of apoptosis (1, 2, 3, 4). Cyt c has been shown to trigger formation of the apoptosome in the apoptotic caspase-9 cascade through its interaction with Apaf-9 (5). It remains controversial whether reactive oxygen species (ROS) act as a signaling molecule for apoptosis (6), but they have been observed to induce the release of cyt c into the cytosol (7,8). Exposure to ROS induces the peroxidase activity of cyt c, which results in the oxidation of cardiolipin, leading to the detachment of cyt c from the inner membrane and, ultimately, its escape to the cytosol (2,9, 10, 11, 12).
Cyt c is a small globular heme binding protein of 104 residues. In its native, respiratory role, the heme of cyt c is liganded by H18 on the proximal side and M80 on the distal side. Numerous high-resolution structures and in-solution structural studies have investigated the effects of redox state, ionic strength, pH, cardiolipin binding, and mutations on the structure and dynamics of cyt c. The above biochemical or biophysical factors were observed to perturb the native distal heme iron ligand, M80, replacing it with H26 (13), H33 (13), K72 (14,15), K73 (15,16), or K79 (15,17) or even remaining unliganded (18). With a change of ligation, the heme’s reduction potential is drastically lowered, affecting cyt c’s rate of electron transfer to cyt c oxidase. Nevertheless, perturbations that favor pentacoordinated heme conformations are thought to be needed for peroxidase activity because of the more dynamic and accessible heme environments (19,20). After ROS exposure, cyt c forms an oxoferryl heme species or a side chain tyrosyl radical on the distal side of the heme (21). With continued ROS exposure, several residues on the distal side of the heme become oxidized. This was observed in a recent study by Yin et al. (22) in which time-resolved exposure to physiological concentrations of H2O2 results in the progressive accumulation of oxidatively modified residues (Y67, K73, and M80) that progressively disrupt the distal coordination of the heme. The accumulation of these modifications was seen to correlate with the onset of peroxidase activity. It is generally expected that these modifications result in a progressive opening of cyt c and greater accessibility of the heme binding pocket. However, limited information is available on the effect of these modifications on the secondary and/or tertiary structures of cyt c that accompany this functional transition.
To further study cyt c’s transition into a peroxidase, we will utilize hydroxyl radical protein footprinting (HRPF), which is steadily emerging as an informative structural method to study protein interactions and dynamics. HRPF hinges on the rapid generation of hydroxyl radicals, which in turn label the side chain of proteins as a function of solvent accessibility. There are several methods to generate hydroxyl radicals, among which is fast photochemical oxidation of proteins (FPOP). FPOP produces hydroxyl radicals through laser photolysis of hydrogen peroxide and limits the radical lifetime to a microsecond with the use of a radical scavenger (23). In this case, H2O2 not only is needed for FPOP but also serves as a trigger of the transition of cyt c into its peroxidase-active state.
Coupling FPOP with bottom-up proteomics allows for the identification of oxidized residues and yields structural information in the form of localized surface accessibility. Often, the interpretation of FPOP data is done in the context of a static high-resolution structure (24, 25, 26). However, in solution, proteins experience structural fluctuations, small or large, and generally exist in an array of conformations that make up a conformational ensemble (27). This is particularly the case for a protein undergoing a functional transition. Because of different protein conformers potentially undertaking alternate biological activity, being able to distinguish these protein conformers in solution will provide a more accurate description of protein structure and dynamics as it relates to function.
Like FPOP, most of the commonly used structural methods are typically unable to distinguish individual conformers but, rather, deliver either a high-resolution image of a single conformer or a structural parameter that is averaged over all conformers of the ensemble and/or through time. Recently, ion mobility separation (IMS) has been combined with mass spectrometry (IM-MS) to separate and provide structural information for copopulated protein conformers. In IM-MS, ions are propelled through the ion mobility cell and decelerated by passing through a neutral gas (28), separating gas-phase ions based on size and shape. This provides the ability to quasi-instantaneously separate protein conformers from a single charge-state ion. The separation of IM-MS based on shape has proven useful to distinguish large conformational variations and yield a global structural parameter in the form of collision cross-sectional areas (29, 30, 31). By combining FPOP with IM-MS, we can study individual conformers that coexist in solution, thus increasing the depth of structural information on a protein system. Because a functional transition is likely to incur significant structural changes, we have used FPOP, IMS, bottom-up MS/MS, and top-down MS/MS to tease out the conformational landscape of bovine cyt c as it transitions from an electron carrier to its peroxidase-active state. With the combination of FPOP, IM-MS, and peroxidase functional assays, we observe cyt c shift to a peroxidase-active compact conformer after H2O2 exposure.
Materials and Methods
FPOP
Cyt c (C3131; Sigma-Aldrich, St. Louis, MO) was resuspended in 100 mM ammonium acetate, pH 6.6. Cyt c was subjected to FPOP as previously described (32,33). An H2O2 control sample without laser irradiation was collected to monitor H2O2-induced modifications. Each FPOP and H2O2 sample consisted of 14.7 μM of cyt c, 20 mM glutamine (Thermo Fisher Scientific, Waltham, MA), and—immediately before infusion—7.5 mM H2O2 (Thermo Fisher Scientific). Maximal exposure to H2O2 before laser irradiation was 2 min. For the FPOP samples, a KrF excimer laser (GAM Laser, Orlando, FL) was used to produce laser irradiation with an energy between 115 and 125 mJ/pulse and a frequency of 10 Hz. With or without laser irradiation, the samples were passed through a 2.59-mm laser irradiation window at a flow rate of 34.33 μL/min. This produced a 20% exclusion fraction, allowing space between each bolus of irradiation to account for hydroxyl radical and protein diffusion. After the irradiation window, each sample was collected in a quench containing a final concentration of 20 mM N,N'-Dimethylthiourea (DMTU) (Acros Organics; Thermo Fisher Scientific, Fair Lawn, NJ) and 20 mM methionine (Thermo Fisher Scientific). Afterward, the samples were split into three aliquots, one each for intact MS, bottom-up proteomics, and native IMS/top-down proteomics.
Intact MS analysis
Intact MS analysis was completed using a nanoAcquity UPLIC (Ultra Performance Liquid Chromatography) (Waters, Milford, MA) coupled to a Q Exactive HF (Thermo Fisher Scientific) mass spectrometer. Protein was loaded onto a MassPREP Micro Desalting column (Waters) and eluted at 60% acetonitrile and 0.1% formic acid for 9 min at a rate of 10 μL/min. The mass spectra were deconvoluted using Unidec (34).
Bottom-up analysis
For bottom-up proteomics, samples were digested as previously described (32). Briefly, the samples were vacuum-centrifuged to dryness and resolubilized in 8 M urea and 100 mM Tris, pH 8.5. The samples were reduced with 10 mM tris (2-carboxyethyl) phosphine (Sigma-Aldrich), alkylated with 20 mM iodoacetamide (Sigma-Aldrich), quenched with 10 mM dithiothreitol (Sigma-Aldrich), and then subjected to an overnight tryptic digest (Thermo Fisher Scientific). The digestion was quenched with 5% formic acid (Thermo Fisher Scientific), desalted using NuTip C18-media-packed Zip Tips (Glygen, Columbia, MD), vacuum centrifuged to dryness, and resuspended in 10% acetonitrile with 0.1% formic acid (Thermo Fisher Scientific).
MS/MS analysis was completed using a nanoAcquity UPLC (Waters) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Samples were loaded on an Acquity UPLC C18 Trap Column (Waters) and washed for 10 min at 15 μL/min with 1% acetonitrile and 0.1% formic acid. Samples were separated on an in-house packed column (20 cm × 75 μM) containing 5-μm C18 particles (Phenomenex, Torrance, CA). The gradient was ramped from 15% acetonitrile and 0.1% formic acid to 45% acetonitrile and 0.1% formic acid over 70 min, followed by 15 min at 100% acetonitrile and 0.1% formic acid for cleaning, and finally equilibrated to 3% acetonitrile and 0.1% formic acid for 11 min. For MS1, the AGC target was set to 3e6, and for data-dependent MS2, it was set to 1e5. Samples were searched using Proteome Discoverer 2.2 (Thermo Fisher Scientific) with Sequest HT (Matrix Sciences) against a bovine FASTA database with all possible FPOP modifications. The extent of oxidation was determined according to Eq. 1:
| (1) |
where “EIC area modified” is the chromatographic area of a peptide with a specific modified residue, and “EIC area” is the total chromatographic area (modified and unmodified) of that peptide.
Native IMS/top-down analysis
Native electrospray ionization (ESI) MS was performed on three cyt c conditions: native cyt c, H2O2 sample, and FPOP sample. Each condition underwent a 24-h dialysis in 100 mM ammonium acetate, pH 6.6, to remove glutamine, DMTU, and methionine. Each sample was sprayed using a gold-coated capillary (Waters), with the voltage set to 1.8 kV and cone voltage at 30 V. The capillary voltage, sample cone, trap bias, and step wave were all ramped from the softest conditions to harsher conditions to test the protein conformer distributions observed and tune for softest parameters. After probing various conditions, we settled on an IMS wave velocity of 600 m/s and IMS wave height of 40 V. After IMS, cyt c was fragmented for top-down analysis, with the collision-induced dissociation (CID) energy set to 90 eV in the transfer cell. Spectra were deconvoluted using MaxEnt3 (Waters), and ions were matched with a 100-ppm mass tolerance using ProSight Light (35).
Peroxidase functional assay
The oxidation of guaiacol (Sigma-Aldrich) was used to monitor the peroxidase activity of cyt c. The final concentration of guaiacol was 10 mM, the final concentration of protein was 14.7 μM, and the final concentration of hydrogen peroxide was 7.5 or 1 mM. Guaiacol oxidation forms tetraguaiacol, which absorbs at 470 nm. A measurement at 470 nm was taken every 10 s for 1 h immediately after H2O2 exposure using a 96-well plate in a BioTek Gen5 plate reader. A blank omitting cyt c was collected to control for basal levels of absorption.
Salt dependence experiment
To examine how salt affects the conformation of cyt c, samples underwent a 24-h dialysis in 100 mM ammonium acetate, pH 6.6, to remove residual salts present in the lyophilized protein. After dialysis, cyt c was exposed to various salt conditions (no salt, 10 mM KCl and NaCl, 80 mM KCl and NaCl, and 150 mM KCl and NaCl). FPOP, digestion, MS analysis, and MS/MS analysis were conducted as described above.
Results
FPOP coupled with bottom-up proteomics
After FPOP, the intact MS of H2O2 samples shows the relative intensity of the singly modified protein (+16 Da) at ∼22% of the unmodified protein. After laser irradiation, the singly modified protein increases to ∼55% of the unmodified protein (Fig. 1, A and B). A higher-than-usual background oxidation is observed but is attributed to the modifications often observed during cyt c’s peroxidase activation by H2O2. The difference between the background oxidation and FPOP oxidation (∼33%) is typical of FPOP experiments.
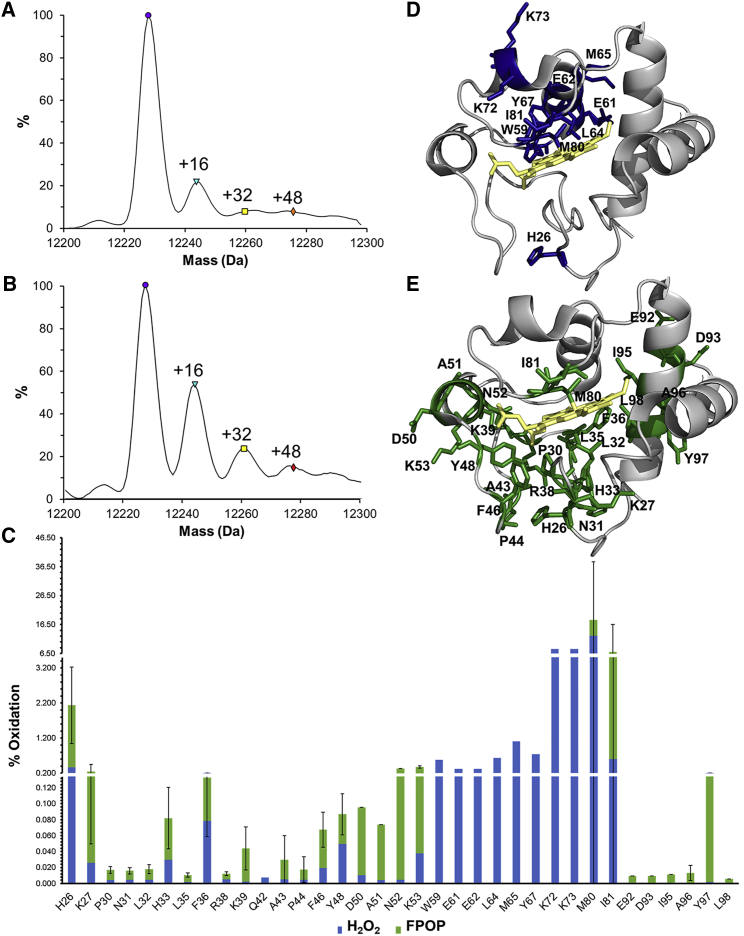
Figure 1.
Oxidative modifications of cyt c. (A and B) Intact analysis of cyt c of the (A) H2O2 control and (B) FPOP sample is shown. (C) The extent of modification calculated for each residue is shown (mean of three experiments ± standard error). The extent of oxidation in the H2O2 sample is represented in blue. The additional modifications detected in the FPOP sample are shown in green. (D) Shown is the crystal structure of bovine cyt c (PDB: 2B4Z) highlighting residues with oxidation attained from H2O2 greater than 0.3%. (E) The crystal structure with all residues showing FPOP modifications is depicted. To see this figure in color, go online.
For the bottom-up analysis, the extent of modification was quantified for the H2O2 samples and FPOP samples. Residue-level quantitative analysis reveals a total of 35 oxidatively modified residues (Fig. 1 C). The extent of modification and modifications detected are listed in Table S1. Without laser irradiation, H2O2 alone modified 29 residues, among which nine residues do not show further FPOP modification. An additional six residues are only modified by the free hydroxyl radical generated by FPOP (Fig. 1 C). In cyt c, modifications resulting from H2O2 exposure are thought to result from the formation of an oxoferryl and/or a tyrosyl radical generated on the distal side of the heme’s iron center (21). As expected, residues with a high extent of modification induced by H2O2 are correlated with their close proximity to the heme (Table S1). Mapping the most abundant modifications clearly reveals that H2O2-induced modifications localize on the distal side of the heme and near the iron center (Fig. 1 D). These modifications are largely in agreement with Yin et al.’s previous studies observing H2O2-induced modifications on cyt c, including the recently reported lysine carbonylation (22,36). Interestingly, K73 and H26 are among these H2O2-induced modifications. In the crystal structure of native respiratory cyt c, these residues are the most distant from the heme iron center among all the H2O2-induced modifications (Fig. 1 D), but they are known distal ligands after an alkaline transition (37) or from cardiolipin-induced extended conformations (38). Our detection of these modifications indicates that they must come in close proximity to the heme. This is an indication of large conformational heterogeneity either before or concurrent with H2O2 exposure. On the other hand, FPOP modifications reflect on the solvent accessibility of side chains to free OH⋅ generated by laser photolysis of H2O2. Consequently, FPOP modifications mapped on the structure are less localized than the H2O2-induced modifications (Fig. 1 E). The extent of FPOP modifications over all residues was compared with high-resolution structures through the quantitative correlation of measured protection factor versus calculated solvent accessible surface area (26). Data suggest poor correlations between the in-solution FPOP and any single high-resolution structure that was tested (Fig. S1). The limited correlation between the FPOP modification and high-resolution structures can potentially be ascribed to many reasons. One reason could be significant conformational heterogeneity of cyt c in solution, in particular after exposure to 7.5 mM H2O2, causing the initiation of the transition to a peroxidase-active state.
Native coordinating M80 is perhaps most illustrative of the conformational heterogeneity of cyt c. It is the most abundantly modified residue, with 12.7% modification in H2O2 and an additional 5.4% oxidation after irradiation (Fig. 1 C). Although, methionine residues are known to be significantly reactive, the high extent of modification for both M65 and M80 in H2O2 reflects their close proximity to the heme. Conversely, the high extent of M80 in the FPOP sample also suggests the presence of a conformer in which M80 is displaced from the heme and accessible to free OH٠ after exposure to H2O2. To further assess the conformational heterogeneity and characterize any differential oxidation pattern of conformers, IMS was used to separate copopulated conformers, followed by CID fragmentation to distinguish conformer-specific FPOP modifications.
FPOP coupled with native IMS and top-down proteomics
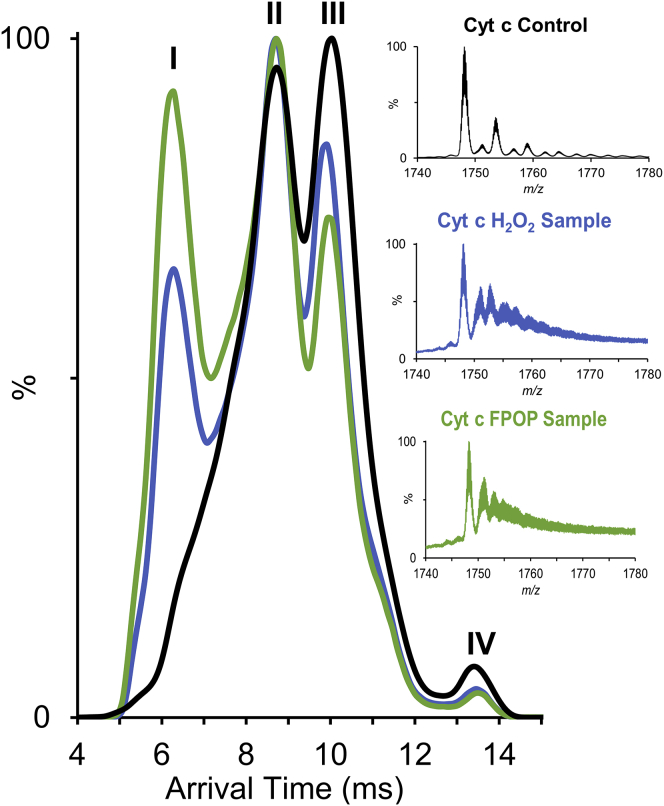
Native ESI MS (parameters found on Table S2) was performed on three cyt c conditions (native cyt c, H2O2 exposed, and FPOP sample), and typical charge states of native cyt c were observed (39,40). The most abundant charge state (7+) was selected for IMS (Fig. S2). By selecting the m/z window of 1740–1780 for native cyt c, three distinct conformers (conformers II–IV) were detectable (Fig. 2). This is in contrast with recent work that observed mostly one conformer with minor shoulders by IMS for bovine cyt c (39). An array of conditions (capillary voltage, sample cone, collisional energy, trap bias, and step wave) were tested to rule out ionization and/or gas-phase artifacts or activation. All three conformers were present under all conditions tested (Fig. S3). If the observed arrival time distribution is reflective of solution-phase structures, these multiple conformational species detected in IMS may reflect on the conformations that result in H2O2-induced modifications of K73 and H26 observed in the bottom-up analysis and could explain the poor correlation between the solvent accessible surface area and the bottom-up FPOP data.
Figure 2.
Ion mobility separation of cyt c before and after H2O2 exposure. Shown is the arrival time distribution of cyt c control (black), cyt c H2O2 control (blue), and cyt c FPOP sample (green) with the m/z region selected shown in the inset. To see this figure in color, go online.
In addition, we observed a shift toward a more compact conformer in the conformational ensemble of cyt c resulting from oxidative modifications (FPOP or H2O2 induced). Softer conditions were tested to help rule out gas-phase stabilization after H2O2 exposure, and a similar accumulation of the compact conformer is observed (Fig. S4). Such compaction was unexpected considering the gradual opening that was thought to happen when transitioning to a peroxidase-active state (41). However, Amacher et al. reported a more compact structure of cyt c mutant T78C/K79G (Protein Data Bank, PDB: 4Q5P) with the heme in a Lys73-ligated ferric state (42). In this mutant, M80 is displaced from coordination, causing the coordination loop to form a tight β hairpin. This allows the heme pocket to be more open but the overall protein structure to be more compact. They report that in this more compact state, cyt c has higher peroxidase activity compared with when M80 (PDB: 2YCC) and water (PDB: 4MU8) are ligated to the heme.
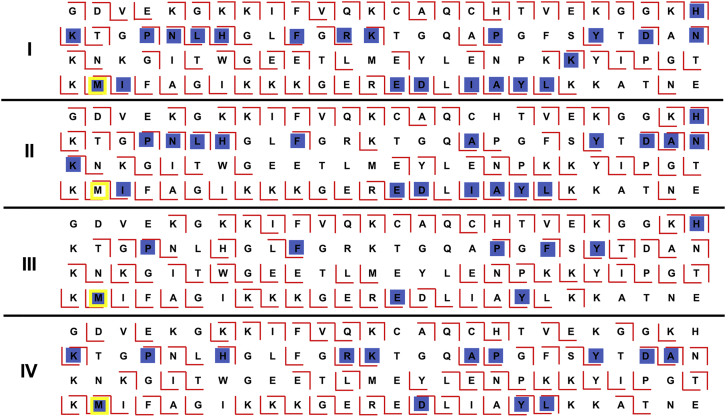
Performing IMS of FPOP-treated cyt c opens the possibility of a conformer-specific top-down characterization of FPOP oxidation. By performing CID fragmentation after IMS, fragment ions retain the arrival time distribution of the parent ion. Examples of fragment spectra obtained can be found in Fig. S5. Consequently, it is possible to determine whether FPOP modifications at specific residues differentially populate one conformer over another, offering conformer-specific higher-resolution structural information. Using the selection window found in Fig. S6 and a mass tolerance of 100 ppm, each conformer reaches 66–70% coverage (Fig. 3). The 35 FPOP modifications observed in the bottom-up analysis were used to focus the modification search in the top-down data analysis. Of the 35 amino acids with FPOP modifications calculated from the bottom-up data, 26 residues were observed with FPOP modifications in the top-down data, with 22 modifications in the most compact conformer (conformer I) followed by 19 in conformer II, 9 in conformer III, and 14 in conformer IV. The fewer modifications detected in conformer III and conformer IV are inconsistent with the more extended conformation suggested by their arrival time distribution. Rather, it may reflect the limit of detection of this method and instrumental platform. Indeed, after exposure to H2O2, conformer III and conformer IV are the least abundant species. In addition, it is unclear whether fragmentation efficiency is maintained for late-arriving conformer III and IV, as they retain abundant parent ions (Fig. S5). With the added complexity of even-lower-abundance FPOP-modified fragment, we cannot rule out that FPOP modification of conformer III and IV has fallen below the limit of detection, rendering quantitative analysis of conformer-specific FPOP modifications difficult.
Figure 3.
Top-down proteomics detecting FPOP modifications from individual conformers. After IMS separation, each conformer (I–IV) undergoes CID fragmentation in the transfer cell with the energy set to 90 eV. The coverage of each conformer is between 66 and 70%. Red ticks represent the detected b and y ions, and the residues with hydroxyl radical modifications are highlighted in blue. A biologically important residue, M80, is accented to show the ability of FPOP combined with native IMS and top-down in providing important structural information. To see this figure in color, go online.
Regardless, qualitative analysis of conformer-specific FPOP modification is highly informative. Looking at the most abundant modification, we do not observe a redistribution of modification across conformers after FPOP and before IMS. Indeed, after FPOP, cyt c could continue to interconvert between its conformational species, resulting in a redistribution of FPOP modifications between each conformer before entering the gas phase. With closer examination of the top-down data, modification of M80 was detected in conformers I (with a mass confidence of 2.5 ppm), III (−3.4 ppm), and IV (−40.3 ppm) but not in conformer II. Hydroxyl radical modification of methionine leads to a sulfoxide. After the sulfoxide modification, M80 would be unable to enter back into coordination with the iron center of the heme, thus hindering the species to convert back to conformer II. Furthermore, the absence of M80 FPOP modification in conformer II suggests that it remains coordinated to the iron center of heme, i.e., the native electron transport conformer. Conversely, the modification of M80 on conformers I, III, and IV suggests that M80 was displaced from heme ligation. It is noteworthy that there exist crystal structures of both more compact (42) and more expanded conformers (18,43,44) with the M80 displaced. We cannot completely rule out that the expanded conformers III and IV may be due to ionization and gas-phase artifacts; it seems unlikely that the appearance of the compact conformer I may be due to such an artifact. If reflective of solution-phase structures, conformers III and IV may possibly reflect on conformers from the alkaline transition or cardiolipin-induced conformers often observed for cyt c’s peroxidase activity with a more open structure and M80 displaced from heme ligation (37).
Correlating cyt c’s peroxidase activity with changes in IMS
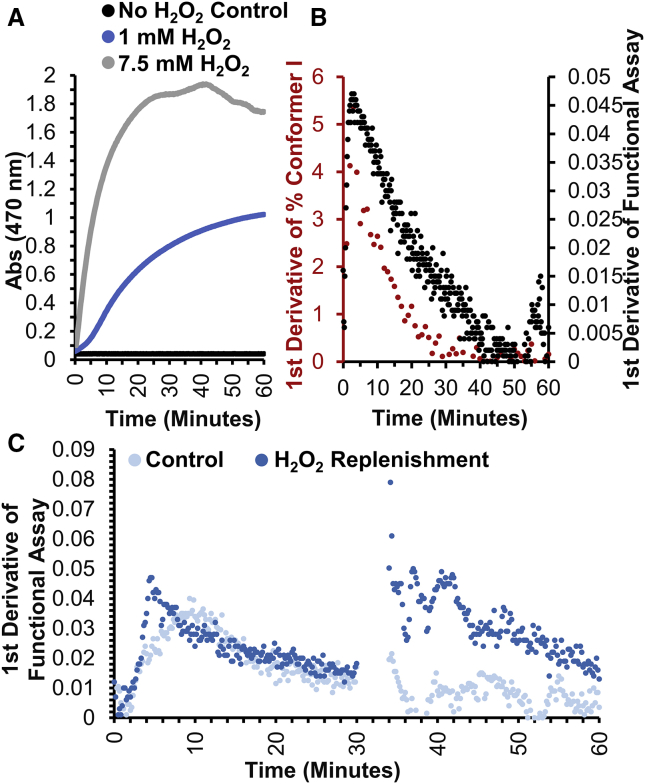
To correlate cyt c’s structural transition to its functional transition, a functional assay monitoring the oxidation of guaiacol into tetraguaiacol was performed (Fig. 4 A). In the absence of cyt c, no accumulation of tetraguaiacol was observed, whereas a steady increase of oxidative product was observed when 14.7 μM cyt c was included. Traditionally, guaiacol kinetics are observed in three stages: lag phase, steady-state phase, and deactivation phase (45,46). The lag phase is the time it takes to change heme coordination necessary for the peroxidase activity of cyt c. The steady-state phase is used to calculate the reactivity of cyt c. Finally, the deactivation phase was believed to be the slow degradation of cyt c (22). Because of the high reactivity of cyt c in the presence of 7.5 mM H2O2 (condition required for FPOP), the lag phase was not observed. The reaction quickly moved to the steady-state phase and continued for ∼10 min and was followed by a very slow deactivation phase. Lowering the H2O2 concentration to 1 mM, the reaction was slowed, allowing all three phases to be observed.
Figure 4.
Tracking conformer compaction to peroxidase activity. (A) The oxidation of 10 mM guaiacol was spectroscopically monitored at 470 nm to measure the peroxidase activity of 14.7 μM cyt c exposed to 1 mM and 7.5 mM H2O2. The control omits cyt c to observe any background oxidation that can take place from H2O2. (B) Shown in red is the rate of accumulation of conformer I with 14.7 μM cyt c exposed to 1 mM H2O2, and black is the rate of accumulation of guaiacol detected in the peroxidase activity using the same concentrations. (C) The change in the rate of cyt c’s peroxidase activity with and without replenishing H2O2 is shown. The light-blue curve is the control, whereas the dark-blue curve is the substrate replenished sample. In each condition, 14.7 μM cyt c was exposed to 1 mM H2O2 for 30 min. After 30 min, 10 μL of ammonium acetate was spiked in the control, whereas 10 μL of 1 mM H2O2 was spiked into the substrate replenished sample. To see this figure in color, go online.
To correlate the structural changes observed by IMS and the onset of the peroxidase activity, native IMS of cyt c was collected for 1 h after 1 mM H2O2 exposure. After a 3-min dead time, native IMS spectra were acquired in 30-s intervals each minute for 60 min. Initially, the compact conformer (conformer I) was a small shoulder protruding from conformer II. After 4 min, conformer I increased in intensity, gaining resolution from conformer II (Fig. S7). The slow accumulation of conformer I correlates with the lag phase observed for the peroxidase activity of cyt c using 1 mM H2O2 (Fig. 4 B, 0–5 min). This was followed by a steady increase in the relative area of conformer I for the next 5 min, consistent with cyt c’s steady-state phase during its peroxidase activity (Fig. 4 B, 5–10 min). And finally, the remainder of the acquisition showed a progressive decrease in the accumulation of conformer I, which was in parallel with the deactivation phase of cyt c’s peroxidase activity (Fig. 4 B, 10–60 min).
To determine whether the deactivation phase resulted from the degradation of cyt c or was due to H2O2 depletion, cyt c’s peroxidase activity was reacquired with an H2O2 replenishment step after 30 min of activity (Fig. 4 C). After the addition of more H2O2, a second steady-state phase of cyt c’s peroxidase activity was observed, showing that cyt c has ∼60% higher peroxidase activity after 30 min of H2O2 exposure. The IMS spectrum at 30 min shows that the majority of cyt c present was in its more compact state, suggesting that the compact conformer of cyt c is peroxidase active and the deactivation phase is due to H2O2 depletion.
The effect of salt on cyt c’s structure
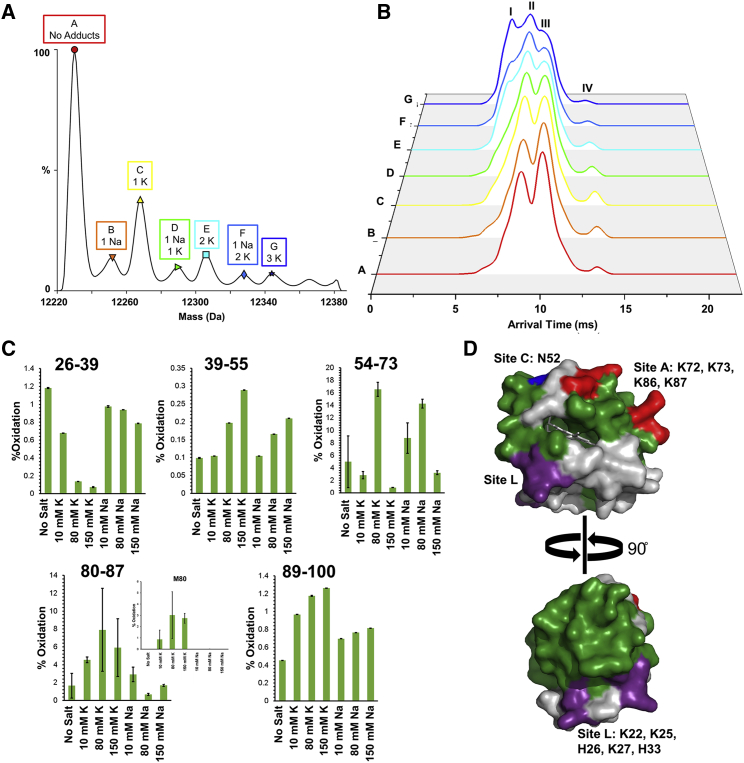
Additional inspection of the native MS and IMS revealed a separate phenomenon. Salt adducts were observed to alter the arrival time distribution of unmodified cyt c in a cation-dependent manner (Fig. 5, A and B). Formation of K+ adducts leads to the appearance and incremental accumulation of conformer I (yellow, cyan, and dark-blue traces in Fig. 5 B). On the other hand, Na+ adducts seem to induce minimal changes to the arrival time distribution (orange, green, and light-blue traces in Fig. 5 B). Whereas ionic strength (47, 48, 49) and anions (9,48) have been shown to modulate its structure, there is little evidence of a cation-specific effect on the structure of cyt c. Functionally, K+ ions have been shown to play a large role in the regulation of apoptotic function of cyt c, in particular during the release from mitochondria and the inhibition of cyt c and apaf1 association (50,51).
Figure 5.
Salt’s effects on cyt c’s conformation. (A) The native cyt c charge state 7+ with labeled salt adducts is shown. (B) The corresponding arrival time distribution of native cyt c for each labeled salt adduct ion shows the appearance of conformer I with increased K+ adducts. (C) The extent of FPOP oxidation for each peptide of cyt c after dialysis (no salt) and with 10, 80, and 150 mM of added KCl and NaCl is shown (mean of three experiments ± standard error). The inset on peptide 80–87 shows the extent of FPOP modification on residue Met80, indicating the significant increase in oxidation with increased K+. (D) The crystal structure (PDB: 2B4Z) of cyt c with the FPOP-labeled peptides highlighted in green, site A highlighted in red, site C highlighted in blue, and site L highlighted in purple is shown. To see this figure in color, go online.
To investigate whether the cation dependence observed in IMS bears any solution-phase relevance, we performed bottom-up FPOP as a function of salt concentration using KCl or NaCl. We observed not only ionic strength but also cation-specific differences in the extent of FPOP modifications. For instance, peptides 89–100 and 39–55 were observed to have increased FPOP modifications as a function of salt concentration irrespective of the type of cation, even though it was more pronounced in K+, indicative of an increase in surface accessibility as a function of ionic strength (Fig. 5 C). On the other hand, peptide 80–87, which comprises native coordinating residue M80, displayed a gradual increase in FPOP modifications as a function of KCl but not NaCl. Within that peptide, examination of residue-level FPOP modifications reveals that M80 makes up a large percentage of the modifications, suggesting that surface accessibility of M80 may be modulated by K+. Also, a drastic decrease in oxidation on peptide 26–39 resulting from increased K+ is observed. This peptide is a part of the site L region of cyt c. Site L helps cyt c bind to the mitochondrial membrane, in particular cardiolipin, through electrostatic interactions using residues K22, K25, K27, H26, and H33 (41). Observing protection of this peptide with increased K+ is indicative of a structural change that could disrupt the electrostatic interactions at site L. Downstream, this could affect its ability to properly stay bound to the membrane. The detachment of cyt c is imperative for cyt c to move into the cytosol for apoptosis. Interestingly, previous observations of an influx of K+ to the mitochondria is linked to cyt c release into the cytosol for the onset of apoptosis (52). These observations are consistent with the cation-specific phenomenon observed with the arrival time distribution of salt adducts and, altogether, suggest some biophysical role for K+ during the transition from mitochondrial to cytosolic functions.
Discussion
Explaining cyt c’s structural transition from the electron transport chain to its peroxidase-active state upon exposure to ROS and its implication for apoptosis has been a subject of much interest. This transition is always accompanied by an increased volume of the heme pocket, oftentimes thought to result from an overall expansion of cyt c’s structure (18,43,44), with the exception of one study observing an overall compaction (42). To characterize this structural transition, we used a common protein footprinting method, FPOP, to gain information on the solvent accessibility of residues, which can then be related to protein structure. However, using FPOP with bottom-up proteomics alone results in a poor correlation to any individual cyt c crystal structure. By combining information from multiple MS-based structural methods, more defined structural information on cyt c can be obtained. Using FPOP with native IMS, we were able to detect significant conformational heterogeneity in cyt c and, more specifically, the progressive accumulation of a compact species upon exposure to H2O2. Subsequent top-down analysis suggested that, in this conformer, M80 is significantly solvent exposed and away from heme coordination, a hallmark of transition from respiratory function to peroxidase function. In parallel with time-resolved native IMS, a time-resolved peroxidase assay revealed that this conformer is highly peroxidase active and not a degradation product of cyt c.
The heterogeneity observed in the ion mobility distribution of unmodified cyt c’s +7 charge state and its relevance to the solution-phase structure has been the subject of previous studies (53, 54, 55, 56, 57). It is possible that some of the late-arriving conformers observed may be the result of partial gas-phase unfolding, and our data do not address this question directly. Regardless, the known susceptibility of cyt c to generating these conformers in the gas phase may also be reflective of the structural changes that it may undergo as it transitions to a peroxidase. Surely, the detection of highly modified residues K73 and H26 upon exposure to H2O2 suggest that these two residues come in close proximity from the distal side of the heme in solution, potentially even in coordination. Such conformations are thought to populate the transition from the native electron transport chain function to its peroxidase apoptotic function (41) and would require significant conformational changes from the respiratory M80 ligated conformation. Similarly, the compaction observed upon exposure to oxidative species may result from modification-induced stabilization from gas-phase unfolding. However, the fact that conformer-specific modification of M80 was observed in postmobility top-down analysis suggests the solution-phase origin of this structural transition. Moreover, at the lowest trap bias energy probed (32 V), a shift in arrival time distribution was still observed upon exposure to H2O2. It is worth noting that recent studies using IMS have also noted the compaction of cyt c in the presence of cardiolipin (57), contrary to what was previously observed (38).
Finally, the compact conformer also preferentially forms adducts with K+ ions rather than Na+ upon ionization. This was corroborated by a K+-specific structural effect detected by bottom-up FPOP in critical structural motifs (distal ligand M80, cardiolipin binding site). The functional implications of the cation-specific observations are unclear. Potassium plays an integral role in the advent of apoptosis with not only an influx into the mitochondria but also an efflux from the cytoplasm (58). The influx of K+ into the mitochondria was seen to be concomitant with the release of cyt c. It is therefore possible that K+-induced structural effects may play a role in the perturbation of the electrostatic interactions between cardiolipin and cyt c, mediated by the L site. In addition, K+ has also been described as inhibiting the first step of the apoptosome complex formation (50,51), the interaction of Apaf1 and cyt c in the cytoplasm. The efflux of K+ from the cytoplasm is thought to help drive the formation of the apoptosome. How the structural effect observed is relevant to the inhibition of the Apaf1-cyt c remains unclear.
Altogether, a combination of FPOP, bottom-up, IMS, and top-down analyses allows us to tease out conformational details in the structural heterogeneity of cyt c, in particular during the peroxidase activation. With further development of FPOP-IMS, we aim to extract quantitative information from the top-down data further correlating FPOP modifications with copopulated conformers in solution.
Author Contributions
L.M.J. supervised the project. E.E.C. acquired the data. E.E.C., D.J.D., and L.M.J. discussed and interpreted the data. E.E.C., D.J.D., and L.M.J. wrote the article.
Acknowledgments
This work was supported by the NSF (MCB1701692). Special thanks to the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP 1841-IQB2014).
Editor: Elizabeth Komives.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.11.011.
Supporting Material
References
- 1.Jiang X., Wang X. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 2.Ow Y.P., Green D.R., Mak T.W. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 3.Li P., Nijhawan D., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 4.Zou H., Li Y., Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 5.Platoshyn O., Zhang S., Yuan J.X. Cytochrome c activates K+ channels before inducing apoptosis. Am. J. Physiol. Cell Physiol. 2002;283:C1298–C1305. doi: 10.1152/ajpcell.00592.2001. [DOI] [PubMed] [Google Scholar]
- 6.Muschel R.J., Bernhard E.J., Koch C.J. Induction of apoptosis at different oxygen tensions: evidence that oxygen radicals do not mediate apoptotic signaling. Cancer Res. 1995;55:995–998. [PubMed] [Google Scholar]
- 7.Chandra J., Samali A., Orrenius S. Triggering and modulation of apoptosis by oxidative stress. Free Radic. Biol. Med. 2000;29:323–333. doi: 10.1016/s0891-5849(00)00302-6. [DOI] [PubMed] [Google Scholar]
- 8.Vacca R.A., Valenti D., Marra E. Cytochrome c is released in a reactive oxygen species-dependent manner and is degraded via caspase-like proteases in tobacco Bright-Yellow 2 cells en route to heat shock-induced cell death. Plant Physiol. 2006;141:208–219. doi: 10.1104/pp.106.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido C., Galluzzi L., Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Paggi D., Hannibal L., Murgida D.H. Multifunctional cytochrome c: learning new tricks from an old dog. Chem. Rev. 2017;117:13382–13460. doi: 10.1021/acs.chemrev.7b00257. [DOI] [PubMed] [Google Scholar]
- 11.Belikova N.A., Vladimirov Y.A., Kagan V.E. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagan V.E., Tyurin V.A., Borisenko G.G. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 13.Colón W., Wakem L.P., Roder H. Identification of the predominant non-native histidine ligand in unfolded cytochrome c. Biochemistry. 1997;36:12535–12541. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 14.Nold S.M., Lei H., Bowler B.E. Effect of a K72A mutation on the structure, stability, dynamics, and peroxidase activity of human cytochrome c. Biochemistry. 2017;56:3358–3368. doi: 10.1021/acs.biochem.7b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosell F.I., Ferrer J.C., Mauk A.G. Proton-linked protein conformational switching: definition of the alkaline conformational transition of yeast iso-1-ferricytochrome c. J. Am. Chem. Soc. 1998;120:11234–11245. [Google Scholar]
- 16.Banci L., Bertini I., Turano P. pH-dependent equilibria of yeast Met80Ala-iso-1-cytochrome c probed by NMR spectroscopy: a comparison with the wild-type protein. Chem. Biol. 1995;2:377–383. doi: 10.1016/1074-5521(95)90218-x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrer J.C., Guillemette J.G., Mauk A.G. Identification of Lys79 as an iron ligand in one form of alkaline yeast iso-1-ferricytochrome c. J. Am. Chem. Soc. 1993;115:7507–7508. [Google Scholar]
- 18.McClelland L.J., Mou T.C., Bowler B.E. Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity. Proc. Natl. Acad. Sci. USA. 2014;111:6648–6653. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vladimirov Y.A., Proskurnina E.V., Alekseev A.V. Molecular mechanisms of apoptosis. structure of cytochrome c-cardiolipin complex. Biochemistry (Mosc.) 2013;78:1086–1097. doi: 10.1134/S0006297913100027. [DOI] [PubMed] [Google Scholar]
- 20.Karsisiotis A.I., Deacon O.M., Worrall J.A. Increased dynamics in the 40-57 Ω-loop of the G41S variant of human cytochrome c promote its pro-apoptotic conformation. Sci. Rep. 2016;6:30447. doi: 10.1038/srep30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence A., Jones C.M., Burkitt M.J. Evidence for the role of a peroxidase compound I-type intermediate in the oxidation of glutathione, NADH, ascorbate, and dichlorofluorescin by cytochrome c/H2O2. Implications for oxidative stress during apoptosis. J. Biol. Chem. 2003;278:29410–29419. doi: 10.1074/jbc.M300054200. [DOI] [PubMed] [Google Scholar]
- 22.Yin V., Shaw G.S., Konermann L. Cytochrome c as a peroxidase: activation of the precatalytic native state by H2O2-induced covalent modifications. J. Am. Chem. Soc. 2017;139:15701–15709. doi: 10.1021/jacs.7b07106. [DOI] [PubMed] [Google Scholar]
- 23.Gau B.C., Sharp J.S., Gross M.L. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones L.M., Zhang H., Gross M.L. Complementary MS methods assist conformational characterization of antibodies with altered S-S bonding networks. J. Am. Soc. Mass Spectrom. 2013;24:835–845. doi: 10.1007/s13361-013-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Gau B.C., Gross M.L. Fast photochemical oxidation of proteins for comparing structures of protein-ligand complexes: the calmodulin-peptide model system. Anal. Chem. 2011;83:311–318. doi: 10.1021/ac102426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W., Ravikumar K.M., Yang S. Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys. J. 2015;108:107–115. doi: 10.1016/j.bpj.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson H.A., McCammon J.A. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000;57:213–218. [PubMed] [Google Scholar]
- 28.Giles K., Pringle S.D., Bateman R.H. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 29.Dixit S.M., Polasky D.A., Ruotolo B.T. Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr. Opin. Chem. Biol. 2018;42:93–100. doi: 10.1016/j.cbpa.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanu A.B., Dwivedi P., Hill H.H., Jr. Ion mobility-mass spectrometry. J. Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 31.Lanucara F., Holman S.W., Eyers C.E. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 32.Chea E.E., Jones L.M. Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins. Protein Sci. 2018;27:1047–1056. doi: 10.1002/pro.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hambly D.M., Gross M.L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Marty M.T., Baldwin A.J., Robinson C.V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fellers R.T., Greer J.B., Thomas P.M. ProSight Lite: graphical software to analyze top-down mass spectrometry data. Proteomics. 2015;15:1235–1238. doi: 10.1002/pmic.201570050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin V., Mian S.H., Konermann L. Lysine carbonylation is a previously unrecognized contributor to peroxidase activation of cytochrome c by chloramine-T. Chem. Sci. (Camb.) 2019;10:2349–2359. doi: 10.1039/c8sc03624a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinkam P., Zimmermann J., Romesberg F.E. Characterization of alkaline transitions in ferricytochrome c using carbon-deuterium infrared probes. Biochemistry. 2008;47:13470–13480. doi: 10.1021/bi801223n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanske J., Toffey J.R., Pletneva E.V. Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theisen A., Black R., Barran P.E. Initial protein unfolding events in ubiquitin, cytochrome c and myoglobin are revealed with the use of 213 nm UVPD coupled to IM-MS. J. Am. Soc. Mass Spectrom. 2019;30:24–33. doi: 10.1007/s13361-018-1992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skilton St.J., Campuzano I., Chen W. Application Notes posted; 2010. Improved Ion Mobility Separation of Protein Conformations in the Gas Phase with SYNAPT G2 HDMS. APNT10174841.https://www.waters.com/waters/library.htm?cid=511436&lid=10174841&locale=en_US [Google Scholar]
- 41.Hannibal L., Tomasina F., Radi R. Alternative conformations of cytochrome c: structure, function, and detection. Biochemistry. 2016;55:407–428. doi: 10.1021/acs.biochem.5b01385. [DOI] [PubMed] [Google Scholar]
- 42.Amacher J.F., Zhong F., Pletneva E.V. A compact structure of cytochrome c trapped in a lysine-ligated state: loop refolding and functional implications of a conformational switch. J. Am. Chem. Soc. 2015;137:8435–8449. doi: 10.1021/jacs.5b01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson C.J., Bowler B.E. pH dependence of formation of a partially unfolded state of a Lys 73--> His variant of iso-1-cytochrome c: implications for the alkaline conformational transition of cytochrome c. Biochemistry. 2000;39:13584–13594. doi: 10.1021/bi0017778. [DOI] [PubMed] [Google Scholar]
- 44.Assfalg M., Bertini I., Gray H.B. Structural model for an alkaline form of ferricytochrome C. J. Am. Chem. Soc. 2003;125:2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 45.Vincelli A.J., Pottinger D.S., Pletneva E.V. Recombinant expression, biophysical characterization, and cardiolipin-induced changes of two Caenorhabditis elegans cytochrome c proteins. Biochemistry. 2013;52:653–666. doi: 10.1021/bi3014938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diederix R.E., Fittipaldi M., Canters G.W. Kinetic stability of the peroxidase activity of unfolded cytochrome c: heme degradation and catalyst inactivation by hydrogen peroxide. Inorg. Chem. 2003;42:7249–7257. doi: 10.1021/ic0343861. [DOI] [PubMed] [Google Scholar]
- 47.Purring-Koch C., McLendon G. Cytochrome c binding to Apaf-1: the effects of dATP and ionic strength. Proc. Natl. Acad. Sci. USA. 2000;97:11928–11931. doi: 10.1073/pnas.220416197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah R., Schweitzer-Stenner R. Structural changes of horse heart ferricytochrome C induced by changes of ionic strength and anion binding. Biochemistry. 2008;47:5250–5257. doi: 10.1021/bi702492n. [DOI] [PubMed] [Google Scholar]
- 49.Trewhella J., Carlson V.A., Heidorn D.B. Differences in the solution structures of oxidized and reduced cytochrome c measured by small-angle X-ray scattering. Biochemistry. 1988;27:1121–1125. doi: 10.1021/bi00404a007. [DOI] [PubMed] [Google Scholar]
- 50.Cain K., Langlais C., Cohen G.M. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J. Biol. Chem. 2001;276:41985–41990. doi: 10.1074/jbc.M107419200. [DOI] [PubMed] [Google Scholar]
- 51.Karki P., Seong C., Park I.S. Intracellular K(+) inhibits apoptosis by suppressing the Apaf-1 apoptosome formation and subsequent downstream pathways but not cytochrome c release. Cell Death Differ. 2007;14:2068–2075. doi: 10.1038/sj.cdd.4402221. [DOI] [PubMed] [Google Scholar]
- 52.Gogvadze V., Orrenius S., Zhivotovsky B. Multiple pathways of cytochrome c release from mitochondria in apoptosis. Biochim Biophys Acta. 2006;1757:639–647. doi: 10.1016/j.bbabio.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 53.Clemmer D.E., Hudgins R.R., Jarrold M.F. Naked protein conformations: cytochrome c in the gas phase. J. Am. Chem. Soc. 1995;117:10141–10142. [Google Scholar]
- 54.Shelimov K.B., Clemmer D.E., Jarrold M.F. Protein structure in vacuo: gas-phase conformations of BPTI and cytochrome c. J. Am. Chem. Soc. 1997;119:2240–2248. [Google Scholar]
- 55.Allen S.J., Eaton R.M., Bush M.F. Structural dynamics of native-like ions in the gas phase: results from tandem ion mobility of cytochrome c. Anal. Chem. 2017;89:7527–7534. doi: 10.1021/acs.analchem.7b01234. [DOI] [PubMed] [Google Scholar]
- 56.Wood T.D., Chorush R.A., McLafferty F.W. Gas-phase folding and unfolding of cytochrome c cations. Proc. Natl. Acad. Sci. USA. 1995;92:2451–2454. doi: 10.1073/pnas.92.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szymkowicz L., Lento C., Wilson D.J. Impact of cardiolipin and phosphatidylcholine interactions on the conformational ensemble of cytochrome c. Biochemistry. 2019;58:3617–3626. doi: 10.1021/acs.biochem.9b00495. [DOI] [PubMed] [Google Scholar]
- 58.Gogvadze V., Robertson J.D., Orrenius S. Mitochondrial cytochrome c release may occur by volume-dependent mechanisms not involving permeability transition. Biochem. J. 2004;378:213–217. doi: 10.1042/BJ20031193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.