Abstract
Background
The genus Trichococcus currently contains nine species: T. flocculiformis, T. pasteurii, T. palustris, T. collinsii, T. patagoniensis, T. ilyis, T. paludicola, T. alkaliphilus, and T. shcherbakoviae. In general, Trichococcus species can degrade a wide range of carbohydrates. However, only T. pasteurii and a non-characterized strain of Trichococcus, strain ES5, have the capacity of converting glycerol to mainly 1,3-propanediol. Comparative genomic analysis of Trichococcus species provides the opportunity to further explore the physiological potential and uncover novel properties of this genus.
Results
In this study, a genotype-phenotype comparative analysis of Trichococcus strains was performed. The genome of Trichococcus strain ES5 was sequenced and included in the comparison with the other nine type strains. Genes encoding functions related to e.g. the utilization of different carbon sources (glycerol, arabinan and alginate), antibiotic resistance, tolerance to low temperature and osmoregulation could be identified in all the sequences analysed. T. pasteurii and Trichococcus strain ES5 contain a operon with genes encoding necessary enzymes for 1,3-PDO production from glycerol. All the analysed genomes comprise genes encoding for cold shock domains, but only five of the Trichococcus species can grow at 0 °C. Protein domains associated to osmoregulation mechanisms are encoded in the genomes of all Trichococcus species, except in T. palustris, which had a lower resistance to salinity than the other nine studied Trichococcus strains.
Conclusions
Genome analysis and comparison of ten Trichococcus strains allowed the identification of physiological traits related to substrate utilization and environmental stress resistance (e.g. to cold and salinity). Some substrates were used by single species, e.g. alginate by T. collinsii and arabinan by T. alkaliphilus. Strain ES5 may represent a subspecies of Trichococcus flocculiformis and contrary to the type strain (DSM 2094T), is able to grow on glycerol with the production of 1,3-propanediol.
Keywords: Comparative genomics; Protein domains; Halophilic; Psychrophilic; 1,3-propanediol
Background
Type strains of existing Trichococcus species have been isolated from diverse and geographically spread ecosystems. Various species derive from waste treatment systems or contaminated sites: T. flocculiformis (activated sludge) [1], T. pasteurii (septic pit sludge) [2], T. collinsii (soil spilled with hydrocarbons) [2], T. ilyis (sulfate reducing anaerobic sludge) [3], T. shcherbakoviae (sludge from low-temperature anaerobic reactor) [4]; while others were isolated from natural environments: T. patagoniensis (guano from penguin, Patagonia) [5], T. palustris (swamp, Russia) [2], and T. paludicola and T. alkaliphilus (high elevation wetland, Tibet) [6].
Trichococcus species share a very high 16S rRNA gene sequence identity, in the range of 98–100% [2–4, 6]. This often impairs the taxonomic classification of new strains within this genus on the basis of 16S rRNA gene sequence identity, and therefore whole genome comparison needs to be performed. This was traditionally done by experimental DNA-DNA hybridisation, but nowadays it is also possible to use genomic information to perform digital DNA-DNA hybridisation (dDDH) [7] or average nucleotide identity (ANI) [8] analyses. Availability of genomic information provides also the opportunity for comparing and analysing gene/function diversity among different species. Functional genome analysis on the level of protein domains can be used to infer potential metabolic functions, thereby connecting genotype and physiology [9, 10].
Trichococcus species are related to the lactic acid bacteria (LAB), and phylogenetically close to the genera Carnobacterium and Aerococcus [11]. Described Trichococcus species can all grow on glucose, cellobiose, D-mannose, fructose and sucrose [1–6]. However, T. pasteurii and Trichococcus strain ES5 are the only strains within the genus capable of converting glycerol to mainly 1,3-PDO [12], with comparable product yields to those of other 1,3-PDO producers, such as Clostridium butyricum and Klebsiella pneumoniae [13, 14]. 1,3-PDO is used as a building block in chemical industry [15], and the discovery of new efficient and resilient biocatalysts for its production are of interest for biotechnological industry. In general, Trichococcus species have a broad temperature range for growth (commonly from 4 °C to 40 °C) [1–6]. T. patagoniensis and T. shcherbakoviae can grow at negative temperatures and tolerate salinities up to 5% (w/v) NaCl [4, 5], which is also the case for several related Carnobacterium species, such as C. funditum, C. alterfunditum and C. pleistocenium [16, 17], but no other Trichococcus species.
The objective of this study was to use functional genome analysis, based on encoded protein domains, for identifying novel metabolic traits in Trichococcus species. Searches were preferentially directed to find properties that can confer versatility to these species in terms of industrial applications such as, types of substrates used, products formed, and resistance to environmental stress.
Results
Comparison of protein domains among Trichococcus species
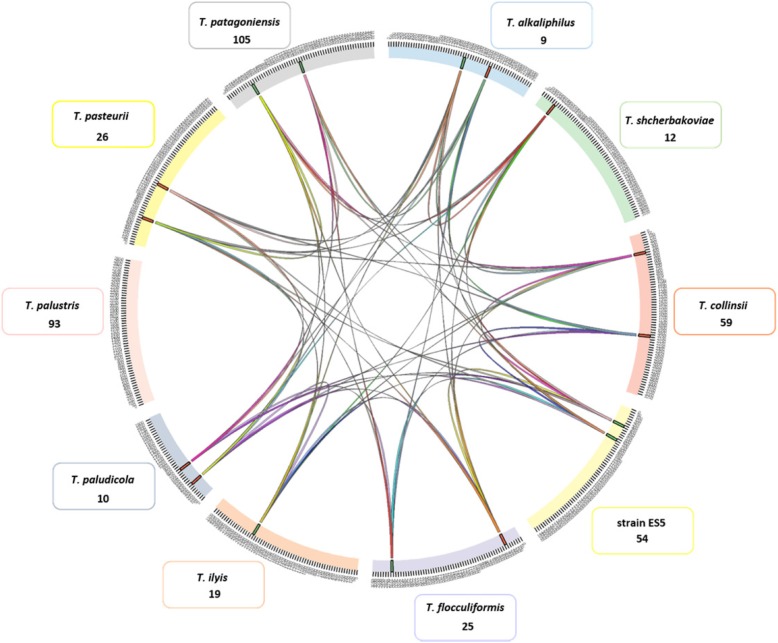
Genome sequences of currently available type strains from the genus Trichococcus – i.e. T. flocculiformis, T. pasteurii, T. palustris, T. collinsii, T. patagoniensis, T. ilyis, T. paludicola, T. alkaliphilus, and T. shcherbakoviae were retrieved from NCBI. In addition, we sequenced the genome of Trichococcus strain ES5, described by Gelder et al. [12]. Strain ES5 is able to convert glycerol to 1,3-PDO, a property that is also found in T. pasteurii, but not in the other Trichococcus species. The Trichococcus species have similar genome sizes (around 3 Mbp), with the exception of T. paludicola that has an estimated genome size of ~ 2 Mbp. However, a completeness assessment of the genomes using BUSCO [18] showed a higher percentage of missing genes in the genome of T. paludicola (i.e. 25.1% missing BUSCOs in T. paludicola and 2.0–2.7% missing BUSCOs in the genomes of the other Trichococcus species) (Additional file 1: Figure S1). Genomes of Trichococcus species and other closely related bacteria (Additional file 1: Table S1) were (re) annotated using the pipeline of Semantic Annotation Platform with Provenance (SAPP) [19], which allows to obtain the predicted genes and protein domain annotations. The resulting matrix with all the domains identified in the different Trichococcus strains is provided in Additional file 2. Among all the analysed strains (T. paludicola was not included in the calculations because of the low number of identified domains), 1424 core protein domains and 1983 pan protein domains could be identified, with multiple protein domains conserved in the different genomes of analysed Trichococcus species (Additional file 2). All Trichococcus genomes shared genomic blocks of 45 kb, except T. palustris (Fig. 1, Additional file 3). In these genomic blocks, 110 domains were identified, with the majority belonging to peptidases, transferases (e.g. acyltransferase, phospholipid/glycerol acyltransferase, phosphatidyltransferase, aminotransferase) and DNA polymerases. Domains of proteins related to carbohydrate metabolism were abundant in the shared genomic blocks among Trichococcus species, which correlates to the ability to degrade multiple sugars.
Fig. 1.
Conserved genomic blocks in the genomes of the ten Trichococcus species compared in this study (represented in the figure are only syntenies larger than 45 kb). Each colour represents a Trichococcus species and coloured lines indicate shared genomic blocks; The majority of the Trichococcus species share two and three 45 kb genomic regions. Note that T. palustris has no shared syntenic regions larger than 45 kb with other Trichococcus species. Numbers indicated below species names indicate the unique protein domains in each of the genomes
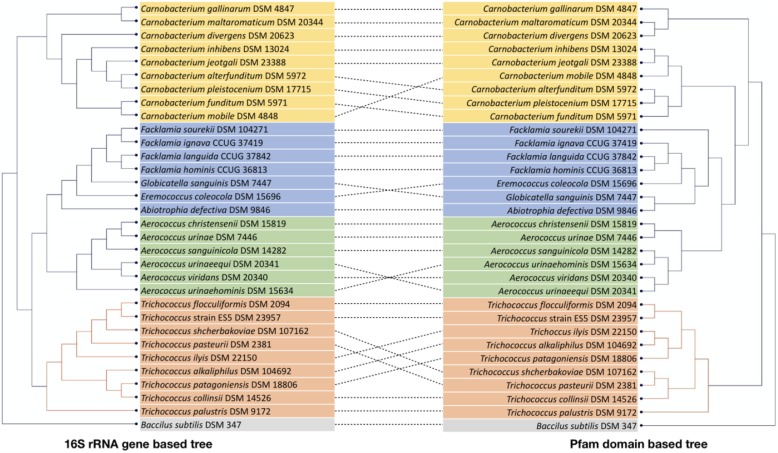
Protein domain-based clustering of Trichococcus species, and other closely related LAB, is shown in Fig. 2 (T. paludicola was not included due to the low number of identified domains). Specifically for the Trichococcus group, it is patent that using protein domains or 16S rRNA genes results in different clustering of the bacteria. This corroborates with the fact that information in the 16S rRNA gene of Trichococcus species is not enough to resolve taxonomy at species level [3, 4, 6], and does not predict the functional relatedness of the different species. 16S rRNA gene and protein domain clustering for the other analysed LAB species is much more conserved (Fig. 2).
Fig. 2.
Dendrograms produced by hierarchical clustering of 16S rRNA gene sequences (left pane) and protein domains (right pane), both showing the Trichococcus strains analysed in this work and closely related lactic acid bacteria (LAB). Bacillus subtilis was used as an outgroup. 16S rRNA gene-based clustering tree was constructed using neighbor-joining algorithm using the software CLC Main Workbench v8.0 (CLC Bio, Aarhus, Denmark). Protein domains are clustered based on presence/ absence in the genomes by applying neighbor-joining method with Dice coefficient using DARwin v6.0 [20]
The SAPP-generated protein domain matrix (Additional file 2) was mined for the identification of metabolic traits in Trichococcus species. A set of metabolic traits (identified in Table 1) was selected for further in vitro testing. One of the most varied aspects among Trichococcus species was the capacity to utilize more substrates than previously described, such as glycerol by T. pasteurii and Trichococcus strain ES5, alginate by T. collinsii and arabinan by T. alkaliphilus (Table 1). Protein domains related to cold adaption and osmoregulation mechanisms, and to defence mechanisms, were identified in all the analysed Trichococcus.
Table 1.
Genes and protein domains highlighted in this study as a result of functional genome analysis of ten Trichococcus strains. Strains (Locus tag_): 1. T. flocculiformis (Tflo_); 2. Trichococcocus strain ES5 (TES5_); 3. T. pasteurii (Tpas_); 4. T. palustris (Tpal_); 5. T. collinsii (Tcol_); 6. T. patagoniensis (Tpat_); 7. T. ilyis (TR210_); 8. T. alkaliphilus (PXZT_); 9. T. paludicola (Ga019_); 10. T. shcherbakoviae (TART1_)
| Feature | Functional genome annotation | Locus tag |
|---|---|---|
| Strain | (Protein domains) | |
| 1,3-PDO production | ||
| 2|3 | Glycerol kinase (IPR005999, IPR018483, IPR018484, IPR018485) | TES5_2082|Tpas_2911 |
| 2|3 | Dihydroxyacetone activator (IPR009057, IPR015893) | TES5_2083|Tpas_2912 |
| 2|3 | Glycerol dehydrogenases (IPR018211, IPR001670) | TES5_2084|Tpas_2913 |
| 2|3 | Phosphoenolpyruvate phosphotransferase (IPR004006, IPR004007, IPR012736, IPR004701, IPR012844) | TES5_2085–2087|Tpas_2914–2916 |
| 2|3 | 1,3-propanediol dehydrogenases (IPR001670, IPR018211) | TES5_2088|Tpas_2917 |
| 2|3 | Dihydroxyacetone (IPR004007, IPR012737) | TES5_2089–2090|Tpas_2918–2919 |
| 2|3 | Glycerol dehydratase (IPR003206, IPR016176, IPR003208, IPR010254, IPR003207 | TES5_2091–2093|Tpas_2920–2922 |
| 2|3 | Glycerol dehydratase activator (IPR028975, IPR030994, IPR003208, IPR010254) | TES5_2094–2095|Tpas_2923–2924 |
| 2|3 | Cobalamin adenosyltransferase (IPR016030, IPR029499) | TES5_2096|Tpas_2925 |
| 2|3 | Hypothetical protein (IPR005624) | TES5_2097,2099|Tpas_2926, 2928 |
| 2|3 | Glycerol uptake facilitator (IPR000425, IPR022357, IPR023271) | TES5_2098|Tpas_2927 |
| Alginate utilisation | ||
| 5 | Alginate lyase (IPR008929) | Tcol_1369,1377,1704 |
| Arabinan utilisation | ||
| 6 | Glycosyl hydrolase (IPR033132) |
Tpat_54,101,590,610,948-949,136,1167,1171,1259,2028,2033, 2527–2528,2577,2585,2682 |
| 6 | Metal-dependent hydrolase (IPR032466) | Tpat_57,88,320,321,954,1043,1060,1227,1247,1391, 1392,2241 |
| 6 | Extracellular endo-alpha-(1- > 5)-L-arabinanase (IPR032291) | Tpat_1197,1296 |
| Psychrotolerance | ||
| All | Cold-shock protein (IPR000194, IPR003593, IPR004665, IPR011112, IPR011113, IPR011129, IPR012340, IPR027417) | TR210_741|Tpas_88|Tpal_285|TES5_627|Tflo_313| Tcol_65|Tpat_494|PXZT01000007.1_99| Ga0192377_1015_35 | TART1_1674 |
| All | Cold shock protein signature (IPR002059, IPR011129, IPR012340, IPR019844) | Tpat_494, 1801, 1802, 1901, 1923|Tcol_65, 532, 554, 1698, 1699|Tflo_313, 455, 458, 688, 980|TES5_627, 827, 849, 1357,1359|Tpal_285,869,1036,1801,1820,1877| Tpas_88,599,1471,1472,2297,2758|TR210_741,1024,1470, 1709, 1819, 1842| PXZT01000016.1_53, 1.1_301, 5.1_152, 4.1_46, 5.1_150| Ga0192377_1004_168, 1008_10–11, 1002_82, 1004_145 | TART1_1477, 1504, 2070, 2071, 2352 |
| Salinity tolerance | ||
| 7 | Glycine betaine transporter OpuD (IPR000060) | TR210_1348 |
| 3|5|6|7|8|9|10 | Betaine binding ABC transporter protein (IPR000515) | Tpas_2814–2815|Tcol_1997|Tpat_1468|TR210_2767–2768,2770 | Ga0192364_3215_54–57 | PXZT01000008. _23–26| TART1_2694–96 |
| 1|2|5|7|8|9|10 | Osmotically activated choline ABC transporter (IPR003439) | TES5_1206–1209|Tcol_773–776|TR210_342–345|Tflo_1131–1134|Ga0192364_2415_1215| PXZT01000003.2_54–57| TART1_266–69 |
| 2|3|6|7 | Choline binding protein A (IPR010126) | TES5_1355|Tpas_1469|Tpat_1570|TR210_2363,1711,2104 |
| 1|2|3|5|6|8|9 | Glycine betaine ABC transport system (IPR003439) | TES5_1660–1662|Tcol_1041–1043|Tpas_2619–21|Tpat_203–05|Tflo_1599–01 | Ga0192356_1653_23–25| PXZT01000006.2_75–77 |
| Bacterial defence | ||
| 6|10 | SNARE associated Golgi protein (IPR032816) | Tpat_1693,1825 | TART1_1950 |
| 4 | Tetracycline resistance (IPR004638) | Tpal_1098,1664,1687 |
| 3 | Toxin antidote HigA (IPR013430) | Tpas_511 |
| 3 | Plasmid system killer (IPR007712) | Tpas_512 |
| 7 | Bacteriocin class Iib (IPR010387, IPR029500) | Tflo_874,878–879 |
| 6 | Cas9 (IPR028629) | Tpat_1430 |
| 1|2|6 | Cas1 (IPR019855) | TES5_196|Tpat_1431|Tflo_184 |
| 6|10 | Cas2 (IPR019199) | TES5_195|Tpat_1432| TART1_1189 |
| 1|2|3|7|10 | Cas3 (IPR006935) | TES5_201|Tpas_1155|TR210_680|Tflo_179|TART1_0176 |
| 1|2|3|6 | Cas5d (IPR021124) | TES5_200|Tpas_1156|TR210_679|Tflo_180 |
| 1|2|6|7 | Casd1 (IPR010144) | TES5_199|Tpas_1157|TR210_675|Tflo_181 |
| 1|2|6|7 | Csd2 (IPR006482) | TES5_198|Tpas_1158| TR210_677|Tflo_182 |
| 1|2|6|7 | Csd4 (IPR022765) | Tpas_1159| TR210_676|Tflo_183 |
Carbohydrate degradation by Trichococcus species
In general, Trichococcus species can utilise cellobiose, sucrose, maltose, and glucose [1–6]. Genes encoding proteins for the Embden-Meyerhof-Parnas (EMP) pathway and pentose phosphate pathway (PPP) were found in the genomes of the ten Trichococcus species analysed here. In addition, genes encoding proteins for the conversion of pyruvate to ethanol, acetate and lactate were found. This is consistent with the products (lactate, formate, acetate and ethanol) formed from glucose fermentation by the tested Trichococcus species (Table 2). Lactate was the main fermentation product, except in cultures of T. patagoniensis. The carbon fraction in lactate in cultures of T. patagoniensis was around 40% (calculated as carbon lactate/carbon all soluble products), while in other Trichococcus cultures lactate corresponded to 60–80% of the carbon detected in the products. Glucose fermentation by T. patagoniensis resulted in a relatively higher formate concentration, which is in agreement with the presence of a pyruvate formate-lyase in the genome of T. patagoniensis (Tpat_2317) and not in others. Ethanol yield in cultures of T. patagoniensis and T. collinsii was 0.2 and 0.1 molethanol/molconsumed glucose, respectively, which is higher than observed for the other Trichococcus species.
Table 2.
Glucose (a) and glycerol (b) fermentation by Trichococcus species. Table shows substrate consumption and product generation (± standard deviation, triplicate assays), measured after 24 h for glucose fermentation experiments and after 40 h for glycerol fermentation experiments. Electron recovery was calculated based on substrate/product consumption/production and excludes electrons used for cellular growth
| (a) Glucose Fermentation | Glucose consumed (mM) | Lactate (mM) | Formate (mM) | Acetate (mM) | Ethanol (mM) | Electron recovery (%) |
| T. flocculiformis (DSM 2094T) | 19.1 ± 0.6 | 21.7 ± 2.1 | 6.9 ± 0.6 | 2.6 ± 0.3 | 3.9 ± 0.2 | 76.5 ± 1.5 |
| Strain ES5 (DSM 23957) | 19.6 ± 0.2 | 22.2 ± 0.7 | 7.5 ± 0.5 | 2.2 ± 0.1 | 4.1 ± 0.2 | 75.0 ± 0.5 |
| T. pasteurii (DSM 2381T) | 18.4 ± 1.0 | 23.8 ± 0.9 | 5.1 ± 0.4 | 1.5 ± 0.0 | 1.9 ± 0.7 | 77.2 ± 1.3 |
| T. palustris (DSM 9172T) | 19.2 ± 0.4 | 16.2 ± 0.9 | 12.6 ± 0.4 | 4.9 ± 0.4 | 6.6 ± 0.2 | 74.1 ± 0.7 |
| T. collinsii (DSM 14526T) | 13.1 ± 0.6 | 20.2 ± 0.6 | 3.3 ± 0.3 | 0.6 ± 0.2 | 1.1 ± 0.1 | 90.4 ± 0.8 |
| T. patagoniensis (DSM 18806T) | 19.1 ± 0.9 | 11.4 ± 1.0 | 18.2 ± 0.9 | 6.8 ± 0.3 | 9.0 ± 0.3 | 75.1 ± 1.0 |
| T. ilyis (DSM 22150T) | 18.9 ± 0.6 | 19.8 ± 0.9 | 8.8 ± 0.5 | 3.2 ± 0.3 | 4.6 ± 0.3 | 75.5 ± 0.8 |
| (b) Glycerol Fermentation | Glycerol consumed (mM) | Lactate (mM) | Formate (mM) | Acetate (mM) | 1,3-PDO (mM) | Electron recovery (%) |
| T. pasteurii (DSM 2381T) | 18.5 | 0.5 ± 0.1 | 0.9 ± 0.5 | 3.6 ± 0.7 | 13.8 ± 0.2 | 99.4 ± 0.6 |
| Strain ES5 (DSM 23957) | 19.0 | 0.5 ± 0.0 | 2.3 ± 0.2 | 4.5 ± 0.2 | 12.3 ± 0.1 | 90.7 ± 0.1 |
T. pasteurii and Trichococcus strain ES5 can ferment glycerol. The most abundant product from glycerol fermentation by T. pasteurii and Trichococcus strain ES5 is 1,3-propanediol (1,3-PDO), which represents about 70–80% of the total carbon detected in products (Table 2). The genomes of these species contain an identical large operon (17 genes organized in identical fashion and with 100% sequence identity), which is involved in glycerol conversion (Table 1). This operon is absent in the other eight studied Trichococcus species that cannot degrade glycerol. Two of the genes in this operon are essential for glycerol conversion to 1,3-PDO: glycerol dehydratase (alpha, beta and gamma subunits) and 1,3-propanediol dehydrogenase. Additional genes in the operon encode for: a glycerol uptake facilitator, a glycerol dehydratase activator (involved in the activation of glycerol dehydratase), and cobalamin adenosyltransferase which is involved in the conversion of cobalamin (vitamin B12) to its coenzyme form, adenosylcobalamin (glycerol dehydratase requires vitamin B12 as a binding co-factor [21]).
T. collinsii has unique domains related to alginate utilisation and encodes three alginate lyases (Table 1). In vitro testing confirmed that T. collinsii utilises alginate (optical density increase of about 0.2 after 72 h incubation).
In the genome of T. patagoniensis, 17 homologous domains of glycoside hydrolases family 1 (includes e.g. glucosidases, galactosidases and hydrolases) were identified, but they all belong to genes encoding hypothetical proteins (Table 1). Metal-dependent hydrolases were identified with 12 homologous genes in the genome of T. patagoniensis. In addition, two copies of the gene encoding for extracellular endo-alpha-(1- > 5)-L-arabinanase are present in the genome. This enzyme catalyses the degradation of arabinan and it is an important enzyme in the degradation of the plant cell wall. To confirm the protein domains prediction, growth of T. patagoniensis on arabinan was tested in vitro. T. patagoniensis could utilise and grow on arabinan (OD of 0.25 ± 0.02 after 96 h incubation).
Growth of Trichococcus species at low temperature
Six cold shock domains (CSD) (IPR011129) were encoded in all Trichococcus genomes (Table 1). One additional CSD was encoded in the genomes of T. palustris and T. ilyis. The conserved CSDs in Trichococcus species were neighbouring genes encoding for domains of the cold-shock DNA-binding site (IPR002059), the nucleic acid-binding OB-fold (IPR012340) and the cold-shock conserved site (IPR019844). One of the CSD is part of a cold shock protein (Table 1), which contains additional domains likely involved in the transcription and regulation of the cold protection mechanisms: ATPase F1 nucleotide-binding (IPR000194), AAA+ ATPase (IPR003593), transcription termination factor Rho (IPR004665), rho termination factor N-terminal (IPR011112), rho termination factor RNA-binding domain (IPR011113), nucleic acid-binding OB-fold domain (IPR012340) and P-loop containing nucleoside triphosphate hydrolase domain (IPR027417). Genomes of twenty-two LAB species closely related to Trichococcus species were analysed for CSDs (complete list of LAB species in Additional file 1: Table S1). A similar cold shock protein to the one encoded in the genomes of Trichococus species was identified in the twenty-two genomes of LAB species, but only seven LAB species contain six to eight additional CSD (Carnobacterium mobile, C. pleistocenium, C. jeotgali, C. inhibens, C. funditum, C. maltaromaticum, C. alterfunditum).
Overall, Trichococcus species can grow at temperatures lower than their optimum growth temperature (25–30 °C) [1–6]. Only four of the Trichococcus species tested in this study were able to grow at 0 °C (on glucose, and over 45 days of incubation): T. pasteurii, T. collinsii, T. patagoniensis and Trichococcus strain ES5 (Additional file 4: Figure S2). At 0 °C, T. patagoniensis and T. palustris had a lag phase of eight days, whereas growth of T. collinsii and Trichococcus strain ES5 was only observed after 23 days of incubation. The recently described T. shcherbakoviae is also able to grow at freezing temperatures [4].
Resistance of Trichococcus to high salinity
Functional genome analysis resulted in the identification of protein domains related to osmoregulation in all the Trichococcus species, except in T. palustris (Table 1). Multiple domains related to glycine and betaine transport systems could be identified. These transport systems are important for living at high salinity because, during osmotic pressure, bacterial cells can increase the concentration of uncharged osmoprotectants (glycine, betaine) in the cytoplasm [22, 23]. In addition, choline transporters were also identified. Glycine and betaine can be formed from choline [24].
Salinity tolerance for the different Trichococcus species was tested. Only T. palustris was sensitive to salinity, and growth was inhibited at 2% NaCl (Additional file 4: Figure S3). All the other tested strains could grow in media with a NaCl concentration of 2%. At 4% salinity and after 6 days, growth was observed for only four of the tested bacteria: T. pasteurii, T. patagoniensis, T. flocculiformis, and Trichococcus strain ES5. After ten days, weak growth was observed at 6% NaCl for T. patagoniensis, T. pasteurii and Trichococcus strain ES5 (Additional file 4: Figure S3). T. paludicola and T. alkaliphilus were previously observed to tolerate NaCl concentrations up to 4.5% [6].
CRISPR and antibiotic resistance genes in Trichococcus species
Recent studies support the effective defence of the CRISPR system in bacteria against viral threats [25]. The CRISPR system contains Cas genes which introduce double strand breaks in foreign DNA in the cells. Cas genes were present in T. flocculiformis, T. pasteurii, T. patagoniensis, T. ilyis, and Trichococcus strain ES5 (Table 1). The CRISPR system in T. patagoniensis can be classified as Cas2, type II-C, while the other studied Trichococcus species encode the class 1 type I-C CRISPR system. Several spacer sequences (i.e. foreign nucleic acid sequences merged in the genome by CRISPR systems) were found in the genomes Trichococcus species: T. pasteurii (115 spacer sequences), T. patagoniensis (88 spacer sequences), Trichococcus strain ES5 (82 spacer sequences), T. ilyis (80 spacer sequences), T. fluccoliformis (27 spacer sequences). The alignment of the spacers sequences from the analysed Trichococcus species resulted in low similarity, likely not containing common foreign DNA.
Alternative defense mechanisms were also found (Table 1). The domain of SNARE associated Golgi protein was encoded in the genomes of T. patagoniensis and T. shcherbakoviae. SNARE proteins can be used for promoting or blocking membrane fusion and act especially against eukaryotic cells [26]. T. palustris contains genes encoding for tetracycline resistance proteins (Table 1), which were not found in the genomes of the other Trichococcus species. Agar plates containing Clostridium medium and increasing concentrations of tetracycline (0.016–256 μg/mL) were used to test resistance to this antibiotic. T. palustris could grow in plates containing 4 μg/mL, whereas T. ilyis and T. palustris did not tolerate tetracycline at this concentration. Genes encoding a toxin antidote protein HigA and a plasmid system killer were found in T. pasteurii (Table 1). The two genes are associated with bacterial toxin-antitoxin (TA) proteins and regulate the tolerance of the cells at environment and chemical stress [27]. The genome of T. flocculiformis contains three homologous genes for the domain bacteriocin class IIb, which is commonly associated with growth inhibition of several microorganisms [28].
Comparison of Trichococcus strain ES5 and T.flocculiformis
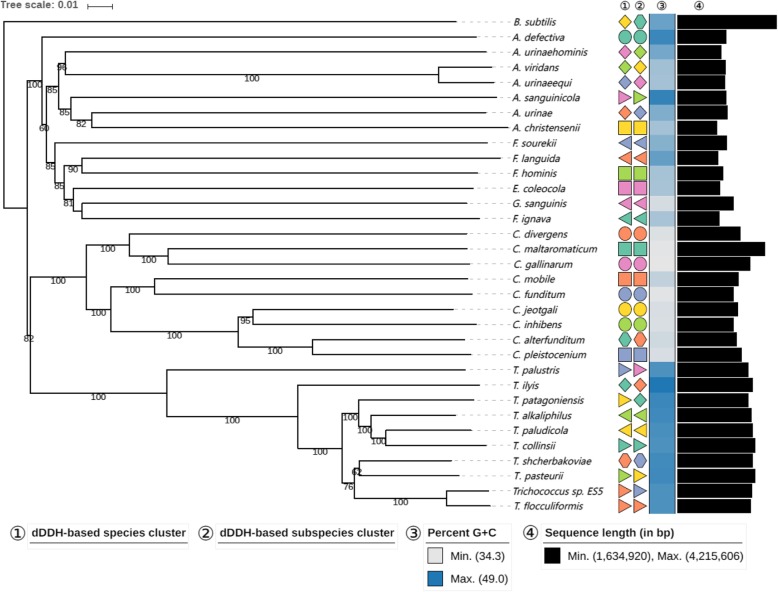
Trichococcus strain ES5 was previously isolated by van Gelder et al. [12]. Based on 16S rRNA gene comparison, strain ES5 was phylogenetically closely related to T. flocculiformis (99%). However, it is known that Trichococcus species have a highly conserved 16S rRNA gene and a correct taxonomic affiliation demands DNA-DNA hybridization [3, 4, 6]. Digital DNA-DNA hybridization (dDDH) between strain ES5 and T. flocculiformis is 71%, with a confidence interval between [68.0–73.9%] (Additional file 5). This value is just above the 70% cut-off value generally recommended for species differentiation [7]. Furthermore, it is below the 79% cut-off value for subspecies delineation [29]. Average Nucleotide Identity (ANI) between strain ES5 and T. flocculiformis is 95.9%, which is above the cut-off value of 95% [8]. Based on these results strain ES5 is a T. flocculiformis strain (Fig. 3; Additional file 5). Nevertheless, strain ES5 has unique physiological properties that are not observed in the type strain, such as the ability to ferment glycerol and an apparent higher tolerance to salinity (could grow at 6% NaCl).
Fig. 3.
Genome-based phylogenomic analysis of Trichococcus species restricted to coding regions. Tree inferred with FastME 2.1.4 from Genome Blast Distance Phylogeny GBDP distances calculated from the ten Trichococcus species, 22 LAB species and B. subtilis genome sequences. The branch lengths are scaled in terms of GBDP distance. The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications, with an average branch support of 88%. Leaf labels are further annotated by their affiliation to species (①, identical symbol shape and color indicate same species clade) and subspecies (②, identical symbol shape and color indicate same subspecies clade) clusters as well as their genomic G + C content ③ and their overall genome sequence length ④
Discussion
The comparative analysis of Trichococcus species described here served two purposes. First, it allowed to identify and predict novel physiological traits within the genus Trichococcus species. Second, a proper taxonomic position of the several analysed Trichococcus strains could be made.
Taxonomic classification of Trichococcus species
The 16S rRNA gene is commonly used for taxonomic classification. However, this gene of Trichoccocus species is highly conserved and thus it cannot be used for taxonomical classification at species level. Therefore, assigning a novel Trichococcus strain to a certain species is more challenging than in other genera. As an example, T. patagoniensis and T. collinsii have a 100% similar 16S rRNA gene sequence and additional tests were needed to show that they belonged to different species [5]. Trichococcus is not the only genus with conserved 16S rRNA genes. Other examples are e.g. Edwardsiella, Clostridium and Mycobacterium [30–32]. Novel omics approaches are helpful in this respect. Previously, the description of two new Trichococcus species (T. ilyis and T. shcherbakovii) was done by complementing 16S rRNA gene analysis with genome-based dDDH [3, 4]. A similar approach was applied for the assignment of T. paludicola and T. alkaliphilus [6], and here we could show that the previously isolate strain ES5 is a T. flocculiformis strain, though some of its physiological properties, such as the ability to grow with glycerol, were different from the type strain. It can be concluded that the use of genomics information (such as dDDH and ANI) can help the taxonomical clustering of novel species in the genus of Trichococcus and in other genera as an efficient and accurate approach.
Extended substrate use of Trichococcus species
The genome-guided approach that was followed in this study shed light on the physiological similarities and differences of Trichococcus species. The presence of genes coding for protein domains related to carbohydrate conversion confirmed the use of previously tested sugar-substrates. Importantly, novel growth substrates can be identified by genomics analysis, and further tested in defined experimental approaches. Usually, laborious substrate tests, based on a somehow random selection, are needed to define which substrates a newly isolated bacterium can use. However, genome analysis can aid in the selection of the most likely substrates to be converted by a specific bacterium. Some members of the genus Trichococcus (T. pasteurii and strain ES5) possess an operon of 17 genes involved in glycerol degradation and 1,3-PDO production and these strains were able to ferment glycerol and produce 1,3-propanediol (1,3-PDO) as a main fermentation product. The strains tested that lacked that operon were not able to ferment glycerol. For both strains in vitro assays showed glycerol fermentation and 1,3-PDO production. Similarly, we identified genes involved in alginate degradation in T. collinsii and involved in arabinan degradation in T. patagoniensis. These two strains tested positive for growth on the respective substrates. It should be noted that when dedicated genes are detected, growth with that particular substrate is not always observed and to ascertain this experimental testing is necessary. For example, genes involved in degradation of tagatose, starch and L-sorbose were present in the genome of T. ilyis, but in vitro bacterial growth with these compounds was not observed [3].
Growth of Trichoccus species at low temperature
Psychrophylic and psychrotolerant microorganisms, due to the extreme environmental conditions, need to adapt and obtain protection mechanisms [33]. All Trichococcus species possess a high number of cold shock domains (CSD), genes related with a psychrotolerant phenotype. However, only five species can grow at 0 °C (i.e. T. pasteurii, T. collinsii, T. patagoniensis and Trichococcus strain ES5, and T. shcherbakoviae). For comparison of CSD, we included 20 lactic acid bacteria (LAB), belonging to the genera of Carnobacterium and Aerococccus. Species of these genera that had been isolated from low temperature had multiple CSDs that resembled those in Trichococcus species. Other possible bacterial adaptation to low temperature is the production of cryoprotectant exopolymeric substances (EPS), which can surround the cells and create a protective layer against cold [34, 35]. A mucoid substance has been observed in T. patagoniensis [5], which is likely related to its capacity to grow at 0 °C. Antifreezing compounds are of potential interest for applications in food bioindustry, agriculture (e.g. incorporation in fertilizers for increasing cold resistance of plants), and medicine (cryopreservation of cells).
Conclusion
Genome-guided characterisation of Trichococcus species resulted in the discovery of novel functional traits within this genus. This approach revealed a large operon that encodes the necessary enzymes for the production of 1,3-PDO from glycerol, which is present in T. pasteuri and Trichococcus strain ES5. It also enabled the identification of genes associated with the degradation of complex molecules, such as alginate and arabinan, in the genomes of some of the analysed Trichococcus species. These metabolic traits of Trichococcus species may set them as possible candidates in biotechnological processes related to the degradation or production of these compounds. Their robust phenotype, ability to grow at low temperature and high salinity, may foster versatile applications (e.g. conversion of organic compounds in high-salinity wastewaters to added-value products). The CRISPR system and the unique defence mechanisms in Trichococcus species provide them against viral attacks, which can confer them higher robustness for industrial applications.
Materials and methods
Source of genomes
The genome of Trichococcus strain ES5 (DSM 23957) was sequenced at the Joint Genome Institute from the US Department of Energy (JGI-DOE) (Walnut Creek, CA) using an Illumina HiSeq2000 platform (Illumina Inc., San Diego, CA). This genome (11,259,926 reads and 151 bp read length) was assembled and annotated as described previously [3]. All the publicly available genome sequences of Trichococcus species, i.e. T. flocculiformis (DSM 2094T), T. pasteurii (DSM 2381T), T. palustris (DSM 9172T), T. collinsii (DSM 14526T), T. patagoniensis (DSM 18806T), T. ilyis (DSM 22150T), T. paludicola (DSM 104691T), T. alkaliphilus (DSM 104692T), and T. shcherbakoviae (DSM 107162T), were obtained from the NCBI Assembly Database [36]. The same database was used to retrieve sequences of twenty-two related lactic acid bacteria (LAB) to Trichococcus species and Bacillus subtilis (outgroup species), for taxonomic hierarchical analysis. A complete list of the LAB used in the comparison is included in (Additional file 1: Table S1).
Functional analysis and genome annotation
Genomes from Trichococcus species (ten), LAB species (twenty-two), and B. subtilis were annotated using the pipeline of Semantic Annotation Platform with Provenance (SAPP) that includes Prodigal v2.6 for predicting coding gene sequences [19, 37]. T. paludicola and T. alkaliphilus locus tags were based on Prodigal v2.6 prediction (T. paludicola: Ga019, T. alkaliphilus: PXZT) for comparison purposes. Functional genome analysis was based on protein Hidden Markov Model domains (HMM) generated by InterProScan v5.17–56.0 based on Pfam domains (−-app pfam) [38–40]. InterPro protein domains matrix was generated for all the Trichococcus, selected LAB, and B. subtilis. B. subtilis was used as an outgroup for the study and was not included in the core and unique protein domain analysis. Core protein domains (present in all compared genomes) and unique protein domains (present in only one of the analysed genomes) were identified. The presence/absence matrix of protein domains from all species was converted to distances by using the dice coefficient method and a neighbour-joining tree was generated. For functional protein domain clustering, the analysis was performed in R and confirmed with DARwin v6.0 [20]. In addition, 16S rRNA gene sequences were extracted from the genomes and aligned using the software CLC Main Workbench v8.0 (CLC Bio, Aarhus, Denmark). A neighbour-joining tree was constructed based on 16S rRNA gene sequences.
Whole-genome based analyses
All pairs of strains were compared using the Genome-to-Genome Distance Calculator 2.1 (GGDC; https://ggdc.dsmz.de) under recommended settings [7] and pairwise digital DNA-DNA hybridisation values (dDDH) were inferred accordingly. Afterwards, the distance matrix was subjected to a clustering using established thresholds for delineating species [7] as well as subspecies [29]. Clustering was done using the OPTSIL clustering program [41].
A genome sequence-based phylogenetic analysis based on the coding regions was conducted using the latest version of the Genome-BLAST Distance Phylogeny (GBDP) method as previously described [42]. Briefly, BLAST+ [43] was used as a local alignment tool and distance calculations were done under recommended settings (greedy-with-trimming algorithm, formula d5, e-value filter 10− 8). A calculation of 100 replicate distances for pseudo-bootstrap support was included. Finally, a balanced minimum evolution tree was inferred using FastME v2.1.4 with SPR post processing [44]. A similar approach was used for the reconstruction of replicate trees and branch support was subsequently mapped onto the tree. Finally, exchanged genomic syntenies were defined with Sibelia v3.0.6 [45] using default parameters, and visualised in circular graph by Circos v0.69 [46].
Microbial growth tests
Growth experiments were conducted with anaerobic basal medium prepared as previously described [47]. 45 mL of medium were dispensed in 120 mL serum bottles, which were sealed with rubber stoppers and aluminium caps. Bottles’ headspace was flushed with N2/CO2 (80/20 v/v) to a final pressure of 1.5 bar. After autoclaving, and before inoculation, medium was supplemented with 0.5 mL of salts solution and 2.5 mL of bicarbonate solution [47]. Yeast extract was added to the medium at a concentration of 0.1 g/L. Substrates were added to the medium from sterile stock solutions. Glucose and glycerol growth assays were done with an initial substrate concentration of 20 mM. Degradation of alginate was tested with a concentration of 5 mM and arabinan (sugar beet, Ara:Gal:Rha:GalUA = 88:3:2:7) with a concentration of 0.4% (v/v). Incubations were in the dark, without stirring and at 30 °C (unless stated otherwise). All tests were done in triplicate. Controls without substrate and blanks without inoculation were also performed.
Antibiotic resistance tests
Antibiotic resistance tests for tetracycline were performed in plates with rich Clostridium medium (Fisher Scientific, PA) and 1% agar. Minimum inhibitory concentration (MIC) tetracycline test stripes were used with a test range of 0.016–256 μg/mL (Liofilchem, Roseto degli Abruzzi, Italy). Plates were incubated at 30 °C in anaerobic containers.
Psychrotolerance and salinity test
Temperature and salinity tests were performed using 20 mM of glucose as substrate and using the anaerobic basal medium previously described [47]. Growth of all members of Trichococcus genus was tested at 0 °C and monitored for 45 days. For salinity tolerance experiments, sodium chloride was used at concentrations of 2, 4, 6, 8, 10% (w/v). Growth of Trichococcus species at different salinities was monitored for ten days.
Analytical measurements
Growth was quantified by optical density (OD 600 nm), measured in a spectrometer (Hitachi U-1500, Labstuff, The Netherlands). Soluble metabolites, such as glucose, glycerol, 1,3-PDO, lactate, ethanol, acetate and formate were measured with Thermo Electron HPLC system equipped with an Agilent Metacarb 67H column (Thermo, Waltham, MA), which had as mobile phase sulphuric acid (5 mM) at a flow rate of 0.8 mL min− 1 and temperature at 45 °C.
Supplementary information
Additional file 1. General genomic information of all species used for in silico analysis in this study.
Additional file 2. SAPP-generated protein domain matrix. XLSX 628 kb.
Additional file 3. Common protein domains in Trichococcus strains. XLSX 28 kb.
Additional file 4. Growth curves of Trichococcus species at 0° and at different salinities (0–10% NaCl (w/v)). DOCX 155 kb.
Additional file 5. Outputs of dDDH and ANI analyses comparing strain ES5 and T. flocculiformis. XLSX 22 kb.
Acknowledgements
Not applicable.
Abbreviations
- 1,3-PDO
1,3-Propanediol
- CSD
Cold Shock Domains
- dDDH
Digital DNA-DNA Hybridisation
- EMP
Embden-Meyerhof-Parnas pathway
- GBDP
Genome-BLAST Distance Phylogeny
- GGDC
Genome-to-Genome Distance Calculator
- HMM
Hidden Markov Model domains
- LAB
Lactic Acid Bacteria
- OD
Optical Density
- PPP
Pentose Phosphate Pathway
- SAPP
Semantic Annotation Platform with Provenance
- TA
Toxin-Antitoxin
Authors’ contributions
AJMS, DZS and NSt conceived the study and designed the experiments. HPK, JMK, MG, NSh and NK contributed with genome sequencing, assembly and annotation of Trichococcus species. NSt and HDN performed laboratory work for physiological characterization of Trichococcus species. NSt and PJS designed and NSt performed bioinformatics analysis. NSt drafted the manuscript, which was revised by DZS and AJMS. All the authors have read and approved the final version of the manuscript.
Funding
This research was supported by the European Research Council under the European Union’s Seventh Framework Programme / ERC Grant Agreement (323009) and by the Gravitation grant (024.002.002) of the Netherlands Ministry of Education, Culture and Science. The work conducted by the U.S. Department of Energy Joint Genome Institute (DOE-JGI), a DOE Office of Science User Facility, was supported by the Office of Science of the DOE under Contract No. DE-AC02-05CH11231. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data from this study are available in the manuscript and additional file. Genomic data are deposited in public databases (accession numbers are provided in Additional File 1: Table S1).
The genomic sequence data of Trichococcus strain ES5 that supports the findings of this study have been deposited in GenBank with accession codes GCA_900067165.1, GCF_900067165.1.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nikolaos Strepis, Email: nikolaos.str@protonmail.com.
Henry D. Naranjo, Email: orion_hn@hotmail.com
Jan Meier-Kolthoff, Email: jan.meier-kolthoff@dsmz.de.
Markus Göker, Email: markus.goeker@dsmz.de.
Nicole Shapiro, Email: nrshapiro@lbl.gov.
Nikos Kyrpides, Email: nckyrpides@lbl.gov.
Hans-Peter Klenk, Email: Hans-Peter.Klenk@newcastle.ac.uk.
Peter J. Schaap, Email: peter.schaap@wur.nl
Alfons J. M. Stams, Email: fons.stams@wur.nl
Diana Z. Sousa, Email: diana.sousa@wur.nl
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6410-x.
References
- 1.Scheff G, Salcher O, Lingens F. Trichococcus flocculiformis gen. Nov. sp. nov. a new gram-positive filamentous bacterium isolated from bulking sludge. Appl Microbiol Biotechnol. 1984;19:114–119. doi: 10.1007/BF00302451. [DOI] [Google Scholar]
- 2.Liu JR, Tanner RS, Schumann P, Weiss N, McKenzie CA, Janssen PH, Seviour EM, Lawson PA, Allen TD, Seviour RJ. Emended description of the genus Trichococcus, description of Trichococcus collinsii sp. nov., and reclassification of Lactosphaera pasteurii as Trichococcus pasteurii comb. nov. and of Ruminococcus palustris as Trichococcus palustris comb. nov. in the low-G+C gram-positive bacteria. Int J Syst Evol Microbiol. 2002;52(Pt 4):1113–1126. doi: 10.1099/00207713-52-4-1113. [DOI] [PubMed] [Google Scholar]
- 3.Strepis N, Sanchez-Andrea I, van Gelder AH, van Kruistum H, Shapiro N, Kyrpides N, Goker M, Klenk HP, Schaap P, Stams AJM, Sousa DZ. Description of Trichococcus ilyis sp. nov. by combined physiological and in silico genome hybridization analyses. Int J Syst Evol Microbiol. 2016;66(10):3957–3963. doi: 10.1099/ijsem.0.001294. [DOI] [PubMed] [Google Scholar]
- 4.Parshina SN, Strepis N, Aalvink S, Nozhevnikova AN, Stams AJM, Sousa DZ. Trichococcus shcherbakoviae sp. nov., isolated from a laboratory-scale anaerobic EGSB bioreactor operated at low temperature. Int J Syst Evol Microbiol. 2019;69(2):529–534. doi: 10.1099/ijsem.0.003193. [DOI] [PubMed] [Google Scholar]
- 5.Pikuta EV, Hoover RB, Bej AK, Marsic D, Whitman WB, Krader PE, Tang J. Trichococcus patagoniensis sp. nov., a facultative anaerobe that grows at −5 °C, isolated from penguin guano in Chilean Patagonia. Int J Syst Evol Microbiol. 2006;56(Pt 9):2055–2062. doi: 10.1099/ijs.0.64225-0. [DOI] [PubMed] [Google Scholar]
- 6.Dai YM, Zhang LL, Li Y, Li YQ, Deng XH, Wang TT, Yang F, Tian YQ, Li N, Zhou XM, et al. Characterization of Trichococcus paludicola sp. nov. and Trichococcus alkaliphilus sp. nov., isolated from a high-elevation wetland, by phenotypic and genomic analyses. Int J Syst Evol Microbiol. 2018;68(1):99–105. doi: 10.1099/ijsem.0.002464. [DOI] [PubMed] [Google Scholar]
- 7.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter M, Rosselló-Móra R, Glöckner FO, Peplies J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2015;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koehorst JJ, van Dam JC, van Heck RG, Saccenti E, Dos Santos VA, Suarez-Diez M, Schaap PJ. Comparison of 432 Pseudomonas strains through integration of genomic, functional, metabolic and expression data. Sci Rep. 2016;6:38699. doi: 10.1038/srep38699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B, Lee D. Protein comparison at the domain architecture level. BMC Bioinformatics. 2009;10(Suppl 15):S5. doi: 10.1186/1471-2105-10-S15-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pikuta EV, Hoover RB. The genus Trichococcus. In: Lactic Acid Bacteria: Biodiversity and Taxonomy. John Wiley & Sons, Ltd; 2014.
- 12.van Gelder AH, Aydin R, Alves MM, Stams AJ. 1,3-Propanediol production from glycerol by a newly isolated Trichococcus strain. Microb Biotechnol. 2012;5(4):573–578. doi: 10.1111/j.1751-7915.2011.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatzifragkou A, Papanikolaou S, Dietz D, Doulgeraki AI, Nychas GJ, Zeng AP. Production of 1,3-propanediol by Clostridium butyricum growing on biodiesel-derived crude glycerol through a non-sterilized fermentation process. Appl Microbiol Biotechnol. 2011;91(1):101–112. doi: 10.1007/s00253-011-3247-x. [DOI] [PubMed] [Google Scholar]
- 14.Cheng KK, Liu DH, Sun Y, Liu WB. 1,3-Propanediol production by Klebsiella pneumoniae under different aeration strategies. Biotechnol Lett. 2004;26(11):911–915. doi: 10.1023/B:bile.0000025902.44458.f4. [DOI] [PubMed] [Google Scholar]
- 15.da Silva GP, Mack M, Contiero J. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27(1):30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Franzmann PD, Hopfl P, Weiss N, Tindall BJ. Psychrotrophic, lactic acid-producing bacteria from anoxic waters in ace Lake, Antarctica; Carnobacterium funditum sp. nov. and Carnobacterium alterfunditum sp. nov. Arch Microbiol. 1991;156(4):255–262. doi: 10.1007/BF00262994. [DOI] [PubMed] [Google Scholar]
- 17.Pikuta EV, Marsic D, Bej A, Tang J, Krader P, Hoover RB. Carnobacterium pleistocenium sp. nov., a novel psychrotolerant, facultative anaerobe isolated from permafrost of the fox tunnel in Alaska. Int J Syst Evol Microbiol. 2005;55(Pt 1):473–478. doi: 10.1099/ijs.0.63384-0. [DOI] [PubMed] [Google Scholar]
- 18.Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35(3):543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehorst JJ, van Dam JCJ, Saccenti E, dos Santos VAP M, Suarez-Diez M, Schaap PJ. SAPP: functional genome annotation and analysis through a semantic framework using FAIR principles. Bioinformatics. 2018;34(8):1401–1403. doi: 10.1093/bioinformatics/btx767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrier X, Jacquemoud-Collet JP. DARwin software. 2006; http://darwin.cirad.fr/.
- 21.Liu JZ, Xu W, Chistoserdov A, Bajpai RK. Glycerol dehydratases: biochemical structures, catalytic mechanisms, and industrial applications in 1,3-propanediol production by naturally occurring and genetically engineered bacterial strains. Appl Biochem Biotechnol. 2016;179(6):1073–1100. doi: 10.1007/s12010-016-2051-6. [DOI] [PubMed] [Google Scholar]
- 22.Boch J, Kempf B, Schmid R, Bremer E. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J Bacteriol. 1996;178(17):5121–5129. doi: 10.1128/JB.178.17.5121-5129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf B, Bremer E. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J Biol Chem. 1995;270(28):16701–16713. doi: 10.1074/jbc.270.28.16701. [DOI] [PubMed] [Google Scholar]
- 24.Canovas D, Vargas C, Kneip S, Moron MJ, Ventosa A, Bremer E, Nieto JJ. Genes for the synthesis of the osmoprotectant glycine betaine from choline in the moderately halophilic bacterium Halomonas elongata DSM 3043. USA Microbiology. 2000;146(Pt 2):455–463. doi: 10.1099/00221287-146-2-455. [DOI] [PubMed] [Google Scholar]
- 25.Sorek R, Kunin V, Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6(3):181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 26.Wesolowski J, Paumet F. SNARE motif: a common motif used by pathogens to manipulate membrane fusion. Virulence. 2010;1(4):319–324. doi: 10.4161/viru.1.4.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 28.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104(18):7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, Rohde C, Rohde M, Fartmann B, Goodwin LA, Chertkov O, Reddy T, Pati A, Ivanova NN, Markowitz V, Kyrpides NC, Woyke T, Göker M, Klenk H-P. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;10:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods. 2016;67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Beye M, Fahsi N, Raoult D, Fournier PE. Careful use of 16S rRNA gene sequence similarity values for the identification of Mycobacterium species. New Microbes New Infect. 2018;22:24–29. doi: 10.1016/j.nmni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi-Tamisier M, Benamar S, Raoult D, Fournier PE. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int J Syst Evol Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 33.D'Amico Salvino, Collins Tony, Marx Jean-Claude, Feller Georges, Gerday Charles. Psychrophilic microorganisms: challenges for life. EMBO reports. 2006;7(4):385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decho AL, Gutierrez T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front Microbiol. 2017;8:922. [DOI] [PMC free article] [PubMed]
- 35.Casillo A, Parrilli E, Sannino F, Mitchell DE, Gibson MI, Marino G, Lanzetta R, Parrilli M, Cosconati S, Novellino E, Randazzo A, Tutino ML, Corsaro MM. Structure-activity relationship of the exopolysaccharide from a psychrophilic bacterium: a strategy for cryoprotection. Carbohydr Polym. 2017;156:364–371. doi: 10.1016/j.carbpol.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, Funk K, Ketter A, Kim S, Kimchi A, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019;gkz899. [DOI] [PMC free article] [PubMed]
- 37.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 40.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 41.Göker M, Garcia-Blazquez G, Voglmayr H, Telleria MT, Martin MP. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4(7):e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Highly parallelized inference of large genome-based phylogenies. Concurr Comput. 2014;26:1715–1729. doi: 10.1002/cpe.3112. [DOI] [Google Scholar]
- 43.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32(10):2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minkin I, Pham H, Starostina E, Vyahhi N, Pham S. C-Sibelia: an easy-to-use and highly accurate tool for bacterial genome comparison. F1000Res. 2013;2:258. doi: 10.12688/f1000research.2-258.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stams AJM, Van Dijk JB, Dijkema C, Plugge CM. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol. 1993;59(4):1114–1119. doi: 10.1128/AEM.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. General genomic information of all species used for in silico analysis in this study.
Additional file 2. SAPP-generated protein domain matrix. XLSX 628 kb.
Additional file 3. Common protein domains in Trichococcus strains. XLSX 28 kb.
Additional file 4. Growth curves of Trichococcus species at 0° and at different salinities (0–10% NaCl (w/v)). DOCX 155 kb.
Additional file 5. Outputs of dDDH and ANI analyses comparing strain ES5 and T. flocculiformis. XLSX 22 kb.
Data Availability Statement
The data from this study are available in the manuscript and additional file. Genomic data are deposited in public databases (accession numbers are provided in Additional File 1: Table S1).
The genomic sequence data of Trichococcus strain ES5 that supports the findings of this study have been deposited in GenBank with accession codes GCA_900067165.1, GCF_900067165.1.