Abstract
Background
Malignant pleural mesothelioma (MPM) is a rare and aggressive cancer that develops in the pleural and outer layer of tissues surrounding the lungs. MPM is primarily caused by occupational exposure to asbestos and results in a poor prognosis. Effective therapeutics as well as early diagnostics for the MPM are still lacking. To identify potential diagnostic biomarkers for MPM, we performed bioinformatics analysis of public database.
Methods
Utilizing databases from Cancer Cell Line Encyclopedia (CCLE) and Gene Expression Omnibus (GEO), we identified several potential candidates that could act as MPM biomarkers. We carried out additional molecular analyses of these potential markers using MPM patient tissue samples via quantitative polymerase chain reaction.
Results
We identified Lysyl oxidase (LOX), Lysyl oxidase homologs 1&2 (LOXL1& LOXL2) Zinc Finger Protein, FOG Family Member 2 (ZFPM2) as potential diagnostic biomarkers for MPM. In this study, we found that the LOX family and ZFPM2 showed comparable diagnostic ability to Fibulin-3 or mesothelin (MSLN) and would be better potential biomarkers than Sulfatase 1 (SULF1), Thrombospondin 2 (THBS2) and Cadherin 11 (CDH11).
Conclusions
LOX family and ZPFM2 were identified as novel MPM diagnostic biomarkers which could strengthen MPM clinical diagnostic capabilities.
Keywords: LOX, ZFPM2, Malignant pleural mesothelioma, Diagnostic biomarker
Background
Malignant pleural mesothelioma (MPM) is a highly malignant tumor which occurs in the pleural mesothelial tissues covering the lung [1]. It is well known that MPM onset is usually due to asbestos exposure in an industrial environment [2]. Although there are various advanced therapeutic approaches, including surgical and chemical therapies to treat MPM, the mean overall survival ranges from six to eighteen months; and the five-year survival rate is < 16% [3–5]. Due to the prolonged latency of asbestos, asbestos mediated-MPM commonly occurs from 20 to 50 years following exposure. Based on exposure rates it is anticipated that the rate of MPM cases will increase over the next several decades [6]. Importantly, due to the high risk of MPM, use of asbestos has recently been banned in many developing countries [7, 8]. Even with considerable efforts to decrease asbestos use, further screening for potential biomarkers is necessary to aid in earlier diagnoses of the disease than is currently achievable with the available MPM biomarkers including Mesothelin, Fibulin-3 and Calretinin 2 (CALB2).
Mesothelin (MSLN) is translated into a pro-protein form, which is subsequently cleaved to into the soluble mesothelin-related protein (SMRP) and megakaryocyte potentiating factor [9, 10]. The SMRP has been detected in the serum of 84% of the MPM patients making it reasonably sensitive and suggesting its potential to act as an MPM diagnostic marker [11].
Fibulin-3 is a glycoprotein encoded by the epidermal growth factor-containing fibulin-like extracellular matrix protein-1 (EFEMP1) gene. Fibulin-3 is known to contribute to cell proliferation and/or migration. As one of the best MPM biomarkers, the sensitivity and specificity of Fibulin-3 have been reported in plasma and pleural effusions as 96 and 84%, and 95 and 93% respectively [12].. Calretinin 2 (CALB2), osteopontin (OPN), and Wilms Tumor 1 Protein (WT-1) have also suggested as potential candidates for an MPM diagnosis [13, 14].
In this study, we aimed to improve the diagnostic potential for MPM patients by establishing a multi-biomarker set comprising of seven individual biomarkers. To that end, we performed bioinformatic screenings using open databases - Cancer cell encyclopedia (CCLE) and Gene expression omnibus (GEO) and found four additional candidate genes including LOX, LOXL1, LOXL2 and ZFPM2 which show high expression levels in MPM patient tissues and cell lines when compared to their normal counterparts. We believe this multi-biomarker set could be further developed into an MPM diagnostic kit to improve the MPM diagnostic power in clinics.
Materials and methods
In silico analysis
Microarray data of cancer cells were obtained from the CCLE (https://portals.broadinstitute.org/ccle). Fourteen malignant pleural mesothelioma and thirty-two lung adenocarcinoma cell lines were selected and analyzed. Based on the collected data, a heat map was generated by the CCLE portal. Microarray data of human tissue was downloaded from the GEO (https://www.ncbi.nlm.nih.gov/geo) portal website. Nine normal and forty MPM tissue samples were included in the “Malignant Pleural Mesothelioma” data (accession number GSE2549) [15]. Collected biomarker candidate genes were analyzed via the following statistical methods.
Statistical analysis
The statistical analyses were performed using Graphpad PRISM 6.0 and IBM SPSS Statistics 24 software. Receiver-operating characteristic (ROC) analyses was carried out to determine the accuracy of the biomarker candidates identified from the Malignant Pleural Mesothelioma dataset. Youden’s method was used for determination of an optimal cutoff point in the ROC curve to maximize sensitivity and specificity. To determine the statistical significance, a two-tailed unpaired t test was performed.
Tissue collection
Human normal or MPM tissue samples were collected from bronchioalveolar lavage (BAL) fluid of non-cancerous or MPM patients. Fluid samples were centrifuged, and RNA was isolated for quantitative real-time PCR (qPCR) assays.
Quantitative real-time PCR
Total RNA was prepared from patient samples using TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA using the qPCR RT Master Mix (Toyobo). mRNA expression levels of genes of interest were determined by quantitative real-time PCR performed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) with SYBR-green real-time PCR master mixes (Life Technologies). Data were analyzed by △△Ct method with the rRNA 18S gene used as a reference [16]. Gene-specific primers are listed in Table 1.
Table 1.
Gene Specific Primer sequence (F: Forward, R: Reverse)
| Gene | Primer sequence |
|---|---|
| 18 s | F: 5′-ACC GCA GCT AGG AAT AAT GGA-3′ |
| R: 5′-GCC TCA GTT CCG AAA ACC A-3’ | |
| Fibulin-3 | F: 5′-GGG AGC AGT GCG TAG ACA TAG-3’ |
| R: 5′-GCT GCC AAT TGA AAC CCA GG-3’ | |
| MSLN | F: 5′-GGA TGA GCT CTA CCC ACA AGG-3’ |
| R: 5′-ACT TGC GAA TGT CCT CAG GG-3’ | |
| CALB2 | F: 5′-CTG CCT GTC CAG GAA AAC TTC-3’ |
| R: 5′-GTA GCC GCT TCT ATC CTT GTC-3’ | |
| LOX | F: 5′-TCT GGC CAG TAC AGC ATA CAG-3’ |
| R: 5′-CTT GGT CGG CTG GGT AAG AA-3’ | |
| LOXL1 | F: 5′-TCT GGC CAG CAC AGC CTA T-3’ |
| R: 5′-GTT GGG GAG GAA GTC TGC TG-3’ | |
| LOXL2 | F: 5′-ACT GCC ACA TAG GTG GTT CC-3’ |
| R: 5′-CGG GGA CAG CTG GTT GTT TA-3’ | |
| ZFPM2 | F: 5′-TGT GTA CAG CAA AGG GGG TC-3’ |
| R: 5′-TGG CAG CTT GTA GCC TTG AG-3’ | |
| THBS2 | F: 5′-TGA GGA CCT GGA CGA GTG TG-3’ |
| R: 5′-GCT GGT TCC CTC TGT ATC GG- 3’ | |
| SULF1 | F: 5′-GCA GTG CAA CCC AAG ACC TA-3’ |
| R: 5′-CCA TCC CAT AAC TGT CCT CTG T-3’ | |
| CDH11 | F: 5′- ACA AGG ATG ACA CGG CCA AT-3’ |
| R: 5′-GCC TGC TGT GTT ATC TCG GT-3’ |
Results
Genetic signatures differentiating MPM from non-MPM cells
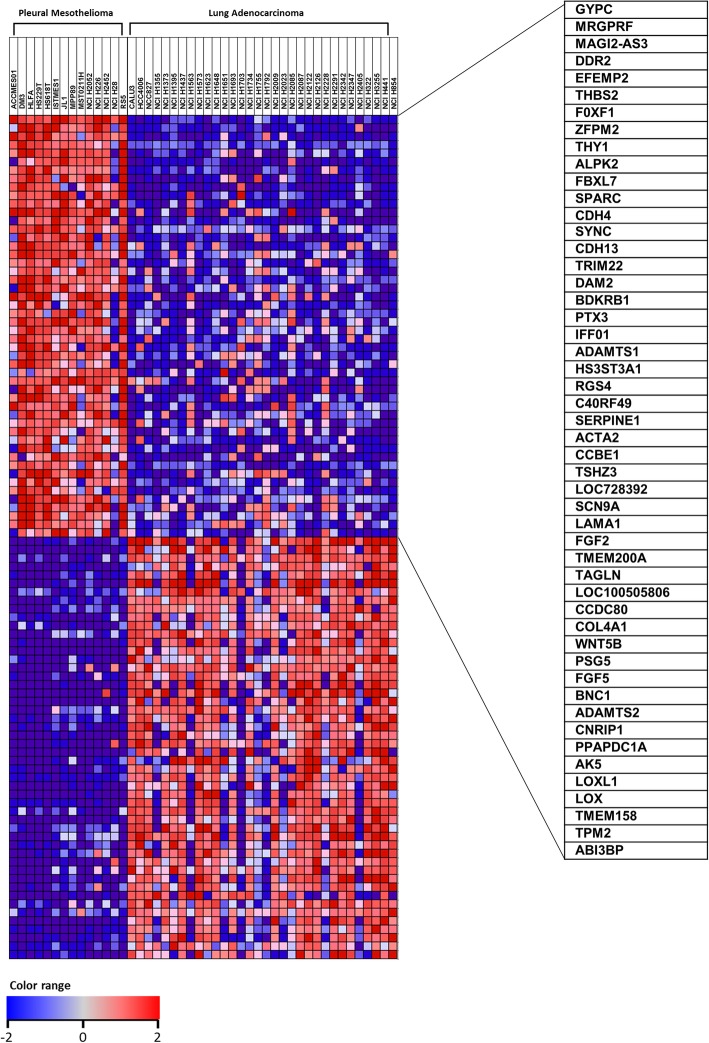
To identify novel MPM biomarkers, we first performed bioinformatic analyses using the CCLE database which provides mRNA expression datasets for > 1100 cancer cell lines [17]. Gene expression patterns were comprehensively analyzed between 14 MPM cell lines and 32 lung adenocarcinoma cell lines using Gene Set Enrichment Analysis (GSEA). Finally, we identified a unique genetic signature which differentiates MPM from lung adenocarcinoma cell lines (Fig. 1, Additional file 1). The genetic signature includes the top 50 genes showing upregulated expression as candidate genes for potential MPM biomarkers. The genetic signature was further analyzed and confirmed as to whether it could specifically represent MPM.
Fig. 1.
Differentiated gene expression between pleural mesothelioma and lung adenocarcinoma. From CCLE database, gene expression signature was compared between 14 MPM and 32 lung adenocarcinoma cell lines. The upper portion in the heat map listed 50 genes showing upregulated expression of mRNA in MPM cell lines. The heat map was generated based on expression score in logarithmic scale
Seven genes as potential MPM biomarker candidates
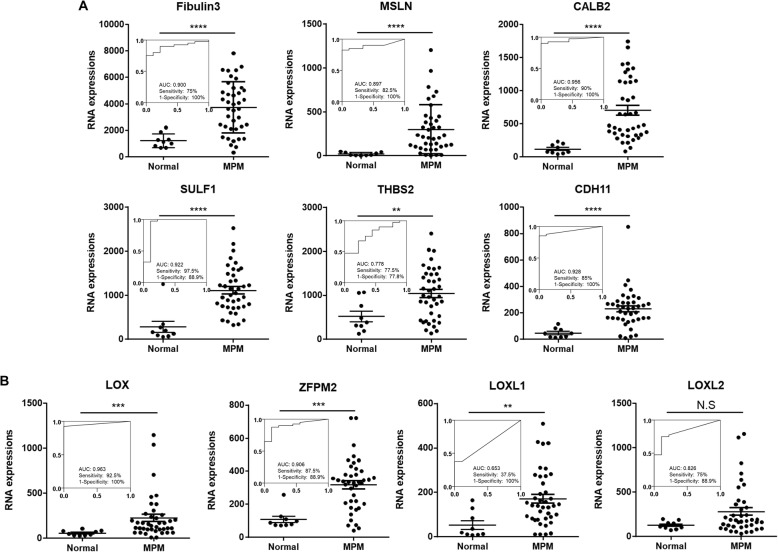
We identified one hundred genes as potential candidates from the in silico analyses and wanted to narrow this down into a manageable number of genes. This was accomplished using multiple independent datasets for confirmation testing. We utilized the GEO database that provides microarray and functional genomic datasets from next-generation sequencing studies. A functional genomics dataset from human MPM tissues [15] was also used to validate the in silico genetic signature. These data were derived from normal pleural or lung specimens (n = 9) and MPM surgical specimens (n = 40). We first validated the reliability of the tissue transcriptomic data using the well-known MPM biomarkers Fibulin-3, MSLN and CALB2 which showed that these genes had significantly higher expression in human MPM tissues when compared with their normal counterparts (Fig. 2a). Fibulin-3 showed the highest sensitivity and specificity in the ROC curve analysis (Fig. 2a). Using the same approach as above, we analyzed the expression of the one-hundred potential MPM candidate genes identified from the GEO dataset (Fig. 1). Interestingly, we found that LOX, LOXL1, LOXL2, ZFPM2, SULF1, THBS2, and CDH11 showed significantly higher expression in the MPM tissues compared to the normal samples (Fig. 2a and b). It is important to reiterate that three of these genes (SULF1, THBS2 and CDH11) have been suggested as MPM biomarkers in previous studies [18]. More significantly, the ROC curve analysis revealed that the LOX family and ZFPM2 showed a high sensitivity and specificity to act as MPM biomarkers (Fig. 2b). Taken together, these data suggest that the seven candidate genes could serve as potential novel MPM biomarkers.
Fig. 2.
Diagnostic potential of LOX family and ZFPM2. Using the GEO database (GSE2549), mRNA expression of known biomarker (a) or candidate genes (b) were analyzed in normal (n = 9) and MPM tissues (n = 40). Each dot represents one sample, values are mean ± SEM of each groups. Statistical analysis between normal and MPM groups was executed using unpaired Student t-test. (**** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05). The In-let represents ROC curves illustrating diagnostic ability of known biomarkers (a) and candidates (b). X and Y axes stands for sensitivity as well as 1-specificity, respectively. Maximal sensitivity and specificity were determined by Youden’s method. AUC (Area Under Curve)
Evaluating the potential of LOX family and ZFPM2 as MPM biomarker candidates in MPM tissues
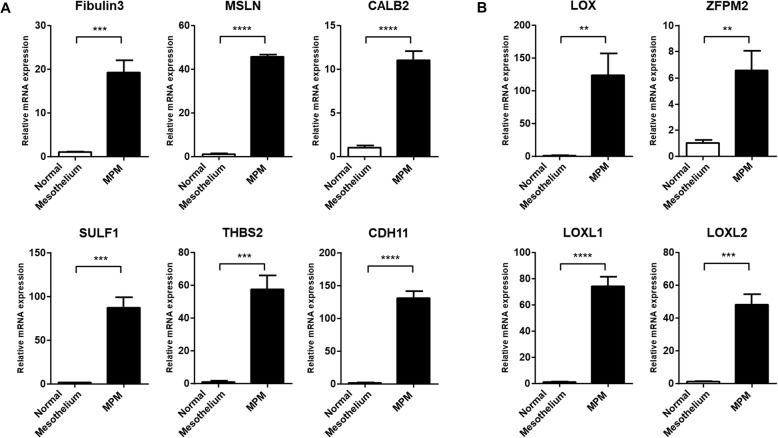
As proposed, a subset of the biomarker candidates for MPM diagnostics was further investigated to determine if their expression signatures could be validated in novel set of clinically confirmed samples. Pleural effusion samples from non-cancer patients and MPM patients were obtained under Institutional Review Board (IRB) approval. Using the qPCR assay, we profiled the mRNA expression of the candidate genes in the tissue samples. Consistent with the data from the GEO analysis, expression of the known MPM biomarkers were significantly higher in the MPM tissue when compared with the normal tissue (Fig. 3a). Following the validation of the known biomarkers, we also checked the mRNA expression levels for the four new proposed MPM biomarker candidates in the same patient samples. Similarly, the expression of the four MPM biomarker candidates LOX, LOXL1, LOXL2 and ZFPM2 were shown to be significantly increased in the MPM tissues when compared to the non-cancer patient samples (Fig. 3b). Taken together, these data suggest that these candidates could be included as members of a panel, together with previously known biomarkers, for improved MPM diagnoses.
Fig. 3.
mRNA expression of the candidate biomarkers in patient samples. Using the qPCR assay, mRNA expression was surveyed for the known (a) and the candidate MPM biomarkers (b) in mesothelium tissues isolated from a non-cancer patient and a MPM patient. Values are mean ± SEM of each groups. The Statistic between normal and MPM groups was analyzed using the unpaired Student t-test. (**** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05)
Discussion
Early detection and treatment of cancer using specific biomarkers helps to reduce disease progression. Thus, multi-biomarker sets rather than a single gene biomarker could increase the diagnostic power and provide patients with a more sensitive diagnostic for mesothelioma [19–24]. In this present study, we utilized the CCLE and GEO databases for a comparative analyses of gene expression patterns between MPM patients and controls. Based on our bioinformatic analyses, we were able to identify a subset of genes which show a higher expression levels in the MPM group when compared to the normal or lung adenocarcinoma group.
The diagnostic potential of the gene candidates was further analyzed together with three previously well-known MPM biomarkers Fibulin-3, MSLN, and CALB2. In particular, we noticed that the diagnostic power of LOX family was comparable to or even greater than that of Fibulin-3 which has shown the most significant diagnostic potential in the literature [12]. We further confirmed the specificity of the MPM biomarkers in the paired set of a normal mesothelium and patient fluid sample. The assembled biomarker panel consisting of seven genes showed significantly increased sensitivity for diagnosing MPM from pleural fluid samples.
One notable finding in this study was that we identified several LOX isoforms as potential MPM biomarker candidates. The LOX gene encodes lysyl oxidase that induces crosslinking of elastin and collagen by catalyzing oxidative deamination on lysine or hydroxylysine residues, and the other LOXL family members do as well. This biochemical function is known to be important in maintaining organismal structure [25–27]. Therefore, dysfunction of LOX and the LOXL family contributes to various types of diseases including liver fibrosis, cardiovascular disease, and cancer. In cancer biology, hypoxia-inducible factor induces LOX expression, which contributes to hypoxic metastasis in breast cancer [28] and is also associated with a poor prognosis and invasion in lung cancer [29]. However, little is known about LOX and LOXL family involvement in mesothelioma. One might think that the MPM specific expression pattern of the LOX and LOXL family needs to be further studied in the MPM cancer biology.
The ZFPM2 gene encodes the Friend of GATA-2 (FOG-2) protein which is a transcription cofactor interacting with GATA family members transcriptionally involved in diverse biological functions including cardiac, pulmonary, gonadal development and hematopoiesis [30–36]. Similarly, we found that ZFPM2 has a high diagnostic potential from the database analyses and this was confirmed via performance of a case-control study.
Conclusions
Taken together, the four biomarker candidates (LOX, LOXL1, LOXL2, ZFPM2) identified in the current study could be combined with the previously proposed biomarkers to strengthen the diagnostic power for MPM patients.
Additional file
Additional file 1. Gene expression list in pleural mesothelioma compared to lung adenocarcinoma. Note that expression logarithmic score was presented to each gene list by comparing 14 MPM and 32 lung adenocarcinoma cell lines from CCLE database.
Acknowledgements
Not applicable.
Abbreviations
- BAL
Bronchioalveolar lavage
- CALB2
Calretinin 2
- CCLE
Cancer cell line encyclopedia
- CDH11
Cadherin 11
- FOG-2
Friend of GATA-2
- GEO
Gene expression omnibus
- GSEA
Gene set enrichment analysis
- IRB
Institutional review board
- LOX
Lysyl oxidase
- MPM
Malignant pleural mesothelioma
- MSLN
Mesothelin
- OPN
Osteopontin
- ROC
Receiver-operating characteristic
- SULF1
Sulfatase 1
- THBS2
Thrombospondin 2
- WT-1
Wilms tumor 1 protein
- ZFPM2
Zinc finger protein, fog family member 2
Author’s contribution
YSJ, SHJ and JWC planned this research and drafted the manuscript. MKK and MRJ performed experiments and analysis. SSO and SJY collected human MPM samples. All authors read and revised the final manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Medical Research Center Program (2017R1A5A2015369), Global Ph.D Fellowship Program (NRF-2016H1A2A1907107 to MK Kim) and the Korea Ministry of Environment under The Environmental Health Action Program (2012001350013) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI17C0039) and the Yonsei University Wonju Campus Future-Leading Research Initiative of 2017-52-0081.
Availability of data and materials
All of data and materials are available from the corresponding author upon reasonable ask.
Ethics approval and consent to participate
Human tissue experiments were reviewed and approved by the Institutional Review Board (IRB) of Yonsei University Wonju College of Medicine with the IRB number YWMR-12-6-026.
Competing interest
The authors have no conflict of interest.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yangsik Jeong, Email: yjeong@yonsei.ac.kr.
Soon-Hee Jung, Email: soonheej@yonsei.ac.kr.
Jong-Whan Choi, Email: biochoi@yonsei.ac.kr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40364-019-0180-0.
References
- 1.Walker AM, Loughlin JE, Friedlander ER, Rothman KJ, Dreyer NA. Projections of asbestos-related disease 1980-2009. J Occup Med. 1983;25(5):409–425. [PubMed] [Google Scholar]
- 2.Boutin C, Schlesser M, Frenay C, Astoul P. Malignant pleural mesothelioma. Eur Respir J. 1998;12(4):972–981. doi: 10.1183/09031936.98.12040972. [DOI] [PubMed] [Google Scholar]
- 3.Musk AW, Olsen N, Alfonso H, Reid A, Mina R, Franklin P, Sleith J, Hammond N, Threlfall T, Shilkin KB, et al. Predicting survival in malignant mesothelioma. Eur Respir J. 2011;38(6):1420–1424. doi: 10.1183/09031936.00000811. [DOI] [PubMed] [Google Scholar]
- 4.Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353(15):1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 5.Rusch VW, Giroux D, Kennedy C, Ruffini E, Cangir AK, Rice D, Pass H, Asamura H, Waller D, Edwards J, et al. Initial analysis of the international association for the study of lung cancer mesothelioma database. J Thorac Oncol. 2012;7(11):1631–1639. doi: 10.1097/JTO.0b013e31826915f1. [DOI] [PubMed] [Google Scholar]
- 6.Lin RT, Chien LC, Jimba M, Furuya S, Takahashi K. Implementation of national policies for a total asbestos ban: a global comparison. Lancet Planet Health. 2019;3(8):e341–e348. doi: 10.1016/S2542-5196(19)30109-3. [DOI] [PubMed] [Google Scholar]
- 7.Haynes RC. Where there is asbestos, there is mesothelioma: filling in the data blanks. Environ Health Perspect. 2011;119(4):A177. doi: 10.1289/ehp.119-a177b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tossavainen A. Global use of asbestos and the incidence of mesothelioma. Int J Occup Environ Health. 2004;10(1):22–25. doi: 10.1179/oeh.2004.10.1.22. [DOI] [PubMed] [Google Scholar]
- 9.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996;93(1):136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholler N, Fu N, Yang Y, Ye Z, Goodman GE, Hellstrom KE, Hellstrom I. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci U S A. 1999;96(20):11531–11536. doi: 10.1073/pnas.96.20.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson BW, Creaney J, Lake R, Nowak A, Musk AW, de Klerk N, Winzell P, Hellstrom KE, Hellstrom I. Mesothelin-family proteins and diagnosis of mesothelioma. Lancet. 2003;362(9396):1612–1616. doi: 10.1016/S0140-6736(03)14794-0. [DOI] [PubMed] [Google Scholar]
- 12.Pass HI, Levin SM, Harbut MR, Melamed J, Chiriboga L, Donington J, Huflejt M, Carbone M, Chia D, Goodglick L, et al. Fibulin-3 as a blood and effusion biomarker for pleural mesothelioma. N Engl J Med. 2012;367(15):1417–1427. doi: 10.1056/NEJMoa1115050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doglioni C, Dei Tos AP, Laurino L, Iuzzolino P, Chiarelli C, Celio MR, Viale G. Calretinin: a novel immunocytochemical marker for mesothelioma. Am J Surg Pathol. 1996;20(9):1037–1046. doi: 10.1097/00000478-199609000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Otterstrom C, Soltermann A, Opitz I, Felley-Bosco E, Weder W, Stahel RA, Triponez F, Robert JH, Serre-Beinier V. CD74: a new prognostic factor for patients with malignant pleural mesothelioma. Br J Cancer. 2014;110(8):2040–2046. doi: 10.1038/bjc.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon GJ, Rockwell GN, Jensen RV, Rheinwald JG, Glickman JN, Aronson JP, Pottorf BJ, Nitz MD, Richards WG, Sugarbaker DJ, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol. 2005;166(6):1827–1840. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadziavdic K, Lekang K, Lanzen A, Jonassen I, Thompson EM, Troedsson C. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One. 2014;9(2):e87624. doi: 10.1371/journal.pone.0087624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012) Nat. 2012;492(7428):290. doi: 10.1038/nature11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melaiu O, Melissari E, Mutti L, Bracci E, De Santi C, Iofrida C, Di Russo M, Cristaudo A, Bonotti A, Cipollini M, et al. Expression status of candidate genes in mesothelioma tissues and cell lines. Mutat Res. 2015;771:6–12. doi: 10.1016/j.mrfmmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Ordonez NG. Application of mesothelin immunostaining in tumor diagnosis. Am J Surg Pathol. 2003;27(11):1418–1428. doi: 10.1097/00000478-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Ordonez NG. Immunohistochemical diagnosis of epithelioid mesothelioma - an update. Arch Pathol Lab Med. 2005;129(11):1407–1414. doi: 10.5858/2005-129-1407-IDOEMA. [DOI] [PubMed] [Google Scholar]
- 21.King J, Thatcher N, Pickering C, Hasleton P. Sensitivity and specificity of immunohistochemical antibodies used to distinguish between benign and malignant pleural disease: a systematic review of published reports. Histopathol. 2006;49(6):561–568. doi: 10.1111/j.1365-2559.2006.02442.x. [DOI] [PubMed] [Google Scholar]
- 22.Suster S, Moran CA. Applications and limitations of immunohistochemistry in the diagnosis of malignant mesothelioma. Adv Anat Pathol. 2006;13(6):316–329. doi: 10.1097/01.pap.0000213064.05005.64. [DOI] [PubMed] [Google Scholar]
- 23.Yaziji H, Battifora H, Barry TS, Hwang HC, Bacchi CE, McIntosh MW, Kussick SJ, Gown AM. Evaluation of 12 antibodies for distinguishing epithelioid mesothelioma from adenocarcinoma: identification of a three-antibody immunohistochemical panel with maximal sensitivity and specificity. Modern Pathol. 2006;19(4):514–523. doi: 10.1038/modpathol.3800534. [DOI] [PubMed] [Google Scholar]
- 24.Marchevsky AM. Application of immunohistochemistry to the diagnosis of malignant mesothelioma. Arch Pathol Lab Med. 2008;132(3):397–401. doi: 10.5858/2008-132-397-AOITTD. [DOI] [PubMed] [Google Scholar]
- 25.Hamalainen ER, Jones TA, Sheer D, Taskinen K, Pihlajaniemi T, Kivirikko KI. Molecular-cloning of human Lysyl oxidase and assignment of the gene to chromosome-5q23.3-31.2. Genomics. 1991;11(3):508–516. doi: 10.1016/0888-7543(91)90057-L. [DOI] [PubMed] [Google Scholar]
- 26.Cenizo V, Andre V, Reymermier C, Sommer P, Damour O, Perrier E. LOXL as a target to increase the elastin content in adult skin: a dill extract induces the LOXL gene expression. Exp Dermatol. 2006;15(8):574–581. doi: 10.1111/j.1600-0625.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 27.Polettini J, Silva MG, Kacerovsky M, Syed TA, Saade GR, Menon R. Screening of lysyl oxidase (LOX) and lysyl oxidase like (LOXL) enzyme expression and activity in preterm prelabor rupture of fetal membranes. J Perinat Med. 2016;44(1):99–109. doi: 10.1515/jpm-2014-0337. [DOI] [PubMed] [Google Scholar]
- 28.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JTA, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nat. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 29.Wilgus ML, Borczuk AC, Stoopler M, Ginsburg M, Gorenstein L, Sonett JR, Powell CA. Lysyl oxidase: a lung adenocarcinoma biomarker of invasion and survival. Cancer. 2011;117(10):2186–2191. doi: 10.1002/cncr.25768. [DOI] [PubMed] [Google Scholar]
- 30.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101(7):729–739. doi: 10.1016/S0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 31.Ackerman KG, Herron BJ, Vargas SO, Huang HL, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1(1):58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnatz AS, Gao Z, Broman M, Martens S, Earley JU, Svensson EC. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev Biol. 2014;395(1):50–61. doi: 10.1016/j.ydbio.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laitinen MP, Anttonen M, Ketola I, Wilson DB, Ritvos O, Butzow R, Heikinheimo M. Transcription factors GATA-4 and GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord-derived ovarian tumors. J Clin Endocrinol Metab. 2000;85(9):3476–3483. doi: 10.1210/jcem.85.9.6828. [DOI] [PubMed] [Google Scholar]
- 34.Smagulova FO, Manuylov NL, Leach LL, Tevosian SG. GATA4/FOG2 transcriptional complex regulates Lhx9 gene expression in murine heart development. BMC Dev Biol. 2008;8:67. doi: 10.1186/1471-213X-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visvader JE, Crossley M, Hill J, Orkin SH, Adams JM. The C-terminal zinc-finger of Gata-1 or Gata-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell-line. Mol Cell Biol. 1995;15(2):634–641. doi: 10.1128/MCB.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19(6):4495–4502. doi: 10.1128/MCB.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Gene expression list in pleural mesothelioma compared to lung adenocarcinoma. Note that expression logarithmic score was presented to each gene list by comparing 14 MPM and 32 lung adenocarcinoma cell lines from CCLE database.
Data Availability Statement
All of data and materials are available from the corresponding author upon reasonable ask.