Abstract
Background
Colistin resistance is considered a serious problem due to a lack of alternative antibiotics. The Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is a resazurin reduction-based technique that relies on the visual detection of bacterial growth in the presence of a defined concentration of colistin. The aim of this study was to evaluate the performance of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test in the detection of colistin susceptibility in common clinical Gram-negative bacteria.
Results
A total of 253 clinical isolates from a teaching hospital, including Acinetobacter baumanii (n = 58, 8 colistin-resistant), Pseudomonas aeruginosa (n = 61, 11 colistin-resistant), Klebsiella pneumoniae (n = 70, 20 colistin-resistant) and Escherichia coli (n = 64, 14 colistin-resistant) were tested in this study. The sensitivity and specificity of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test compared to Broth microdilution method was 100 and 99%, respectively.
Conclusions
Our results suggest that Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test could be used as an accurate detection method for colistin resistance.
Keywords: Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test, Colistin-resistant, Gram-negative bacteria, Rapid diagnosis
Background
Polymyxin E, also known as colistin is a multicomponent polypeptide antibiotic, which belongs to the group of polymyxin [1]. Polymyxin E was discovered in the 1940s; yet, later on, it was abandoned in clinical practice due to its increased nephrotoxicity. However, due to the increase of multidrug resistance (MDR) in Gram-negative bacteria, especially in the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), colistin has been applied in clinical practice for the last few years as the last resort treatment option [2, 3]. Currently, colistin resistance is considered a serious problem, due to a lack of alternative antibiotics [4, 5]. As for now, rapid identification of colistin resistance is considered essential for the effective control of MDR Gram-negative bacteria infection.
Broth microdilution (BMD) is the only reference method that has been recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Clinical and Laboratory Standards Institute (CLSI) for the detection of minimum inhibitory concentrations (MICs) of colistin [6, 7]. Nevertheless, colistin antimicrobial susceptibility testing is very challenging to perform [8, 9]. For example, the operational steps of BMD are complex and time-consuming, making it unsuitable for clinical use [10]. Clinical microbiology laboratories are especially affected by the lack of an accurate, fast and easy-to-conduct method to test the colistin susceptibility [11–13]. Therefore, it is of great significance for clinical anti-infective treatment to develop and promote new, convenient, economical, rapid and accurate colistin sensitivity detection method.
In 2016, Nordmann et al developed the Rapid Polymyxins NP test for Enterobacteriaceae spp [14]. The method can be used to detect bacteria that can grow, metabolize glucose, and produce acid in the presence of polymyxin such as polymyxin B or colistin through color changes of PH indicators. However, one of the significant limitations when using this approach is that it cannot be applied for non-fermentative bacteria such as A. baumannii and P. aeruginosa. More recently, Lescat et al have developed a rapid resazurin-mucoid susceptibility test method called Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test, which can quickly detect the sensitivity of colistin for both Enterobacteriaceae spp and non-fermentative bacteria within 4 h [15]. The method is mainly based on detection of the strain viability by observing the color change of resazurin (an active colorant) from blue to purple or pink in the presence of colistin (3.75 mg/L).
In this study, we analyzed the performance of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test in the detection of colistin susceptibility in 253 nonduplicate clinical Gram-negative isolates aiming to provide a basis for the popularization and application of a new method for rapid screening of colistin-resistant common clinical Gram-negative bacteria.
Results
The colistin MICs of the 253 Gram-negative isolates ranged from ≤0.06 to ≥32 mg/L. BMD results were used as a standard, and 53 colistin-resistant strains and 198 colistin susceptible strains were correctly detected by the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test. Very major errors (VME) and major errors (ME) corresponded to false-susceptible and false-resistant results, respectively [16]. There were only two ME in A. baumannii; details are shown in Tables 1 and 2. The specificity of A. baumannii was 96%; while the sensitivity and specificity of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test to P. aeruginosa, K. pneumoniae and E. coli were 100% (Table 3).
Table 1.
Colistin MICs obtained by broth microdilution and results of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test
| Isolate | Species | Resistant Phenotype | MIC (mg/L) | Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP Test | |
|---|---|---|---|---|---|
| Result | Discrepancies with BMD MIC colistin result | ||||
| BM1539 | A. baumannii | R | 8 | Positive | No |
| BM1579 | A. baumannii | R | 4 | Positive | No |
| BM1595 | A. baumannii | R | 4 | Positive | No |
| BM2349 | A. baumannii | R | 4 | Positive | No |
| BM2370 | A. baumannii | R | 16 | Positive | No |
| BM2412 | A. baumannii | R | 4 | Positive | No |
| BM2431 | A. baumannii | R | 8 | Positive | No |
| BM2622 | A. baumannii | R | 8 | Positive | No |
| TL1671 | P. aeruginosa | R | 4 | Positive | No |
| TL1722 | P. aeruginosa | R | 4 | Positive | No |
| TL1736 | P. aeruginosa | R | ≥32 | Positive | No |
| TL1744 | P. aeruginosa | R | 4 | Positive | No |
| TL2204 | P. aeruginosa | R | 4 | Positive | No |
| TL2294 | P. aeruginosa | R | 4 | Positive | No |
| TL2314 | P. aeruginosa | R | ≥32 | Positive | No |
| TL2917 | P. aeruginosa | R | 4 | Positive | No |
| TL2967 | P. aeruginosa | R | 4 | Positive | No |
| TL3008 | P. aeruginosa | R | 16 | Positive | No |
| TL3086 | P. aeruginosa | R | ≥32 | Positive | No |
| FK20 | K. pneumoniae | R | ≥32 | Positive | No |
| FK26 | K. pneumoniae | R | ≥32 | Positive | No |
| FK150 | K. pneumoniae | R | ≥32 | Positive | No |
| FK169 | K. pneumoniae | R | ≥32 | Positive | No |
| FK171 | K. pneumoniae | R | ≥32 | Positive | No |
| FK591 | K. pneumoniae | R | ≥32 | Positive | No |
| FK610 | K. pneumoniae | R | ≥32 | Positive | No |
| FK1342 | K. pneumoniae | R | ≥32 | Positive | No |
| FK1913 | K. pneumoniae | R | ≥32 | Positive | No |
| FK1986 | K. pneumoniae | R | 8 | Positive | No |
| FK2066 | K. pneumoniae | R | ≥32 | Positive | No |
| FK2166 | K. pneumoniae | R | ≥32 | Positive | No |
| FK2778 | K. pneumoniae | R | ≥32 | Positive | No |
| FK2911 | K. pneumoniae | R | ≥32 | Positive | No |
| FK3789 | K. pneumoniae | R | ≥32 | Positive | No |
| FK3810 | K. pneumoniae | R | ≥32 | Positive | No |
| FK3994 | K. pneumoniae | R | ≥32 | Positive | No |
| FK6556 | K. pneumoniae | R | 32 | Positive | No |
| FK6663 | K. pneumoniae | R | 32 | Positive | No |
| FK6696 | K. pneumoniae | R | 16 | Positive | No |
| DC90 | E. coli | R | 8 | Positive | No |
| DC2562 | E. coli | R | 8 | Positive | No |
| DC3411 | E. coli | R | 4 | Positive | No |
| DC3539 | E. coli | R | 16 | Positive | No |
| DC3599 | E. coli | R | 8 | Positive | No |
| DC3658 | E. coli | R | 8 | Positive | No |
| DC3737 | E. coli | R | 8 | Positive | No |
| DC3802 | E. coli | R | 4 | Positive | No |
| DC3806 | E. coli | R | 8 | Positive | No |
| DC3846 | E. coli | R | 16 | Positive | No |
| DC4887 | E. coli | R | 8 | Positive | No |
| DC5262 | E. coli | R | 8 | Positive | No |
| DC5286 | E. coli | R | 8 | Positive | No |
| DC7333 | E. coli | R | 4 | Positive | No |
| BM1505 | A. baumannii | S | 0.125 | Negative | No |
| BM1506 | A. baumannii | S | 0.5 | Negative | No |
| BM1507 | A. baumannii | S | 0.06 | Negative | No |
| BM1508 | A. baumannii | S | 0.125 | Negative | No |
| BM1509 | A. baumannii | S | 0.125 | Negative | No |
| BM1510 | A. baumannii | S | 0.125 | Negative | No |
| BM1511 | A. baumannii | S | 0.125 | Negative | No |
| BM1512 | A. baumannii | S | 0.25 | Negative | No |
| BM1513 | A. baumannii | S | 0.125 | Negative | No |
| BM1514 | A. baumannii | S | 0.125 | Negative | No |
| BM4151 | A. baumannii | S | 0.25 | Negative | No |
| BM4152 | A. baumannii | S | 0.06 | Negative | No |
| BM4153 | A. baumannii | S | 0.03 | Negative | No |
| BM4154 | A. baumannii | S | 0.125 | Negative | No |
| BM4155 | A. baumannii | S | 0.125 | Negative | No |
| BM4156 | A. baumannii | S | 0.125 | Negative | No |
| BM4158 | A. baumannii | S | 0.125 | Negative | No |
| BM4159 | A. baumannii | S | 0.125 | Negative | No |
| BM4160 | A. baumannii | S | 0.5 | Negative | No |
| BM4161 | A. baumannii | S | 0.125 | Negative | No |
| BM4162 | A. baumannii | S | 0.06 | Negative | No |
| BM4163 | A. baumannii | S | 0.06 | Negative | No |
| BM4164 | A. baumannii | S | 0.06 | Negative | No |
| BM4165 | A. baumannii | S | 0.06 | Negative | No |
| BM4166 | A. baumannii | S | 0.125 | Negative | No |
| BM4167 | A. baumannii | S | 0.125 | Negative | No |
| BM4168 | A. baumannii | S | 0.25 | Negative | No |
| BM4169 | A. baumannii | S | 0.5 | Negative | No |
| BM4170 | A. baumannii | S | 0.125 | Negative | No |
| BM4171 | A. baumannii | S | 0.06 | Negative | No |
| BM4172 | A. baumannii | S | ≤0.06 | Negative | No |
| BM4173 | A. baumannii | S | 0.06 | Negative | No |
| BM4174 | A. baumannii | S | 0.06 | Negative | No |
| BM4175 | A. baumannii | S | 2 | Negative | No |
| BM4176 | A. baumannii | S | 0.06 | Negative | No |
| BM4177 | A. baumannii | S | 0.06 | Negative | No |
| BM4178 | A. baumannii | S | 0.06 | Negative | No |
| BM4179 | A. baumannii | S | 0.25 | Negative | No |
| BM4180 | A. baumannii | S | 0.06 | Negative | No |
| BM4181 | A. baumannii | S | 0.125 | Negative | No |
| BM4182 | A. baumannii | S | 0.25 | Negative | No |
| BM4183 | A. baumannii | S | 1 | Negative | No |
| BM4184 | A. baumannii | S | 1 | Positive | Yes, ME |
| BM4185 | A. baumannii | S | 1 | Negative | No |
| BM4186 | A. baumannii | S | 1 | Negative | No |
| BM4187 | A. baumannii | S | 0.125 | Negative | No |
| BM4188 | A. baumannii | S | 0.5 | Positive | Yes, ME |
| BM4189 | A. baumannii | S | 0.5 | Negative | No |
| BM4190 | A. baumannii | S | 0.125 | Negative | No |
| BM4191 | A. baumannii | S | 0.5 | Negative | No |
| TL2916 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2915 | P. aeruginosa | S | ≤0.06 | Negative | No |
| TL2914 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2913 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2911 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2910 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2908 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2907 | P. aeruginosa | S | 0.5 | Negative | No |
| TL2906 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2905 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2904 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2901 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2899 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2898 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2897 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2895 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2893 | P. aeruginosa | S | ≤0.06 | Negative | No |
| TL2892 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2891 | P. aeruginosa | S | 0.06 | Negative | No |
| TL2890 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2889 | P. aeruginosa | S | 0.5 | Negative | No |
| TL2886 | P. aeruginosa | S | 0.5 | Negative | No |
| TL2885 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2884 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2883 | P. aeruginosa | S | 0.5 | Negative | No |
| TL2882 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2881 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2879 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2878 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2877 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2875 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2874 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2873 | P. aeruginosa | S | 1 | Negative | No |
| TL2872 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2871 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2870 | P. aeruginosa | S | 2 | Negative | No |
| TL2869 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2868 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2867 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2866 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2865 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2864 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2863 | P. aeruginosa | S | ≤0.06 | Negative | No |
| TL2862 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2861 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2858 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2857 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2856 | P. aeruginosa | S | 0.125 | Negative | No |
| TL2855 | P. aeruginosa | S | 0.25 | Negative | No |
| TL2854 | P. aeruginosa | S | 0.125 | Negative | No |
| FK3640 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3642 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3646 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3660 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3671 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3686 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3695 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3696 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3703 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3712 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3719 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3721 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3724 | K. pneumoniae | S | 1 | Negative | No |
| FK3727 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3730 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3732 | K. pneumoniae | S | 0.25 | Negative | No |
| FK3738 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3739 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3740 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3741 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3745 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3746 | K. pneumoniae | S | 1 | Negative | No |
| FK3749 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3758 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3764 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3767 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3771 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3784 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3800 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3803 | K. pneumoniae | S | 0.25 | Negative | No |
| FK3813 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3817 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3824 | K. pneumoniae | S | 0.06 | Negative | No |
| FK3830 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3831 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3838 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3844 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3853 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3878 | K. pneumoniae | S | 0.25 | Negative | No |
| FK3882 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3891 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3927 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3938 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3943 | K. pneumoniae | S | 0.06 | Negative | No |
| FK3946 | K. pneumoniae | S | 0.5 | Negative | No |
| FK3989 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK3990 | K. pneumoniae | S | 1 | Negative | No |
| FK3996 | K. pneumoniae | S | 0.125 | Negative | No |
| FK3999 | K. pneumoniae | S | ≤0.06 | Negative | No |
| FK4002 | K. pneumoniae | S | ≤0.06 | Negative | No |
| DC8640 | E. coli | S | 0.25 | Negative | No |
| DC8641 | E. coli | S | ≤0.06 | Negative | No |
| DC8642 | E. coli | S | 0.125 | Negative | No |
| DC8643 | E. coli | S | 0.125 | Negative | No |
| DC8644 | E. coli | S | 0.5 | Negative | No |
| DC8645 | E. coli | S | ≤0.06 | Negative | No |
| DC8646 | E. coli | S | ≤0.06 | Negative | No |
| DC8647 | E. coli | S | ≤0.06 | Negative | No |
| DC8648 | E. coli | S | ≤0.06 | Negative | No |
| DC8649 | E. coli | S | 0.125 | Negative | No |
| DC8650 | E. coli | S | 0.06 | Negative | No |
| DC8651 | E. coli | S | 0.06 | Negative | No |
| DC8652 | E. coli | S | 0.125 | Negative | No |
| DC8653 | E. coli | S | 0.06 | Negative | No |
| DC8654 | E. coli | S | 0.06 | Negative | No |
| DC8655 | E. coli | S | 0.06 | Negative | No |
| DC8656 | E. coli | S | 0.125 | Negative | No |
| DC8657 | E. coli | S | ≤0.06 | Negative | No |
| DC8658 | E. coli | S | 0.06 | Negative | No |
| DC8659 | E. coli | S | ≤0.06 | Negative | No |
| DC8660 | E. coli | S | ≤0.06 | Negative | No |
| DC8661 | E. coli | S | ≤0.06 | Negative | No |
| DC8663 | E. coli | S | 0.125 | Negative | No |
| DC8664 | E. coli | S | 2 | Negative | No |
| DC8665 | E. coli | S | ≤0.06 | Negative | No |
| DC8666 | E. coli | S | 0.06 | Negative | No |
| DC8667 | E. coli | S | ≤0.06 | Negative | No |
| DC8668 | E. coli | S | ≤0.06 | Negative | No |
| DC8669 | E. coli | S | ≤0.06 | Negative | No |
| DC8670 | E. coli | S | ≤0.06 | Negative | No |
| DC8671 | E. coli | S | ≤0.06 | Negative | No |
| DC8672 | E. coli | S | ≤0.06 | Negative | No |
| DC8673 | E. coli | S | ≤0.06 | Negative | No |
| DC8674 | E. coli | S | 0.06 | Negative | No |
| DC8675 | E. coli | S | ≤0.06 | Negative | No |
| DC8676 | E. coli | S | ≤0.06 | Negative | No |
| DC8677 | E. coli | S | ≤0.06 | Negative | No |
| DC8678 | E. coli | S | ≤0.06 | Negative | No |
| DC8679 | E. coli | S | ≤0.06 | Negative | No |
| DC8680 | E. coli | S | 0.25 | Negative | No |
| DC8681 | E. coli | S | 2 | Negative | No |
| DC8682 | E. coli | S | 0.125 | Negative | No |
| DC8683 | E. coli | S | ≤0.06 | Negative | No |
| DC8684 | E. coli | S | ≤0.06 | Negative | No |
| DC8685 | E. coli | S | ≤0.06 | Negative | No |
| DC8686 | E. coli | S | 0.06 | Negative | No |
| DC8687 | E. coli | S | 0.06 | Negative | No |
| DC8688 | E. coli | S | 0.06 | Negative | No |
| DC8690 | E. coli | S | 0.06 | Negative | No |
| DC8691 | E. coli | S | 0.125 | Negative | No |
ME major error, S susceptible, R resistant
Table 2.
Colistin MICs for 253 Gram-negative isolates
| Organism | Number of isolates | Colistin MIC (mg/L) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | ||
| Total | 253 | 86 | 56 | 27 | 19 | 8 | 4 | 14 | 13 | 5 | 21 |
| A. baumannii | 58 | 15 | 19 | 5 | 6 | 4 | 1 | 4 | 3 | 1 | 0 |
| P. aeruginosa | 61 | 4 | 23 | 17 | 4 | 1 | 1 | 7 | 0 | 1 | 3 |
| K. pneumoniae | 70 | 30 | 6 | 3 | 8 | 3 | 0 | 0 | 1 | 1 | 18 |
| E. coli | 64 | 37 | 8 | 2 | 1 | 0 | 2 | 3 | 9 | 2 | 0 |
Table 3.
Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test results among Gram-negative isolates
| Organism | Susceptibility to polymyxins | Resistance mechanism | Isolates | Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| A. baumannii | Resistant | Mediated by chromosomea | 8 (3.16%) | 8 positive result | 100% | 96% |
| Susceptible | 50 (19.76%) | 48 negative results and 2 positive result | ||||
| P. aeruginosa | Resistant | Mediated by chromosome | 11 (4.35%) | 11 positive result | 100% | 100% |
| Susceptible | 50 (19.76%) | 50 negative results | ||||
| K. pneumoniae | Resistant | Mediated by chromosomea | 20 (7.91%) | 20 positive result | 100% | 100% |
| Susceptible | 50 (19.76%) | 50 negative results | ||||
| E. coli | Resistant | Mediated by plasmid | 14 (5.54%) | 2 positive result | 100% | 100% |
| Susceptible | 50 (19.76%) | 50 negative results |
aUnpublished
Discussion
In this study, we described the diagnostic performance of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test, a phenotypic method for differentiation between colistin-resistant strains and colistin-susceptible strains. Compared with the reference BMD, the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test showed accuracy in detecting the resistance to colistin. Besides, the method was fast, easy to perform, and the obtained data were easy to interpret. Rapid Polymyxin NP test makes up for the limitations of applicability in non-fermenters [14]. In our study, we examined it efficiency in detecting non-fermentative bacteria, but also fermentative bacteria, such as E. coli strains and K. pneumoniae strains. The results showed that the sensitivity and specificity of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test to Enterobacteriaceae were 100%, which was consistent with a previous study [15]. In the present study, there were only two ME in colistin-susceptible A. baumannii strains. The categorical agreement for all tested isolates was 99.2% for the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test. In addition, the sensitivity and specificity were respectively 100 and 99%, which further suggested that this method is suitable for detecting fermentative bacteria.
So far, a number of studies have examined the mechanism of colistin resistance [17, 18]. This study revealed that chromosome mutations of two-component regulatory systems (TCSs) and mcr-1, which were located in plasmid, were the main causes of colistin resistance in 53 strains. In addition, we were able to detect drug resistance without a difference. Therefore, compared with the Rapid Polymyxin NP test, the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is suitable to be used in more scenes.
MicroScan Colistin Well is a newly developed kit for detection of colistin resistance in Gram-negative bacteria [19]. The fundamental principle of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is similar to MicroScan Colistin Well. Both methods can be used to detect living bacteria in the medium with 4 mg/L or 3.75 mg/L of colistin (close to the breakpoint of colistin resistance). Similarly, the MICs cannot be determined utilizing the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test and the MicroScan Colistin Well. Only colistin resistance results or sensitive test results can be obtained by them. However, there are two major differences between the two methods. First, Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is significantly faster compared to MicroScan Colistin Well. For example, the detection of P. aeruginosa by Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test takes maximum 5 h to analyze the results, while MicroScan Colistin Well requires 16 to 18 h. Secondly, in the presence of resazurin reagent PrestoBlue®, the growth of living bacteria of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test can be more clearly observed compared to MicroScan Colistin Well.
The principle of Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is based on the visual detection of the reduction of the resazurin reagent, a viability colorant that is observed by color change (blue to purple or pink). Interestingly, in the current study, no significant color changes were observed in colistin-resistant P. aeruginosa after the addition of the resazurin reagent for 1 h. After prolonging the observation time for another 1 h, the color changed from blue to purple. In other words, the results were not obtained until 2 h later in the study, while very obvious color changes were observed 15 min after the addition of the resazurin reagent in the colistin-resistant strains of A. baumanii, K. pneumoniae and E. coli, including 2 ME. This may be because the growth rate of P. aeruginosa is slower than that of Enterobacteriaceae, thus taking longer to decompose resazurin into fluorescent substance resorufin. It suggested that the observation time of the results of this experiment needed to be optimized according to the strain.
However, the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test still has some limitations. Firstly, the accurate MIC values could not be obtained. Since the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test was not suitable for the study of high-level drug resistant strains, the method could only show whether the colistin resistant was present or not. Secondly, several mcr-harboring isolates with an MIC of 2 mg/L (or even less) to colistin or polymyxin B have been reported [20, 21], while our method could only be used to screen colistin resistant strains with MIC ≥4 mg/L. Thirdly, the reading time of P. aeruginosa results was different from that reported by the inventors, requiring an additional 1 h of observation time.
Conclusion
The Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test has great stability and sensitivity in detection of colistin resistance in Gram-negative bacteria such as A. baumanii, P. aeruginosa, K. pneumoniae and E. coli strains. In addition, this method is fast and easy to perform. It can contribute in selecting more precise therapeutic choices, and optimizing antibiotic stewardship, and preventing the development of outbreaks with multidrug-resistant isolates. Nevertheless, the testing time of P. aeruginosa is longer than that reported by the inventor, so the observation time of this method needs to be further optimized.
Methods
Bacterial strains
A total of 253 nonduplicate clinical Gram-negative isolates including A. baumanii strains (n = 58), P. aeruginosa strains (n = 61), K. pneumoniae strains (n = 70) and E. coli strains (n = 64) were obtained from a teaching hospital in Wenzhou, China. Species identification was performed using the Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry (MALDI-TOF MS, Bruker Daltonics, US). A total of 53 colistin-resistant strains were selected from our previous studies and were detected by BMD, including 8 A. baumanii strains, 11 P. aeruginosa strains, 20 K. pneumoniae strains and 14 E. coli strains. In addition, 50 colistin-susceptible isolates of each four bacterial species mentioned above were randomly selected as the control group. E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as control strains [6].
Antimicrobial susceptibility test
BMD was performed in triplicate. According to the EUCAST/CLSI joined guidelines [6, 7], the clinical breakpoints for colistin provided for P. aeruginosa and A. baumanii were ≤ 2 mg/L (susceptible breakpoint) and ≥ 4 mg/L (resistant breakpoint) and Enterobacteriaceae are ≤2 mg/L (susceptible breakpoint) and > 2 mg/L (resistant breakpoint).
Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test
The experimental procedure was performed according to the previously described protocol [15]. Briefly, the colistin-containing Mueller Hinton broth (MHB, OXOID, UK) solution was prepared with an initial concentration of 4.16 mg/L. Then, a 180 μl colistin-free MHB solution and colistin-containing MHB solution were added to lines A and B of a 96-well polystyrene micro test plate, respectively. For each isolate, 20 μl of the bacterial suspension at a 3.5 McFarland optical density (~ 1 × 109 CFU/mL) was inoculated in parallel into two wells, with and without colistin. The bacterial suspension was mixed with the medium by pipetting up and down. The final concentration of colistin was 3.75 mg/L. In the same way, 20 μl of 0.85% NaCl was used as an aseptic control, 20 μl of the colistin-susceptible isolate (E. coli ATCC 25922 and P. aeruginosa ATCC 27853) suspension was used as negative control; and 20 μl of the colistin-resistant isolate (the clinical isolates of Morgan, inherent resistance to polymyxin) suspension was used as a positive control. After testing several isolates, we ensured that the color-transfer of colistin suspension and the mixing of bacterial suspension in the micro test plate were completed within 15 min. The inoculated tray was incubated at 35 ± 2 °C for 3 h. Then, 22 μl of the resazurin reagent PrestoBlue® (ThermoFisher Scientific, US, final concentration is 10% V/V) was added per well and each well was mixed by pipetting up and down. Finally, the tray was visually inspected every 15 min within 1 h. Susceptibility of colistin is determined by the color changes, where discoloration indicates that the strain is colistin-resistant, while the lack of discoloration indicates that the strain is colistin-susceptible [15]. All experiments were performed in triplicate.
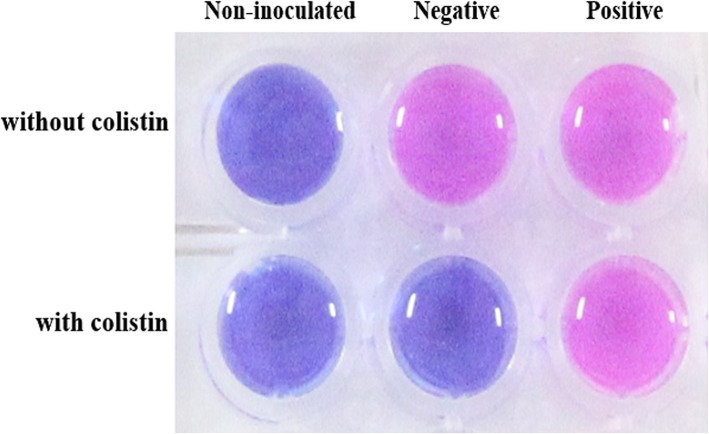
The test was considered to be positive (i.e., purple or pink) if the colistin-resistant isolate was viable in presence of colistin, or negative (i.e., blue) if the colistin-susceptible isolate was not viable in presence of colistin. The Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test interpretation is illustrated in Fig. 1.
Fig. 1.
Representative results of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test. Non-inoculated well is shown as the control of the medium and the color changed (first column). Negative, the tested isolate only grows in the absence of colistin (second column). Positive, the tested isolate grows in the presence and absence of colistin (third column)
Acknowledgements
Not applicable.
Abbreviations
- ATCC
American Type Cultures Collection
- BMD
Broth Microdilution
- CFU
Colony Forming Unit
- CLSI
Clinical and Laboratory Standards Institute
- EUCAST
European Committee of Antibiotic Susceptibility Testing
- MDR
Multidrug resistance
- ME
Major Error
- MHB
Mueller Hinton broth
- MIC
Minimal Inhibitory Concentration
- VME
Very Major Error
Authors’ contributions
HJ conducted the experiments, analyzed the data and wrote the manuscript. RF participated in experiments and writing. JL and XT provided colistin-resistant strains and participated in analysis of results. YZ participated in analysis of results. LC helped design the study. JC and TZ designed the study and corrected the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundations of China (No. 81971986) and the General Scientific Research Project of the Education Department of Zhejiang Province of China (No. Y201942210) and the Planned Science and Technology Project of Wenzhou (no. Y20170204).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianming Cao, Email: wzcjming@163.com.
Tieli Zhou, Email: wyztli@163.com.
References
- 1.Gregoire N, Aranzana-Climent V, Magreault S, Marchand S, Couet W. Clinical pharmacokinetics and pharmacodynamics of Colistin. Clin Pharmacokinet. 2017;56(12):1441–1460. doi: 10.1007/s40262-017-0561-1. [DOI] [PubMed] [Google Scholar]
- 2.Brooks LE, Ul-Hasan S, Chan BK, Sistrom MJ. Quantifying the Evolutionary Conservation of Genes Encoding Multidrug Efflux Pumps in the ESKAPE Pathogens To Identify Antimicrobial Drug Targets. mSystems. 2018;3(3):e00024–18. [DOI] [PMC free article] [PubMed]
- 3.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 4.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44(6):500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Liu Y, Qi X, Wang R, Jin L, Zhao M, Zhang Y, Wang Q, Chen H, Wang H. Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatient and avian isolates from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int J Antimicrob Agents. 2017;50(4):536–541. doi: 10.1016/j.ijantimicag.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2019.
- 7.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0; 2019. http://www.eucast.org.
- 8.Humphries RM. Susceptibility testing of the Polymyxins: where are we now? Pharmacother J Human Pharmacol Drug Ther. 2015;35(1):22–27. doi: 10.1002/phar.1505. [DOI] [PubMed] [Google Scholar]
- 9.Vasoo S, Munson E. Susceptibility testing for the Polymyxins: two steps back, three steps forward? J Clin Microbiol. 2017;55(9):2573–2582. doi: 10.1128/JCM.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphries RM, Hindler JA, Epson E, Horwich-Scholefield S, Miller LG, Mendez J, Martinez JB, Sinkowitz J, Sinkowtiz D, Hershey C, et al. Carbapenem-resistant Enterobacteriaceae detection practices in California: what are we missing? Clin Infect Dis. 2018;66(7):1061–1067. doi: 10.1093/cid/cix942. [DOI] [PubMed] [Google Scholar]
- 11.Satlin MJ. The Search for a Practical Method for Colistin Susceptibility Testing: Have We Found It by Going Back to the Future? J Clin Microbiol. 2019;57(2):e01608–18. [DOI] [PMC free article] [PubMed]
- 12.Matuschek E, Ahman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect. 2018;24(8):865–870. doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Ezadi F, Ardebili A, Mirnejad R. Antimicrobial Susceptibility Testing for Polymyxins: Challenges, Issues, and Recommendations. J Clin Microbiol. 2019;57(4):e01390–18. [DOI] [PMC free article] [PubMed]
- 14.Nordmann P, Jayol A, Poirel L. Rapid detection of Polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis. 2016;22(6):1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescat M, Poirel L, Tinguely C, Nordmann P. A Resazurin Reduction-Based Assay for Rapid Detection of Polymyxin Resistance in Acinetobacter baumannii and Pseudomonas aeruginosa. J Clin Microbiol. 2019;57(3):e01563–18. [DOI] [PMC free article] [PubMed]
- 16.Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect. 2018;24(2):175–179. doi: 10.1016/j.cmi.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Lu H, Wang C, Dong G, Xu C, Zhang X, Liu H, Zhang M, Cao J, Zhou T. Prevalence and Molecular Characterization of Escherichia coli Clinical Isolates Carrying mcr-1 in a Chinese Teaching Hospital from 2002 to 2016. Antimicrob Agents Chemother. 2018;62(9):e02623–17. [DOI] [PMC free article] [PubMed]
- 18.Lin J, Xu C, Fang R, Cao J, Zhang X, Zhao Y, Dong G, Sun Y, Zhou T. Resistance and hetero-resistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob Agents Chemother. 2019;63(10):e00556–19. [DOI] [PMC free article] [PubMed]
- 19.Lutgring JD, Kim A, Campbell D, Karlsson M, Brown AC, Burd EM. Evaluation of the MicroScan Colistin Well and Gradient Diffusion Strips for Colistin Susceptibility Testing in Enterobacteriaceae. J Clin Microbiol. 2019;57(5):e01866-18. [DOI] [PMC free article] [PubMed]
- 20.Chew KL, La M-V, Lin RTP, Teo JWP, Munson E. Colistin and Polymyxin B susceptibility testing for Carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol. 2017;55(9):2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillonetto M, Mazzetti A, Becker GN, Siebra CA, Arend L, Barth AL. Low level of polymyxin resistance among nonclonal mcr-1-positive Escherichia coli from human sources in Brazil. Diagn Microbiol Infect Dis. 2019;93(2):140–142. doi: 10.1016/j.diagmicrobio.2018.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.