Abstract
Background
Chromatin organization is central to precise control of gene expression. In various eukaryotic species, domains of pervasive cis-chromatin interactions demarcate functional domains of the genomes. In nematode Caenorhabditis elegans, however, pervasive chromatin contact domains are limited to the dosage-compensated sex chromosome, leaving the principle of C. elegans chromatin organization unclear. Transcription factor III C (TFIIIC) is a basal transcription factor complex for RNA polymerase III, and is implicated in chromatin organization. TFIIIC binding without RNA polymerase III co-occupancy, referred to as extra-TFIIIC binding, has been implicated in insulating active and inactive chromatin domains in yeasts, flies, and mammalian cells. Whether extra-TFIIIC sites are present and contribute to chromatin organization in C. elegans remains unknown.
Results
We identified 504 TFIIIC-bound sites absent of RNA polymerase III and TATA-binding protein co-occupancy characteristic of extra-TFIIIC sites in C. elegans embryos. Extra-TFIIIC sites constituted half of all identified TFIIIC binding sites in the genome. Extra-TFIIIC sites formed dense clusters in cis. The clusters of extra-TFIIIC sites were highly over-represented within the distal arm domains of the autosomes that presented a high level of heterochromatin-associated histone H3K9 trimethylation (H3K9me3). Furthermore, extra-TFIIIC clusters were embedded in the lamina-associated domains. Despite the heterochromatin environment of extra-TFIIIC sites, the individual clusters of extra-TFIIIC sites were devoid of and resided near the individual H3K9me3-marked regions.
Conclusion
Clusters of extra-TFIIIC sites were pervasive in the arm domains of C. elegans autosomes, near the outer boundaries of H3K9me3-marked regions. Given the reported activity of extra-TFIIIC sites in heterochromatin insulation in yeasts, our observation raised the possibility that TFIIIC may also demarcate heterochromatin in C. elegans.
Keywords: Transcription factor III C (TFIIIC), RNA polymerase III, Caenorhabditis elegans, Insulator, Chromatin, Chromosome, Lamina-associated domain, Nuclear periphery, LEM-2, Histone H3 lysine 9 methylation
Background
Eukaryotic genomes are organized into domains of various chromatin features including actively transcribed regions, transcription factor-bound regions, and transcriptionally repressed regions [1–4]. Demarcation of chromatin domains is central to precise control and memory of gene expression patterns. Several proteins have been proposed to have activity in demarcating chromatin domains by acting as a physical boundary [5, 6], generating nucleosome-depleted regions [7], mediating long-range chromatin interactions [8, 9], or tethering chromatin to the nuclear periphery [10]. Despite intense studies [11–15], how chromatin domains are demarcated remains poorly understood.
The genome of nematode Caenorhabditis elegans has served as a model to study chromatin organization [3, 16–18]. The highest level of chromatin organization in C. elegans is the chromatin feature that distinguishes between the X chromosome and the autosomes. The X chromosome in C. elegans hermaphrodites is organized into large self-interacting domains that have some features shared with topologically associated domains (TADs) seen in other metazoan genomes [19–22]. The five autosomes, however, lack robust self-interacting domains [22]. Instead, each autosome can be subdivided into three, megabase-wide domains, the left arm, the right arm, and the center [23]. The center domains display a low recombination rate [24, 25], a high density of essential genes [26], and low heterochromatin-associated histone modifications [16, 27]. The autosome arms are rich in repetitive elements [23] and heterochromatin-associated histone modifications [16, 27], and are associated with the nuclear membrane [18, 28, 29]. Within these generally euchromatic centers and heterochromatic arms lie kilobase-wide regions of various chromatin states including transcriptionally active and inactive regions [3, 4]. While condensins, a highly conserved class of architectural proteins [30], define the boundaries of TAD-like self-interacting domains in the X chromosome [22], the contribution of condensins to autosomal chromatin organization is unclear [14, 31]. Furthermore, CTCF, another conserved architectural protein central to defining TAD boundaries in vertebrates, is thought to be lost during the C. elegans evolution [32]. How chromatin domains and chromatin states in the C. elegans autosomes are demarcated remains an area of active investigation [4, 28, 33, 34].
The transcription factor IIIC complex (TFIIIC) is a general transcription factor required for recruitment of the RNA polymerase III (Pol III) machinery to diverse classes of small non-coding RNA genes [35]. TFIIIC has also been implicated in chromatin insulation [36, 37]. TFIIIC binds DNA sequence elements called the Box-A and Box-B motifs [35]. When participating in Pol III-dependent transcription, TFIIIC binding to Box-A and Box-B motifs results in recruitment of transcription factor IIIB complex (TFIIIB), including TATA-binding protein (TBP), which then recruits Pol III [35, 38]. By mechanisms that remain unknown, however, TFIIIC is also known to bind DNA without further recruitment of TBP and Pol III [37]. These so-called “extra-TFIIIC sites”, or “ETC”, have been identified in various organisms including yeast [39, 40], fly [41], mouse [42], and human [43]. In Saccharomyces cerevisiae and Schizosaccharomyces pombe, extra-TFIIIC sites exhibit chromatin boundary functions both as heterochromatin barriers and insulators to gene activation [40, 44, 45]. In addition, extra-TFIIIC sites in these yeast species have been observed at the nuclear periphery, suggesting a contribution to spatial organization of chromosomes [40, 46]. In fly, mouse, and human genomes, extra-TFIIIC sites were found in close proximity to architectural proteins including CTCF, condensin, and cohesin [41–43, 47]. These studies collectively suggest a conserved role for extra-TFIIIC sites in chromatin insulation and chromosome organization. However, whether extra-TFIIIC sites exist in the C. elegans genome is unknown.
In this study, we unveiled extra-TFIIIC sites in the C. elegans genome. Extra-TFIIIC sites were highly over-represented within a subset of autosome arms that presented a high level of heterochromatin-associated histone H3K9 trimethylation (H3K9me3). Extra-TFIIIC sites formed dense clusters in cis and were embedded in the lamina-associated domains. Despite the heterochromatin environment of extra-TFIIIC sites, the individual clusters of extra-TFIIIC sites were devoid of and resided near the boundaries of H3K9me3-marked regions. Our study thus raised the possibility that, like extra-TFIIIC sites in other organisms, C. elegans extra-TFIIIC sites may have a role in demarcating chromatin domains.
Results
Half of C. elegans TFIIIC binding sites lack Pol III co-occupancy
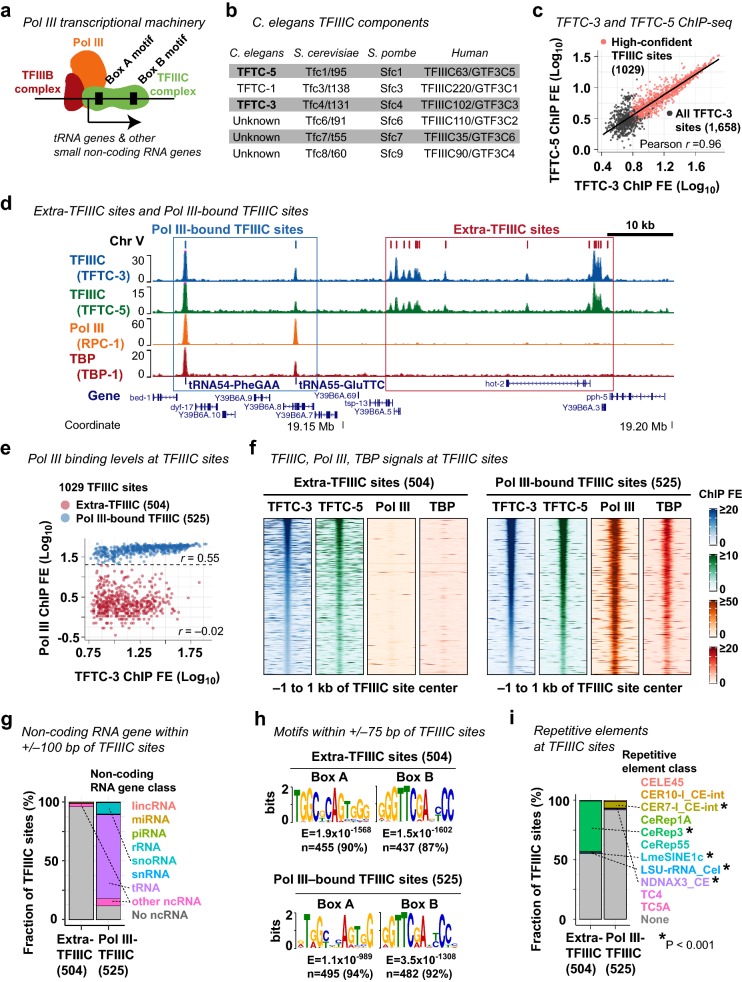
The TFIIIC complex is a general transcription factor required for the assembly of the RNA polymerase III (Pol III) machinery at small non-coding RNA genes such as tRNA genes (Fig. 1a). Extra-TFIIIC sites are TFIIIC-bound sites lacking Pol III co-occupancy, and are implicated in insulating genomic domains and spatially organizing chromosomes [48]. To determine whether the C. elegans genome includes extra-TFIIIC sites, we analyzed the ChIP-seq data published in our previous study [49] for TFIIIC subunits, TFTC-3 (human TFIIIC63/GTF3C3 ortholog) and TFTC-5 (human TFIIIC102/GTF3C5 ortholog) (Fig. 1b); the Pol III catalytic subunit RPC-1 (human POLR3A ortholog, “Pol III” hereafter); and the TFIIIB component TBP-1 (human TBP ortholog, “TBP” hereafter) in mixed-stage embryos of the wild-type N2 strain C. elegans. We identified 1029 high-confidence TFIIIC-bound sites exhibiting strong and consistent enrichment for both TFTC-3 and TFTC-5 (Fig. 1c). tRNA genes were strongly enriched for TFTC-3, TFTC-5, Pol III, and TBP as expected (Fig. 1d). We also observed numerous TFIIIC-bound sites with low or no Pol III and TBP enrichment (Fig. 1d). Of the 1029 TFIIIC-bound sites (Additional file 1), we identified 504 sites (49%) with no or very low Pol III and TBP enrichment (Fig. 1e, f), which we referred to as extra-TFIIIC sites, following the nomenclature in literature [39–43]. We also identified 525 TFIIIC-bound sites with strong Pol III enrichment (51%), which we referred to as Pol III-bound TFIIIC sites (Fig. 1e, f).
Fig. 1.
Identification of extra-TFIIIC sites in the C. elegans genome. a A schematic of the RNA polymerase III transcriptional machinery. b C. elegans TFIIIC complex proteins and their yeast and human orthologs for reference. c Correlation between TFTC-3 and TFTC-5 ChIP-seq fold enrichment (FE) scores at the 1658 TFTC-3-binding sites. The 1029 high-confident TFIIIC sites are indicated. d A representative genomic region showing extra-TFIIIC sites and Pol III-bound TFIIIC sites. e TFTC-3 and Pol III (RPC-1) FE scores at the 1029 TFIIIC sites. r, Pearson correlation coefficient within the TFIIIC subclasses. f TFTC-3, TFTC-5, Pol III (RPC-1), TBP (TBP-1) ChIP-seq FE signals at extra- and Pol III-bound TFIIIC sites. g Fraction of TFIIIC sites harboring the transcription start site of non-coding RNA genes within ± 100 bp of the TFIIIC site center. Dotted lines indicate non-coding RNA gene classes found in ≥ 10 TFIIIC sites. Pol III-TFIIIC, Pol III-bound TFIIIC sites. h DNA sequence motifs found in ± 75 bp of TFIIIC site centers. i Fraction of TFIIIC sites overlapping repetitive elements. P, empirical P-value based on 2000 permutations of the extra- or Pol III-bound TFIIIC sites within chromosomes. Dotted lines indicate repetitive element classes with P < 0.001
The lack of Pol III and TBP binding in extra-TFIIIC sites may represent a premature Pol III preinitiation complex assembled at Pol III-transcribed non-coding RNA genes [50]. Alternatively, C. elegans extra-TFIIIC sites could be unrelated to Pol III transcription and similar to extra-TFIIIC sites reported in other organisms. To distinguish these two possibilities, we examined the presence of transcription start sites (TSSs) of non-coding RNA genes near TFIIIC-bound sites. We observed that only 4% of extra-TFIIIC sites (20/504) harbored TSSs of non-coding RNA genes within 100 bp of the TFIIIC-bound site center (Fig. 1g). In contrast, almost all Pol III-bound TFIIIC sites (464 of 525 sites, 88%) harbored TSSs of non-coding RNA genes within 100 bp, and the vast majority of these genes encoded tRNAs (376 sites, 72%) or snoRNAs (52 sites, 10%) (Fig. 1g) as expected [35]. Thus, extra-TFIIIC sites in the C. elegans genome are unlikely to participate in local Pol III-dependent transcription, a characteristic behavior of extra-TFIIIC sites reported in other organisms [37].
C. elegans extra-TFIIIC sites possess strong Box-A and Box-B motifs
The TFIIIC complex binds the Box-A and Box-B DNA motifs [35] (Fig. 1a). However, the majority of extra-TFIIIC sites in yeast and human possess only the Box-B motif [39, 40, 43]. To determine whether C. elegans extra-TFIIIC sites contain Box-A and Box-B motifs, we performed de novo DNA motif analyses at extra-TFIIIC sites. Almost all of the 504 extra-TFIIIC sites in C. elegans harbored both the Box-A and Box-B motifs (90% with Box-A, E = 1.9 × 10−1568; 87% with Box-B, E = 1.5 × 10−1602; Fig. 1h). The pervasiveness of these motifs in extra-TFIIIC sites was comparable to that in Pol III-bound TFIIIC binding sites (94% with Box-A, E = 1.1 × 10−589; and 92% with Box-B, E = 3.5 × 10−1308) (Fig. 1h).
Because the Box-A and Box-B motifs constitute the gene-internal promoter for tRNA genes in eukaryotic genomes [35] (Fig. 1a), we hypothesized that extra-TFIIIC sites correspond to genetic elements similar to tRNA genes. In C. elegans, a class of interspersed repetitive elements called CeRep3 has been suspected as tRNA pseudogenes [51]. To determine whether extra-TFIIIC sites coincide with repetitive elements, we surveyed the overlap between extra-TFIIIC sites and all annotated repetitive elements (Fig. 1i). Strikingly, 44.6% (225 sites) of extra-TFIIIC sites overlapped repetitive elements (permutation-based empirical P < 0.001), and almost all of the overlapped elements (95.1%; 214 sites) were the CeRep3 class of repetitive elements (Fig. 1i). In contrast, although significant, only 8.2% (43 sites) of Pol III-bound TFIIIC sites overlapped repetitive elements of any class (permutation-based empirical P < 0.001). Therefore, unlike extra-TFIIIC sites in yeast and humans, C. elegans extra-TFIIIC sites harbored both the Box-A and Box-B motifs; furthermore, a large fraction of these sites corresponded to a class of putative tRNA pseudogenes CeRep3.
C. elegans extra-TFIIIC sites are not associated with regulatory elements for protein-coding genes
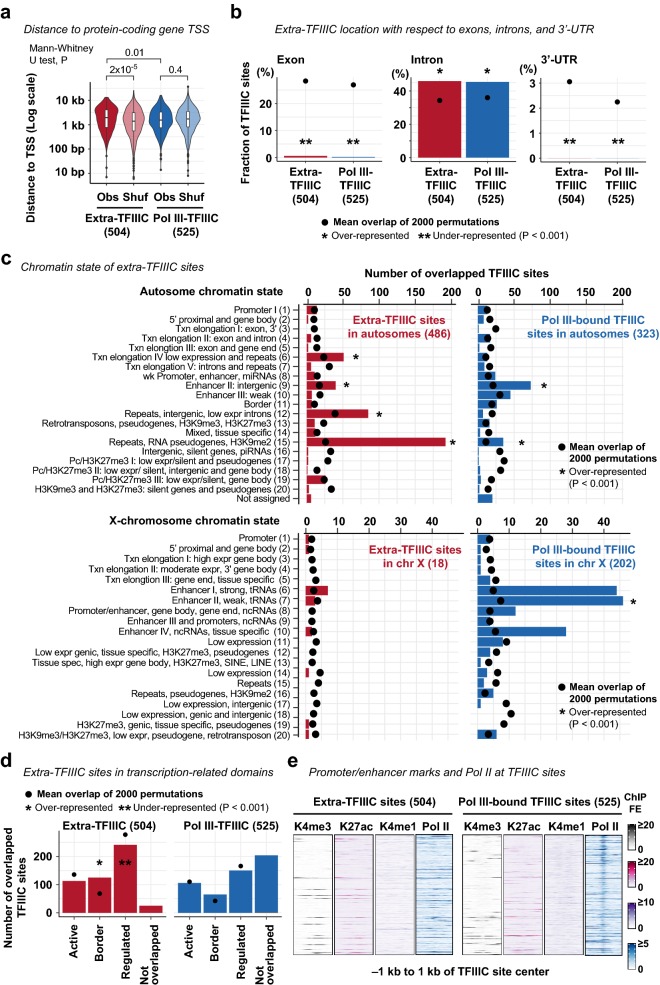
Previous studies in human and S. cerevisiae reported that extra-TFIIIC sites are overrepresented near protein-coding gene promoters, proposing a role in regulating RNA polymerase II (Pol II)-dependent transcription [39, 43]. To determine whether C. elegans extra-TFIIIC sites were located near protein-coding genes, we measured the distance from extra-TFIIIC sites to TSSs of nearest protein-coding genes. C. elegans extra-TFIIIC sites were not located near protein-coding gene TSSs compared with Pol III-bound TFIIIC sites (Mann–Whitney U test, P = 0.01) or with randomly permutated extra-TFIIIC sites (Mann–Whitney U test, P = 2 × 10−5; Fig. 2a). Extra-TFIIIC sites were overrepresented in introns and underrepresented in exons and 3′-UTRs, as were Pol III-bound TFIIIC sites (permutation-based empirical P < 0.001; Fig. 2b). Thus, extra-TFIIIC sites were not differentially represented near protein-coding gene TSSs or in exons, introns, and 3′-UTRs compared with Pol III-bound TFIIIC sites.
Fig. 2.
Extra-TFIIIC sites do not reside in protein-coding gene promoters or enhancers. a Distance between TFIIIC site center and the transcription start site (TSS) of protein-coding genes. Obs the observed distance. Shuf, the distance of a permutated set of TFIIIC sites (one permutation within chromosomes) to TSS. b Fraction of TFIIIC sites located in exons, introns, and 3′-UTRs of protein-coding genes. Permutations of TFIIIC sites were performed within chromosomal domains (i.e., center and left/right arms). P, empirical P-value based on the 2000 permutations. c Chromatin state annotation of TFIIIC sites. The chromatin state annotation reported by Evans et al. [4] is used. Permutations and P-value computation are performed as in b. d Number of TFIIIC sites resided in the three transcription-related domains reported by Evans et al. [4]. Permutations and P-value computation are performed as in b. d H3K4me3, H3K27ac, H3K4me1, and RNA polymerase II (Pol II) fold-enrichment scores (FE) at TFIIIC sites
To further investigate the relationship between extra-TFIIIC sites and cis-regulatory elements for protein-coding genes, we examined the chromatin states defined by a combination of histone modifications in early embryos [4]. Extra-TFIIIC sites were not overrepresented within “promoter” regions (14 sites, 2.8%, permutation-based empirical P = 0.4; Fig. 2c), consistent with the distance-based analysis. Instead, extra-TFIIIC sites were overrepresented among the chromatin states associated with repetitive elements including “transcription elongation IV: low expression and repeats” (51 sites, 10.5%), “Repeats, intergenic, low expression introns” (85 sites, 17.5%), and “repeat, RNA pseudogenes, H3K9me2” (192 sites, 39.5%) (permutation-based empirical P < 0.001; Fig. 2c), consistent with CeRep3 repeat overrepresentation at extra-TFIIIC sites. Extra-TFIIIC sites were also overrepresented in “enhancer II, intergenic” (40 sites, 8.2%; permutation-based empirical P < 0.001; Fig. 2c) and “borders” between “active” and “regulated” chromatin domains known to harbor gene-distal transcription factor binding sites [4] (125 sites, 25%; permutation-based empirical P < 0.001; Fig. 2d). However, extra-TFIIIC sites were devoid of histone modifications associated with active enhancers (H3K27ac), poised enhancers (H3K4me1), active promoters H3K4me3, or of Pol II enrichment (Fig. 2e). Pol III-bound TFIIIC sites were also not marked by H3K27ac, H3K4me1, or H3K4me3, but showed enrichment of Pol II, similar to previous observations [52]. Collectively, these results suggest that C. elegans extra-TFIIIC sites do not present features of active cis-regulatory elements for Pol II-dependent transcription.
C. elegans extra-TFIIIC sites are densely clustered in the distal arms of autosomes
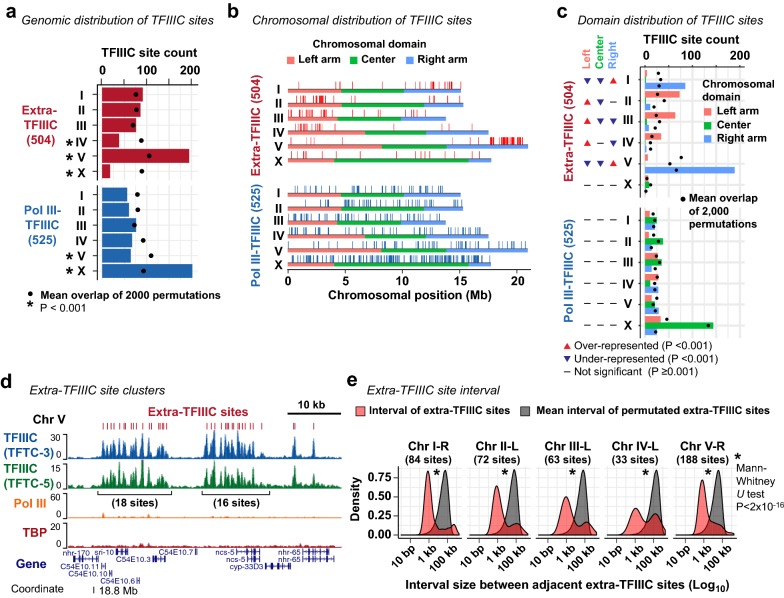
The lack of robust association with local regulatory features of Pol II and Pol III transcription led us to hypothesize that extra-TFIIIC sites were related to large-scale organization of chromosomes as in the case of yeasts [40, 53]. To test this hypothesis, we examined the distribution of extra-TFIIIC sites in the genome. We observed that extra-TFIIIC sites were highly overrepresented in chromosome V (195 of the 504 sites, 39%; permutation-based empirical P < 0.001), but strongly under-represented in the X chromosome (18 of the 504 sites, 4%; permutation-based empirical P < 0.001; Fig. 3a). In contrast, Pol III-bound TFIIIC sites were highly over-represented in the X chromosome (202 of the 525 sites, 38%; permutation-based empirical P < 0.001) consistent with tRNA gene overrepresentation in the X chromosome [23].
Fig. 3.
Extra-TFIIIC sites are clustered in autosomal arms. a Distribution of TFIIIC sites across chromosomes. Permutations of TFIIIC sites were performed across the genome. P, empirical P-value based on the 2000 permutations. b Distribution of TFIIIC sites within chromosomes. Horizontal color bars indicate chromosomal domains defined by Rockman and Kruglyak [24]. c Fraction of TFIIIC sites within chromosomal domains. Permutations of TFIIIC sites were performed within chromosomes. P, empirical P-value based on the 2000 permutations. d A representative genomic region harboring clusters of extra-TFIIIC sites. e Red, distribution of the interval size between two adjacent extra-TFIIIC sites. Gray, distribution of the mean of the interval sizes of permutated TFIIIC sites (2000 means representing 2000 permutations). Permutations were performed within chromosomal domains (i.e., center and left/right arms)
C. elegans autosomes can be subdivided into three domains of similar size (left arm, center, and right arm) based on repetitive element abundance, recombination rates, and chromatin organization [17, 23, 25]. We found that most extra-TFIIIC sites were located in autosome arms (486 of the 504 sites, 96%; Fig. 3b). In addition, extra-TFIIIC sites were overrepresented in only one of each autosome’s two arms that harbors the meiotic pairing centers [54] (overrepresented in the right arm of chromosome I; left arm of chromosome II; left arm of chromosome III; left arm of chromosome IV; right arm of chromosome V; permutation-based empirical P < 0.001; Fig. 3c). Furthermore, within autosomal arms, extra-TFIIIC sites were locally densely clustered (Fig. 3d), with a median interval between neighboring extra-TFIIIC sites of 1207 bp (Mann–Whitney U test vs. within-arm permutations P = 2 × 10−16; Fig. 3e). Among the autosome arms, the chromosome V right arm contained the largest number of extra-TFIIIC sites (188 sites) with extensive clusters (Fig. 3b, d, e). Thus, C. elegans extra-TFIIIC sites were fundamentally different from Pol III-bound TFIIIC sites in their genomic distribution and highly concentrated at specific locations within autosomal arm domains.
C. elegans extra-TFIIIC sites intersperse H3K9me3-marked heterochromatin domains
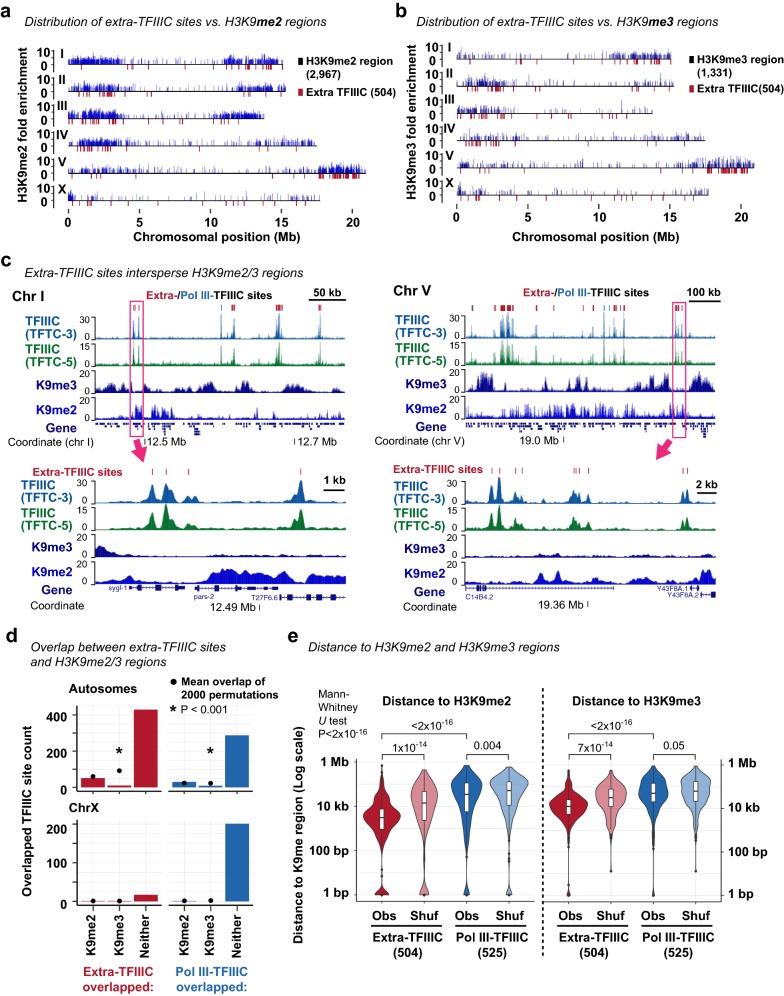
The autosome arms in C. elegans exhibit high levels of H3K9me2 and H3K9me3, histone modifications associated with constitutive heterochromatin [16, 17]. Furthermore, on each autosome, H3K9me2 and H3K9me3 signals are known to be stronger in one arm than the other [16, 27]. We hypothesized that extra-TFIIIC sites were located near H3K9me2 or H3K9me3-marked regions because extra-TFIIIC sites have been implicated in heterochromatin insulation [55, 56]. To test this hypothesis, we compared the locations of TFIIIC-bound sites with the locations of H3K9me2 and H3K9me3-enriched regions identified in early embryos [3]. Strikingly, the chromosome arms in which extra-TFIIIC sites were overrepresented coincided with the arms that exhibited strong H3K9me2 and H3K9me3 enrichment (Fig. 4a, b). However, at the local level, extra-TFIIIC sites did not reside in H3K9me3-enriched or H3K9me2-enriched regions (Fig. 4c), and were strongly underrepresented in H3K9me3-enriched regions (only 2% in H3K9me3-enriched regions; permutation-based P < 0.001; Fig. 4d). Instead, extra-TFIIIC sites were located significantly closer to H3K9me2-enriched regions (median distance 3.1 kb) and H3K9me3-enriched regions (median distance 12.5 kb) compared with Pol III-bound TFIIIC sites (H3K9me2, median distance 34.8 kb, Mann–Whitney U test P < 2 × 10−16; H3K9me3, median distance 50.1 kb, P < 2 × 10−16) or extra-TFIIIC sites permutated within autosomal arms (H3K9me2, median distance 11.3 kb, Mann–Whitney U test P = 1 × 10−14; H3K9me3, median distance 28.9 kb, P = 7 × 10−14) (Fig. 4e). Our analysis thus revealed that C. elegans extra-TFIIIC sites were located close to, but not overlapped with, H3K9me2 and H3K9me3-enriched regions within autosomal arm domains.
Fig. 4.
Extra-TFIIIC sites are located adjacent to H3K9me3-enriched regions. a Distribution of TFIIIC sites and H3K9me2-enriched regions. b Distribution of TFIIIC sites and H3K9me3-enriched regions. c (top) Two representative genomic regions showing extra-TFIIIC sites interspersing H3K9me3-enriched regions. (bottom) Genomic regions shown in rectangles in the top panels. d Fraction of TFIIIC sites overlapping H3K9me2/H3K9me3-enriched regions. Because the H3K9me2/H3K9me3-enriched regions are infrequent in the X chromosome, the X chromosome is plotted separately. Permutations of TFIIIC sites were performed within chromosomal domains (i.e., center and left/right arms). P, empirical P-value based on the 2000 permutations. e Distance between TFIIIC site center and H3K9me2- (left) or H3K9me3-enriched (left) regions. Obs the observed distance. Shuf, distance between a permutated set of TFIIIC sites and H3K9me2-enriched regions. Permutation was performed within chromosomal domains
C. elegans extra-TFIIIC sites are located within nuclear membrane-associated domains
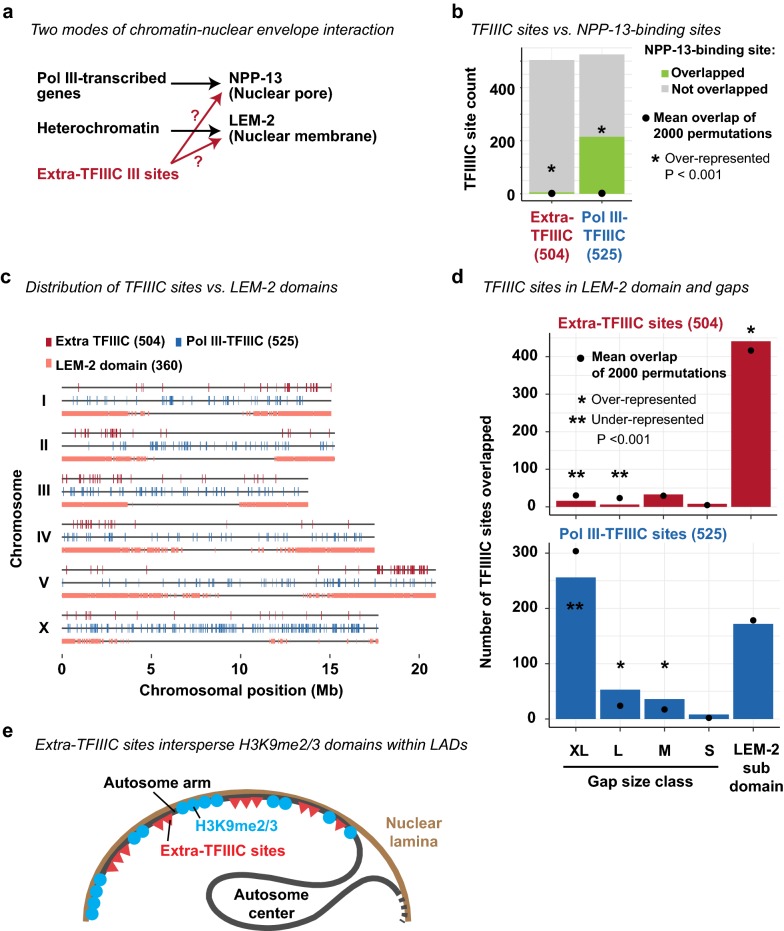
In S. pombe and S. cerevisiae, extra-TFIIIC sites are localized at the nuclear periphery and thought to regulate spatial organization of chromosomes [40, 46]. In C. elegans, Pol III-transcribed genes including tRNA genes are associated with the nuclear pore component NPP-13 [49], similar to tRNA genes in S. pombe associated with the nuclear pores [57] (Fig. 5a). We hypothesized that extra-TFIIIC sites are associated with NPP-13, given the similarity of extra-TFIIIC sites to Pol III-transcribed genes. To test this hypothesis, we compared the locations of extra-TFIIIC sites with those of NPP-13-bound sites identified in mixed-stage embryos [49]. We observed that only 6 of the 504 extra-TFIIIC sites (1.2%) overlapped NPP-13-bound sites (Fig. 5b), in contrast to a large fraction of Pol III-bound TFIIIC sites (215 sites, 41%) that overlapped NPP-13-bound sites (permutation-based P < 0.001; Fig. 5b). Thus, extra-TFIIIC sites were not likely to be associated with the nuclear pore.
Fig. 5.
Extra-TFIIIC sites reside in LEM-2-associated domains. a Two hypothetical scenarios in which extra-TFIIIC sites associate with the nuclear periphery. b Fraction of TFIIIC sites overlapping 223 NPP-13-binding sites (± 500 bp of NPP-13 site center) defined by Ikegami and Lieb [49]. Permutations of TFIIIC sites were performed within chromosomal domains (i.e., center and left/right arms). P, empirical P-value based on the 2000 permutations. c Distribution of TFIIIC sites and LEM-2 subdomains defined by Ikegami et al. [28]. d Fraction of TFIIIC sites overlapping LEM-2 subdomains and gaps of various sizes between LEM-2 subdomains. Permutations and P-value computation are performed as in b. e Model for chromosomal organization where extra-TFIIIC sites are clustered in the autosome arms corresponding LEM-2 domains and intersperse H3K9me3-enriched regions
Another mode of chromatin–nuclear envelope interactions in C. elegans is mediated by nuclear membrane-anchored, lamin-associated protein LEM-2 [28] (Fig. 5a). LEM-2 associates with the large genomic regions called “LEM-2 subdomains” that occupy 82% of the autosome arms [28]. Between LEM-2 subdomains are non-LEM-2-associated “gap” regions of various sizes [28]. We therefore investigated the locations of extra-TFIIIC sites with respect to those of LEM-2 subdomains and gaps (Fig. 5c). Strikingly, 441 of the 504 extra-TFIIIC sites (88%) were located within LEM-2 subdomains, demonstrating overrepresentation in a statistical test that accounted for the overrepresentation of extra-TFIIIC sites within autosome arms (permutation-based P < 0.001, permutation performed within chromosomal center/arm domains) (Fig. 5c, d). In contrast, Pol III-bound TFIIIC sites were underrepresented within LEM-2 associated domains (permutation-based P < 0.001, permutation performed within chromosomal domains). Together, our results suggest that C. elegans extra-TFIIIC sites are localized at the nuclear periphery and intersperse H3K9me3-marked heterochromatin regions (Fig. 5e).
Discussion
In this paper, we identified genomic sites bound by transcription factor III C (TFIIIC), the basal transcription factor for RNA polymerase III (Pol III), that do not participate in Pol III-dependent transcription in C. elegans. Similar TFIIIC-bound sites devoid of Pol III-dependent transcription, termed extra-TFIIIC sites, have been reported in yeast [39, 40], fly [41], mouse [42], and human [43]. Our data demonstrated that half of all TFIIIC-bound sites in C. elegans embryos lack Pol III binding, TBP binding, and nearby non-coding RNA genes, revealing pervasive extra-TFIIIC sites in the C. elegans genome.
Previous studies have suggested that extra-TFIIIC sites act as genomic insulators by blocking enhancer activity or heterochromatin spreading [40, 56, 58], or mediating three-dimensional genome interactions [41, 58]. Some of the genomic and chromatin features of C. elegans extra-TFIIIC sites reported in this paper resemble characteristics of extra-TFIIIC sites participating in chromatin insulation. First, C. elegans extra-TFIIIC were densely clustered in cis, similar to clusters of TFIIIC-bound sites capable of insulating heterochromatin and enhancer activities in S. pombe and human cells [40, 58]. Second, C. elegans extra-TFIIIC sites were located close to, but not within, H3K9me3-marked regions, similar to the observation that some extra-TFIIIC sites are located at the boundaries of heterochromatin [40, 56, 58]. Third, C. elegans extra-TFIIIC sites coincided with genomic regions known to be associated with nuclear membrane protein LEM-2 [28], similar to yeast extra-TFIIIC sites localized to the nuclear periphery [40, 46]. These observations raise the possibility that C. elegans extra-TFIIIC sites may act as a chromatin insulator at the nuclear periphery.
There are also differences between C. elegans extra-TFIIIC sites and those in other organisms. First, C. elegans extra-TFIIIC sites possess both the Box-A and Box-B motifs, unlike yeast and human extra-TFIIIC sites that only possess the Box-B motif [39, 43]. Second, C. elegans extra-TFIIIC sites were neither located near gene promoters nor associated with chromatin features of Pol II-dependent regulatory regions, unlike human, fly, and mouse extra-TFIIIC sites that are located near regulatory elements for RNA polymerase II (Pol II) transcription [41–43]. Third, C. elegans extra-TFIIIC sites are unrelated to CTCF binding, unlike human and mouse extra-TFIIIC sites that are located near CTCF binding sites [42, 43], because the C. elegans genome does not encode CTCF [32]. Our study could thus offer an opportunity for a comparative analysis of extra-TFIIIC functions across eukaryotic species.
The molecular mechanisms underlying the chromatin organization of C. elegans autosomes remain poorly understood. Unlike the X chromosome, which is organized into large self-interacting domains that have some features shared with topological associated domains (TADs) reported in other organisms, the autosomes do not present strong and pervasive self-interacting domains [22]. The condensin binding sites that could create boundaries between self-interacting domains in the X chromosome did not do so in the autosomes [14]. Several mechanisms for autosome chromatin organization have been proposed. These mechanisms include the antagonism between H3K36 methyltransferase MES-4 and H3K27 methyltransferase RPC2 that defines active versus repressed chromatin boundaries [4, 34]; small chromatin loops emanating from the nuclear periphery that allow active transcription within heterochromatin domains [28]; and active retention of histone acetylase to euchromatin that prevents heterochromatin relocalization [33]. Our observation that extra-TFIIIC sites are highly overrepresented in autosome arms and cluster in cis near the boundaries of H3K9me3-marked regions warrant future investigation of whether TFIIIC proteins participate in chromatin organization in C. elegans autosomes.
How strategies to demarcate chromatin domains have evolved in eukaryotes remain unclear. In vertebrates, CTCF has a central role in defining TAD boundaries and is essential for development [11, 59, 60]. In D. melanogaster, CTCF is essential but does not appear to define TAD boundaries, and instead acts as a barrier insulator [61–63]. In the non-bilaterian metazoans, some bilaterian animals (such as C. elegans), plants, and fungi, CTCF orthologs are absent [32, 64]. In contrast to CTCF, the TFIIIC proteins are conserved across eukaryotes [65] and extra-TFIIIC sites have been reported in human, [43], mouse [42], fly [41], C. elegans (this study), and yeast [39, 40]. Whether extra-TFIIIC is an evolutionary conserved mechanism for demarcating chromatin domains in eukaryotes, including those lacking a CTCF ortholog, will be an interesting subject of future studies.
Conclusions
We identified TFIIIC-bound sites that do not participate in RNA polymerase III-dependent transcription in the C. elegans genome. These “extra-TFIIIC” sites were highly over-represented in the arm domains of the autosomes interacting with the nuclear lamina. Extra-TFIIIC sites formed dense clusters in cis near the outer boundaries of individual H3K9me3-marked heterochromatin regions. These genomic features of C. elegans extra-TFIIIC sites resemble extra-TFIIIC sites reported in other organisms that have activities in insulating heterochromatin. Our study warrants future investigation of whether TFIIIC proteins participate in heterochromatin insulation in C. elegans.
Methods
ChIP-seq dataset
ChIP-seq of TFTC-3, TFTC-5, RPC-1, and TBP-1 was performed in chromatin extracts of the mixed-stage N2-strain embryos in duplicates and have been reported in our previous publication [49]. These data sets are available at Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE28772 (TFTC-3 ChIP and input), GSE28773 (TFTC-5 ChIP and input), GSE28774 (RPC-1 ChIP and input), and GSE42714 (TBP-1 ChIP and input). ChIP-seq datasets for H3K4me3, H3K4me1, H3K27ac, H3K9me2, and H3K9me3, performed in chromatin extracts of early-stage N2-strain embryos [3], were downloaded from ENCODE website (https://www.encodeproject.org/comparative/chromatin/).
Reference genome
The ce10 reference sequence was used throughout. The chromosomal domains (left arm, center, and the right arm) defined by recombination rates [24] were used.
Gene annotation
The genomic coordinates and the types of C. elegans transcripts were downloaded from the WS264 annotation of WormMine. The WS264 genomic coordinates were transformed to the ce10 genomic coordinates using the liftOver function (version 343) with the default mapping parameter using the ce11ToCe10.over.chain chain file downloaded from the UCSC genome browser.
TFIIIC site definition
MACS2 [66] (version 2.1.0) identified 1658 TFTC-3-enriched sites. Of those, sites that had the TFTC-3 fold-enrichment (FE) score greater than 5, harbored TFTC-5-binding sites within 100 bp, and were located in the nuclear chromosomes were considered “high-confidence” TFIIIC-bound sites (1029 TFIIIC sites). The “center” of each TFIIIC site was defined by the position of the base with the largest TFTC-3 FE score. Of the 1029 TFIIIC sites, those with the maximum Pol III (RPC-1) FE greater than 20 within ± 250 bp of the site center were defined as “Pol III-bound TFIIIC” sites (525) and the remaining sites were defined as “extra-TFIIIC” sites (504). The genomic coordinates for Pol III-bound TFIIIC sites and extra-TFIIIC sites are listed in Additional file 1.
Heatmap
For the heatmaps around TFIIIC sites, a set of 20-bp windows with a 10-bp offset that covered a 2-kb region centered around the center of TFIIIC sites was generated for each site. For each window, the mean of fold-enrichment score was computed from replicate-combined input-normalized fold enrichment bedgraph files. The signals were visualized using the ggplot2’s geom_raster function (version 2.2.1) in R.
Non-coding RNA genes
The genomic location and classification of non-coding RNA genes was described in “Gene annotation” section. For each TFIIIC site extended ± 100 bp from the site center, whether the region contained the transcription start site (TSS) of non-coding RNA genes was assessed using the Bedtools intersect function [67] (version 2.26.0).
DNA motif analysis
To find DNA motifs de novo, 150-bp sequences centered around the center of the TFIIIC-bound sites were analyzed by MEME (version 4.11.3) [68] with the following parameters: minimum motif size, 6 bp; maximum motif size, 12 bp; and the expected motif occurrence of zero or one per sequence (-mod zoops) and with the 1st-order Markov model (i.e., the dinucleotide frequency) derived from the ce10 genome sequence as the background.
Genomic intersection and permutation
Unless otherwise noted, the overlap between the 1-bp center of each of the TFIIIC sites and genomic features of interest (with size ≥ 1 bp) was assessed using the Bedtools intersect function [67] (version 2.26.0). To estimate the probability of observing the overlap frequency by chance given the frequency, location, and size of the features of interest and TFIIIC sites, the TFIIIC sites were permutated using the Bedtools shuffle function (version 2.26.0). The TFIIIC sites were shuffled across the genome, or within the chromosomes, or within chromosomal domains in which they reside, as described in each analysis section. After each permutation, the permutated set of TFIIIC sites were assessed for the overlap with the features of interest. This permutation was repeated 2000 times to assess the frequency at which the number of intersections for the permutated set of the TFIIIC sites was greater or less than the number of intersections for the observed TFIIIC sites. If none of the 2000 permutations resulted in the number of overlaps greater or less than the observed number of overlaps, the observed degree of overlaps was considered overrepresented or underrepresented, respectively, with the empirical P value cutoff of 0.001. The mean number of overlaps after 2000 permutations was computed for visualization.
Repetitive element analysis
The ce10 genomic coordinate and classification of repetitive elements, compiled as the “RepeatMasker” feature, were downloaded from the UCSC genome browser. The intersection between repetitive elements and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomes in which they resided.
Protein-coding gene distance
The genomic location of protein-coding genes was described in “Gene annotation” section. For each TFIIIC site, the absolute distance between the center of the TFIIIC site and the closest TSS of a protein-coding gene was obtained using the Bedtools closest function [67] (version 2.26.0). To assess the probability of observing such distance distribution by chance given the frequency and location of the TSSs and TFIIIC sites, the TFIIIC sites were permutated once using the Bedtools shuffle function (version 2.26.0) such that the TFIIIC sites were shuffled within the chromosomes, and the distance between the permutated TFIIIC sites and closest protein-coding gene TSS was obtained. Mann–Whitney U test, provided by the wilcox.test function in R, was used to assess the difference of the distribution of the TFIIIC–TSS distances between groups.
TFIIIC sites in exons, introns, and 3′-UTRs
The genomic coordinates of exons, introns, and 3′-UTRs were extracted from WS264 gff3 file downloaded from wormbase. Exons, introns, and 3′-UTRs that belong to protein-coding genes (i.e., transcript type is “coding_transcript”) were processed. The genomic coordinates were transformed to the ce10 genomic coordinates as described above. The intersection between these features and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomal domains (see “Reference genome”).
Chromatin state analysis
The chromatin state annotations and annotations of “active”, “regulated”, and “border” domains are reported previously [4]. The intersection between chromatin state annotations and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomal domains (see “Reference genome”) to account for the difference of the chromatin state representation among different chromosomal domains.
Chromosomal distribution of TFIIIC sites
The number of TFIIIC sites in each chromosome was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed across the genome. The number of TFIIIC sites in each chromosomal domain (see “Reference genome”) was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within chromosomes.
Extra-TFIIIC site interval
For each chromosomal domain, the genomic distance between every pair of two neighboring extra-TFIIIC sites (center-to-center distance) was computed in R. To estimate the degree of closeness between extra-TFIIIC sites only explained by the frequency of extra-TFIIIC sites within chromosomal domains, the extra-TFIIIC sites were permutated 2000 times within chromosomal domains as described in “Genomic intersection and permutation” section. In each of the 2000 permutations, the genomic distance between two neighboring permutated extra-TFIIIC sites was computed, and the mean of the distances was computed. The distribution of the 2000 means (by 2000 permutations) was compared with the distribution of observed distribution of TFIIIC interval sizes by Mann–Whitney U test.
Analysis of H3K9me2 and H3K9me3 regions
To define H3K9me2-enriched and H3K9me3-enriched regions, the genome was segmented into 1-kb windows, and the mean fold-enrichment score of H3K9me2 and H3K9me3 (see ChIP-seq dataset) was computed for each window using the Bedtools map function (version 2.26.0). Windows with the mean fold-enrichment score greater than 2.5 (1.5× standard deviation above mean for H3K9me2; and 1.3× standard deviation above mean for H3K9me3) were considered enriched for H3K9me2 or H3K9me3 and merged if located without a gap. This yielded 2967 H3K9me2-enriched regions (mean size, 2.1 kb) and 1331 H3K9me3-enriched regions (mean size, 4.9 kb).
The intersection between H3K9me2-enriched or H3K9me3-enriched regions and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomal domains.
For each TFIIIC site, the absolute distance between the center of the TFIIIC site and the closest H3K9me2-enriched and H3K9me3-enriched regions was obtained using the Bedtools closest function [67] (version 2.26.0). To assess the probability of observing such distance distribution by chance given the frequency, location, and size of H3K9me2-enriched and H3K9me3-enriched regions and TFIIIC sites, the TFIIIC sites were permutated once using the Bedtools shuffle function (version 2.26.0). For the analysis of the distance to H3K9me2-enriched regions, this permutation was performed within the chromosomal domains. For the analysis of the distance to H3K9me3-enriched regions, permutation was performed with the chromosomal domains, but excluding the H3K9me3-enriched regions themselves because the TFIIIC sites were strongly underrepresented in the H3K9me3-enriched regions.
TFIIIC sites in NPP-13-binding sites
The 223 NPP-13-binding sites identified in mixed-stage N2-stage embryos are previously reported [49]. The genomic coordinates were converted to the ce10 genomic coordinates using the UCSCtools liftOver function (version 343). The intersection between NPP-13-binding sites (± 500 bp of NPP-13 binding site center) and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomal domains.
TFIIIC sites in LEM-2 subdomain and gaps
The LEM-2 subdomains and gaps between LEM-2 subdomains identified in mixed-stage N2-stage embryos are previously reported [28]. The genomic coordinates were converted to the ce10 genomic coordinates using the UCSCtools liftOver function (version 343). The intersection between LEM-2 subdomains or gaps of variable size classes and TFIIIC sites was assessed as described in “Genomic intersection and permutation” section. The permutation of TFIIIC sites was performed within the chromosomal domains.
Supplementary information
Additional file 1. TFIIIC-binding sites.
Acknowledgements
We thank Jason D. Lieb, Sevinc Ercan, and Sebastian Pott for discussion.
Abbreviations
- C. elegans
Caenorhabditis elegans
- D. melanogaster
Drosophila melanogaster
- S. cerevisiae
Saccharomyces cerevisiae
- S. pombe
Schizosaccharomyces pombe
- ChIP-seq
chromatin immunoprecipitation followed by sequencing
- Pol II
RNA polymerase II
- Pol III
RNA polymerase III
- TBP
TATA-binding protein
- TFIIIB
transcription factor III B
- TFIIIC
transcription factor III C
- TSS
transcription start site(s)
- H3K9me2
histone H3 lysine 9 dimethylation
- H3K9me3
histone H3 lysine 9 trimethylation
- H3K27ac
histone H3 lysine 27 acetylation
- H3K4me1
histone H3 lysine 4 monomethylation
- H3K4me3
histone H3 lysine 4 trimethylation
- FE
fold enrichment
- TAD
topologically associated domain
Authors’ contributions
KI conceived the study. KI, AS, AL, and VB analyzed the data. KI, AS, AL, and VB wrote the manuscript. All authors read and approved the final manuscript.
Funding
K.I., A.S., and V.B. are supported by National Institutes of Health grant R21/R33 AG054770. A.S. was supported in part by National Institutes of Health grant R25 GM5533619. A.L. was supported by the Princeton University Program in Quantitative and Computational Biology and the Lewis-Sigler Richard Fisher ‘57 Fund.
Availability of data and materials
The datasets supporting the conclusions of this article are available in Gene Expression Omnibus with accession number GSE28772, GSE28773, GSE28774, and GSE42714 at https://www.ncbi.nlm.nih.gov/geo/. The datasets supporting the conclusions of this article are also included within the article (Additional file 1).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alexis V. Stutzman, April S. Liang and Vera Beilinson contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13072-019-0325-2.
References
- 1.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho JWK, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–452. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans KJ, Huang N, Stempor P, Chesney MA, Down TA, Ahringer J. Stable Caenorhabditis elegans chromatin domains separate broadly expressed and developmentally regulated genes. Proc Natl Acad Sci USA. 2016;113:E7020–E7029. doi: 10.1073/pnas.1608162113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun F-L, Elgin SCR. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/S0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 7.Lhoumaud P, Hennion M, Gamot A, Cuddapah S, Queille S, Liang J, et al. Insulators recruit histone methyltransferase dMes4 to regulate chromatin of flanking genes. EMBO J. 2014;33:1599–1613. doi: 10.15252/embj.201385965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/S1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 10.van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, et al. The insulator protein SU (HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169(930.e22):944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet. 2019;51:1263–1271. doi: 10.1038/s41588-019-0466-z. [DOI] [PubMed] [Google Scholar]
- 14.Anderson EC, Frankino PA, Higuchi-Sanabria R, Yang Q, Bian Q, Podshivalova K, et al. X Chromosome domain architecture regulates caenorhabditis elegans lifespan but not dosage compensation. Dev Cell. 2019 doi: 10.1016/j.devcel.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174(744–57):e24. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung M-S, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 19.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160:1049–1059. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 22.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 24.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes TM, Kohara Y, Codson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141:159. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 27.Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119:73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González-Aguilera C, Ikegami K, Ayuso C, de Luis A, Íñiguez M, Cabello J, et al. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol. 2014;15:R21. doi: 10.1186/gb-2014-15-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson DF, Marshall KM, Earnshaw WC. Condensin: architect of mitotic chromosomes. Chromosome Res. 2009;17:131–144. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- 31.Kranz A-L, Jiao C-Y, Winterkorn LH, Albritton SE, Kramer M, Ercan S. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013;14:R112. doi: 10.1186/gb-2013-14-10-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heger P, Marin B, Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabianca DS, Muñoz-Jiménez C, Kalck V, Gaidatzis D, Padeken J, Seeber A, et al. Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature. 2019;569:734–739. doi: 10.1038/s41586-019-1243-y. [DOI] [PubMed] [Google Scholar]
- 34.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 36.Noma K-I, Kamakaka RT. The human Pol III transcriptome and gene information flow. Nat Struct Mol Biol. 2010;17:539–541. doi: 10.1038/nsmb0510-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donze D. Extra-transcriptional functions of RNA polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene. 2012;493:169–175. doi: 10.1016/j.gene.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Ramsay EP, Vannini A. Structural rearrangements of the RNA polymerase III machinery during tRNA transcription initiation. Biochim Biophys Acta Gene Regul Mech. 2018;1861:285–294. doi: 10.1016/j.bbagrm.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noma K-I, Cam HP, Maraia RJ, Grewal SIS. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 41.Van Bortle K, Nichols MH, Li L, Ong C-T, Takenaka N, Qin ZS, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrière L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:270–283. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiraga S-I, Botsios S, Donze D, Donaldson AD. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol Biol Cell. 2012;23:2741–2754. doi: 10.1091/mbc.e11-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuen KC, Slaughter BD, Gerton JL. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci Adv. 2017;3:e1700191. doi: 10.1126/sciadv.1700191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Bortle K, Corces VG. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012;3:277–284. doi: 10.4161/trns.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell. 2013;51:840–849. doi: 10.1016/j.molcel.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EPS. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 51.Stricklin SL, Griffiths-Jones S, Eddy SR. C. elegans noncoding RNA genes. WormBook. 2005;25:1–7. doi: 10.1895/wormbook.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwasaki O, Tanaka A, Tanizawa H, Grewal SIS, Noma K-I. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010;21:254–265. doi: 10.1091/mbc.e09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11:934–942. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkland JG, Raab JR, Kamakaka RT. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim Biophys Acta. 2013;1829:418–424. doi: 10.1016/j.bbagrm.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, Gartenberg MR, et al. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE. 2011;6:e21923. doi: 10.1371/journal.pone.0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 60.Carmona-Aldana F, Zampedri C, Suaste-Olmos F, Murillo-de-Ozores A, Guerrero G, Arzate-Mejía R, et al. CTCF knockout reveals an essential role for this protein during the zebrafish development. Mech Dev. 2018;154:51–59. doi: 10.1016/j.mod.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz YB, Cavalli G. Three-dimensional genome organization and function in Drosophila. Genetics. 2017;205:5–24. doi: 10.1534/genetics.115.185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell. 2017;67(837–52):e7. doi: 10.1016/j.molcel.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci USA. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsutani S. Evolution of the B-block binding subunit of TFIIIC that binds to the internal promoter for RNA polymerase III. Int J Evol Biol. 2014;2014:609865. doi: 10.1155/2014/609865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. TFIIIC-binding sites.
Data Availability Statement
The datasets supporting the conclusions of this article are available in Gene Expression Omnibus with accession number GSE28772, GSE28773, GSE28774, and GSE42714 at https://www.ncbi.nlm.nih.gov/geo/. The datasets supporting the conclusions of this article are also included within the article (Additional file 1).