Abstract
The novel nucleotide oligomerization domain (NOD)-like receptor (NLR) with a caspase activation and recruitment domain (CARD) 3 (NLRC3) protein belongs to the NLR family of cytosolic pathogen recognition receptors. NLRC3 has the characteristic NOD and leucine-rich repeat configuration with a less well defined CARD. T lymphocytes are known to have high NLRC3 expression, which may be involved in suppression of T cell activation. Here, we report that NLRC3 is a cytoplasmic protein that negatively regulates pro-IL-1β maturation. Among well-known inflammasome components, NLRC3 can interact with apoptosis-associated speck-like protein containing a CARD (ASC) and caspases 1 and 5. Transient transfection of NLRC3 into stable EGFP-ASC-expressing HEK293FT cells reduces NLR family, pyrin domain-containing 3 (NLRP3)/cryopyrin-induced formation of ASC specks in a dose- and time-dependent manner. This suggests that NLRC3 can regulate ASC speck formation, caspase-1 activation and IL-1β maturation. We show for the first time that inflammasome-like complexes assemble when caspase-1 and ASC are cotransfected together with NLRC3 in HEK293FT cells. However, overexpression of NLRC3 with NLRP3/cryopyrin inflammasome components suppresses pro-caspase-1 cleavage and IL-1β processing. Our study suggests that NLRC3 negatively regulates inflammatory responses.
Key Words: NLRC3, Inflammasome, Anti-inflammatory, NF-κB
Introduction
The cells of innate immunity recognize microbes via a limited number of germ line-encoded proteins known as pattern recognition receptors (PRRs). PRRs are able to discriminate between noninfectious self and infectious nonself by sensing pathogen-associated molecular patterns expressed by pathogens and conserved among microorganisms. Pathogen-associated molecular patterns include many different molecules ranging from lipopolysaccharide (LPS), peptidoglycan, lipoproteins and proteins to nucleic acids [1, 2]. Accumulating evidence indicates that PRRs are also responsible for recognizing endogenous molecules released from damaged cells, termed damage-associated molecular patterns [3, 4, 5].
The nucleotide oligomerization domain (NOD)-like receptor (NLR) family with its 23 members has been determined to be a novel class of PRRs [6, 7]. Family members share three characteristic domains, as follows: a leucine-rich repeat at the C terminus, the central NOD and N-terminal protein-protein interaction domains. These domains play a role in ligand sensing, autoregulation and regulation of inflammatory responses in the signaling cascades, respectively [1, 2, 3, 6]. Several NLR members can assemble multiprotein platforms known as inflammasome complexes. In the NLR family, pyrin domain-containing (NLRP) 3/cryopyrin inflammasome, the interaction between NLRP3/cryopyrin and the adaptor protein apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (CARD; ASC) bridges the association of pro-caspase-1 to NLRP3/cryopyrin. This regulates the activation of caspase-1, leading to the secretion of biologically active inflammatory cytokines such as IL-1β, IL-18 and IL-33 [8, 9, 10, 11, 12, 13].
NLR family members regulate inflammatory responses in two different ways. Several NLRs, such as NOD1, NOD2 and NLRP3/cryopyrin, trigger the secretion of proinflammatory cytokines, whereas others, such as PYPAF2, PYPAF3, NLRP10/PYNOD, NLR with a CARD (NLRC) 5 and NLRP12/Monarch-1/PYPAF7, inhibit ASC-mediated NF-κB activation and caspase-1-dependent proinflammatory cytokine secretion [7, 14, 15, 16, 17, 18, 19, 20, 21]. Therefore, these two groups can be classified as proinflammatory and anti-inflammatory NLRs, respectively. Anti-inflammatory NLRs can attenuate inflammatory responses in different ways. To illustrate, interaction of NLRC5 with IKKα and IKKβ blocks their phosphorylation and leads to the inhibition of NF-κB-dependent responses [15, 22].
NLRC3 has a typical nucleotide binding domain-leucine-rich repeat configuration at its central and C-terminal domains. NLRC3 has a characteristic death domain fold composed of a 6-α-helical bundle at the amino terminus, suggesting that the NLRC3 effector domain is structurally similar to CARD or PYRIN domains [23]. Recently, this death fold domain was termed a CARD domain [24].
Previously, NLRC3 was shown to be highly expressed in T lymphocytes and in other immune cells, and it was suggested that NLRC3 functions as a suppressor of T cell activation in response to anti-CD3 and anti-CD28 antibodies or PMA and ionomycin treatment [25]. Recently, it has been reported that NLRC3 downregulates Toll-like receptor signaling via ubiquitinylation of the adaptor TRAF6, suppressing the activity of the NF-κB pathway [24].
Here, we describe a putative role for NLRC3 in the inflammasome framework. We show that NLRC3 is cytoplasmic, where it can colocalize with ASC and caspases 1 and 5. This physical interaction with inflammasome components leads to a reduction in caspase-1 activation and IL-1β release. Hence, we propose that NLRC3 negatively regulates NLR-mediated inflammatory responses.
Materials and Methods
Maintenance of Cell Lines
Human embryonic kidney cell lines HEK293FT and HEK293T and human T cell leukemia Jurkat cells were grown in DMEM and RPMI 1640, respectively, supplemented with 10% FBS, 1× L-glutamine, 1× penicillin/streptomycin and 1× MEM Non-Essential Amino Acids (Gibco BRL, Grand Island, N.Y., USA). Cell lines were grown at 37°C in 5% CO2. Cells were frozen with culture medium containing 10% DMSO at −80°C, when needed.
RNA Isolation and cDNA Synthesis
Total RNA isolation from human blood cells was performed with a Roche High Pure RNA Tissue Kit (Roche, Germany). cDNA synthesis followed the process of total RNA isolation. Firstly, 1 µg of total RNA was prepared in each microcentrifuge tube, 1 µl of poly-dT primer was added and the RNA-primer mixes were incubated at 72°C for 5 min. Then, the mixture was immediately chilled in ice water for at least 5 min. In the master mix, 4 µl of 5× reaction buffer (Promega, Madison, Wisc., USA), 5 µl of MgCl2, 1 µl of dNTP mix, 0.5 µl of RNase inhibitor and 1 µl of reverse transcriptase were added. Then, 11.5 µl of master mix was distributed to each microcentrifuge tube. The reaction took place at 42°C for 1 h, and as a final step, the reverse transcriptase enzyme was inactivated by heating at 70°C for 15 min.
Cloning of NLRC3 into pcDNA3-MYC/FLAG/HA
Total RNA from lymphocytes was used to clone the Nlrc3 gene. The primers used for cloning were as follows: Nlrc3 forward primer, 5′-TAAGTCTAGAAGGAAGCAAGAGGTGCGG-3′, and reverse, 5′-GATAGACGCGGCCGCTCACATTTCAACAGTGCA-3′ (MGH DNA Core Facility, Cambridge, Mass., USA). To clone Nlrc3 cDNA, high-fidelity Taq polymerase (Finzymme, Finland) was used to amplify the coding sequences using the primers shown above. The expected PCR products were extracted from the gel and purified with a Qiagen Agarose Gel Extraction kit (Qiagen, Germany). The multiple cloning site of the pcDNA3 vector had been previously modified to express FLAG, HA or MYC tags after the starting codon (gift of Prof. Gabriel Nunez, University of Michigan, Ann Arbor, Mich., USA). The 5′XbaI- and 3′NotI-digested PCR product was ligated to 5′XbaI- and 3′PspOMI-digested pcDNA3-MYC/FLAG or HA plasmids. Ligation reactions were transformed via heat shock into Top10 chemically competent Escherichia coli.
Generation of EGFP-NLRC3 Fusion Protein
pEGFP C3 plasmid (Clontech, Mountain View, Calif., USA) was used to generate EGFP-NLRC3 fusion protein. The Nlrc3 gene was subcloned from pET30a(+) NLRC3 plasmid into pEGFP C3 vector using BgIII and NotI restriction sites.
Caspase-5 was subcloned into pEGFP C3 to generate EGFP-caspase-5 fusion protein.Both pcDNA3-MYC-caspase-5 and pEGFP C3 plasmids were restriction digested with XhoI and XmaI, and the digested caspase-5 fragment was ligated into XhoI-XmaI-digested pEGFP C3.
NLRC3 was subcloned into pTagRFP C3 to generate TagRFP-NLRC3 fusion protein. pET30 NLRC3 plasmid was digested with KpnI and NotI enzymes. pTagRFP C3 plasmid was restriction digested with KpnI and PspOMI. KpnI-NotI-digested Nlrc3 fragment was ligated into KpnI-PspOMI-digested plasmid. For all cloning processes, NEB restriction and ligation enzymes were used (Ipswich, Mass., USA). Colonies were scanned with CVM forward primer (5′-CGCAAATGGGCGGTAGGCGTG-3′) and Nlrc3 cloning reverse primer. Following the diagnostic digestions, all positive plasmids were sequenced (Macrogen, Seoul, South Korea) and the predicted cloning sequences were verified.
Speck Assay
A total of 6 × 105 HEK293FT cells stably expressing ASC-EGFP were transfected with appropriate amounts of plasmids in 6-well plates. Two independent samples were prepared for each condition and were transfected into separate wells. DNA concentration was equalized between wells by using empty pcDNA3 vector. Five hours after transfection, medium was changed. Pictures were taken 24 and 48 h after transfection under an inverted fluorescent microscope. Speck number was counted in 4 randomly chosen fields per well (8 fields per condition), and Student's t test for independent samples was performed with SPSS software. Equality of variance was determined with Levene's test, and corresponding two-tailed p values were used.
Ca3(PO4)2 Transfection
HEK293FT cells were seeded at 1 × 106 cells/well in a 6-well plate and incubated overnight at 37°C in 5% CO2. The medium was replaced with fresh complete DMEM after 8–12 h to remove chloroquine. For co-immunoprecipitation experiments, transfection assays were performed with 10 ml of DMEM, 61 µl of 2 M CaCl2 and 500 µl of 2× HBS (HEPES-buffered saline) buffer in a 10 cm2 plate.
Western Blotting
A total of 1 × 106 HEK293T cells were plated in 6-well plates, and after 24 h, 250 or 500 ng or 1 μg of pcDNA3-MYC-NLRC3 was cotransfected with 1 μg of pro-IL-1β-IRES2-EGFP, pFLAG-ECFP-pro-caspase-1 and pTagRFP C3-ASC or without ASC via Ca3(PO4)2. The total amount of transfected plasmid DNA was 5 μg/well, and for each condition, the total amount of transfected DNA was brought to 5 μg/well by adding empty pcDNA3 vector DNA. Six hours after transfection, cell medium was exchanged with 1 ml of fresh medium. Then, 24 h after transfection, the cells were lysed in RIPA buffer (Sigma, St. Louis, Mo., USA) containing protease/phosphatase inhibitors (Roche). Before Western blotting, total protein amounts were quantified with BCA (Pierce-Thermo Scientific, Rockford, Ill., USA), and 20 μg/well total protein was loaded into 12% SDS-PAGE (Fermentas, Canada). The following primary antibodies were used: rabbit anti-human NLRC3 from Abcam (Cambridge, Mass., USA, ab77817; 1:1,000), rabbit anti-human caspase-1 from Santa Cruz (Santa Cruz, Calif., USA, sc56036; 1: 1,000), rabbit anti-mouse caspase-1 from Santa Cruz (sc514; 1: 1,000), rabbit anti-human IL-1β from Cell Signaling Technologies (Danvers, Mass., USA, 2021; 1:1,000), mouse anti-human β-actin from Santa Cruz (sc47778; 1:1,000), rabbit anti-EGFP from Abcam (ab290), monoclonal mouse anti-EGFP from Clontech (632592; 1:2,000), rabbit anti-human ASC from Novus (St. Charles, Mo., USA, NB100-94244; immunoprecipitation: 1/250; Western blotting: 1:1,000), rabbit anti-FLAG from Cell Signaling Technologies (2368; immunoprecipitation: 1/250; Western blotting: 1/1,000) and rabbit anti-MYC from Cell Signaling Technologies (2272; immunoprecipitation: 1/250; Western blotting: 1:1,000).
Confocal Analysis
One day before transfection, a cover slip (Isolab Laborgeräte, Germany) was put into each well and 106 HEK293FT cells/well were plated into 6-well plates. Cells were transfected with respective combinations of plasmids, and 24 h later, the cells were washed twice with 1 ml of 1× PBS and fixed in 1 ml of 4% paraformaldehyde by incubating for 5 min at room temperature. Slides were visualized using a Leica TCS SP5 II upright confocal microscope. All of the images used for the experiments were taken under identical conditions. Cells were monitored with a ×20 objective and ×20 magnification (755 × 755 μm visual field) at locations with 100% cell confluency (pinhole = 30.5 μm). RFP or DsRED, EGFP and ECFP were monitored by sequential scan in which EGFP is excited by the 488-nm line and ECFP by the 458-nm line of the argon laser. To capture the images, 525- to 600-nm and 465- to 475-nm windows were used for EGFP and ECFP, respectively.
Co-Immunoprecipitation
HEK293FT cells were seeded at 5 × 106 cells/10 cm2 culture plates and incubated overnight at 37°C in 5% CO2. These cells were cotransfected with 2.5 μg of candidate plasmid and 10 μg of pcDNA3-MYC/FLAG/HA-NLRC3. The negative control plate was transfected with empty vector, 3 μg of pcDNA3-FLAG/MYC and pEGFP C3. Cells were transfected with respective plasmids, and 24 h later, they were harvested. The pellet of cells was washed 3 times with cold 1× PBS. The pellets were resuspended in 700 µl of 0.5% NP40 lysing buffer. Resuspended pellets were incubated on ice for 1 h and vortexed gently and periodically. After 1 h, cell lysates were centrifuged at 13,000 rpm at 4°C for 30 min. The supernatant was removed, and 100 µl was stored as total lysate sample. Then, 15 µl of protein A (Pierce-Thermo Scientific) and 15 µl of protein G (Thermo Scientific, Rockford, Ill., USA) beads were washed with 0.5% NP40 lysing buffer 3 times. Subsequently, 600 µl of supernatant was incubated with the mixture of washed protein A/G beads on the rotator in the cold room for 30 min. In parallel, 25 µl of protein A and 25 µl of protein G beads were washed with 0.5% NP40 lysis buffer 3 times. At the end of the incubation period, the beads-supernatant mixture was centrifuged at 13,000 rpm at 4°C for 1 min. The supernatant was collected and added to the clean beads. Primary antibody (2 µl) was added to the supernatant-beads and incubated in the cold room on a rotator overnight. Beads-supernatant-antibody mixture was centrifuged at 10,000 rpm at 4°C for 1 min, and the beads were washed with isotonic lysis buffer (10 mM NaH2PO4, 50 mM NaCl, 3 mM MgCl2, 0.5% NP40, 1 tablet protease inhibitor cocktail, pH 8.0) 3 times. Each time, beads-lysis buffer mixture was centrifuged at 10,000 rpm at 4°C for 1 min. In the final step, the beads were resuspended in 50 µl of sample buffer, and Western blotting analyses were performed. For some of the co-immunoprecipitation experiments, the hosts of the primary and secondary antibodies were the same animal species, therefore the IgG heavy chain (50 kDa) was also detected on the membranes after Western blotting.
Luciferase Reporter Assay for NF-κB Activity
A total of 1 × 106 HEK293FT cells were plated in 6-well plates and cotransfected with pBVIx-Luc firefly luciferase reporter plasmid (50 ng), pRL-TK-Luc renilla luciferase (5 ng) as an internal control, pcDNA3-MYC-NLRC3 (10/50/100 ng), pcDNA3-HA-NOD1 (100 ng) and pEGFP C3 (150 ng) via the calcium phosphate method. The total amount of DNA (750 ng) was kept constant by inclusion of empty pcDNA3 vector. Then, 24 h after transfection, NF-κB-dependent transcription was determined using a Dual-Glo Luciferase Assay System (Promega). pRL-TK-Luc renilla luciferase was used for normalizing transcription efficiencies. Luminescence was measured with Fluoraskan Ascent FL and Fluoroskan Ascent (Thermo Scientific). Assays were performed in triplicate. Student's t test was used for statistical analysis of the results.
Results
NLRC3 Is a Cytoplasmic Protein
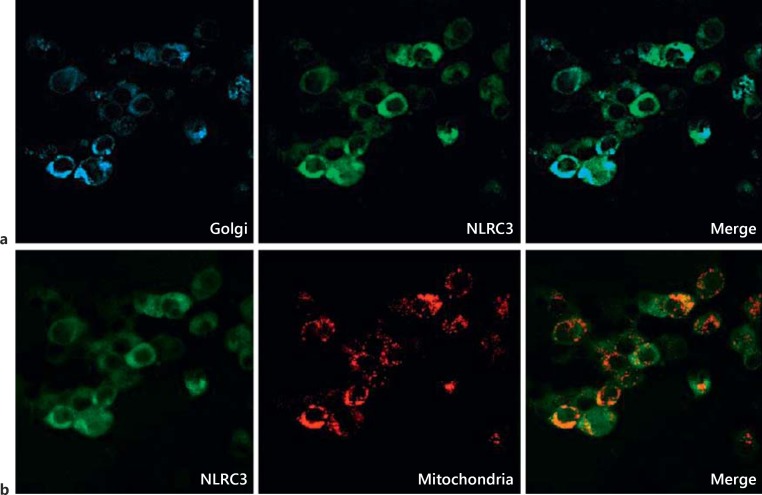
NLR family members are generally localized in the cytoplasm. However, several can be found in other intracellular compartments, e.g. NLRX1 in mitochondria and CIITA and NLRC5 in the nucleus [26, 27, 28]. Bioinformatics analyses predict that the probability of NLRC3 being found in the cytosol is 48%, and in mitochondria 4% [29]. To clarify the subcellular localization of NLRC3, the EGFP-NLRC3 fusion protein was generated and expressed alone or coexpressed with specific organelle markers tagged with different fluorescence proteins. After 24 h, transfected HEK293FT cells were analyzed under the confocal microscope, and EGFP-NLRC3 was largely detected in the cytoplasm (fig. 1). Based on these results, we conclude that the NLRC3 protein is mainly localized in the cytosol and can be partially localized in mitochondria (22%; fig. 1b) and the Golgi complex (12%; fig. 1a).
Fig. 1.
NLRC3 is a cytoplasmic protein. Shown are the cytosolic distribution of NLRC3 protein and subcellular colocalization of EGFP-NLRC3 with ECFP-Golgi and DsRED-MTS mitochondrial markers in HEK293FT cells. A total of 1 × 106 HEK293FT cells were transfected with 2 μg of pEGFP C3-NLRC3 alone or cotransfected with 1.5 μg of DsRED-MTS or ECFP-Golgi plasmid. Paraformaldehyde-fixed slides were analyzed with a confocal microscope. Three fields with similar cell counts per field were used for quantification. For each organelle, cotransfected (EGFP-positive and DsRED- or ECFP-positive) cells were firstly counted and then divided by the total number of transfected cells. Thereby, the given colocalization ratio was calculated. a EGFP-NLRC3 protein was colocalized with CFP-tagged Golgi marker. b EGFP-NLRC3 and DsRED-tagged mitochondrial protein were coexpressed.
Interaction of NLRC3 with Inflammasome Components: NLRC3 Can Weakly Interact with ASC
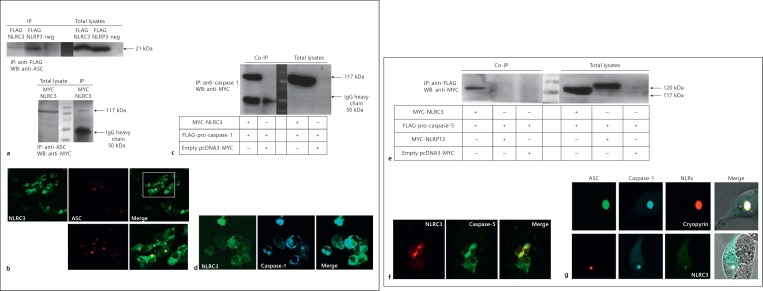
In the NLRP3/cryopyrin inflammasome, the adaptor protein ASC bridges the association of caspase-1 to NLRP3/cryopyrin. The ASC adaptor possesses an N-terminal PYRIN and C-terminal CARD domain. NLRC3 shares a putative CARD domain with ASC [24]. To test the possible interaction between NLRC3 and ASC, pcDNA3-ASC and pcDNA3-FLAG-NLRC3 were cotransfected into HEK293FT cells, and immunoprecipitation was carried out to pull down NLRC3 or our positive control NLRP3/cryopyrin and any interaction partners. Immunoblotting was performed on the immunoprecipitated samples using an anti-ASC antibody. The results suggest that NLRC3 can interact with ASC when overexpressed. However, NLRC3 interacts with ASC only weakly, when compared to NLRP3/cryopyrin (fig. 2a). To verify the interactions, we performed the reciprocal pull down and obtained the same result (fig. 2a, online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000363602).
Fig. 2.
NLRC3 interacts with inflammasome components ASC and caspases 1 and 5 and forms inflammasome-like structures. a Upper panel: NLRC3 can weakly interact with ASC. HEK293FT cells were cotransfected with 2.5 μg of pcDNA3-human ASC and 10 μg of pcDNA3-FLAG-NLRC3 plasmids. As a positive control, 10 μg of pcDNA3-FLAG-NLRP3/cryopyrin was also transfected with pcDNA3-human ASC into HEK293FT cells. Rabbit anti-FLAG was used for immunoprecipitation, and immunoblotting was performed with rabbit anti-human ASC. Lower panel: this is a reciprocal experiment of the upper panel. Proteins were precipitated with rabbit anti-human ASC and then immunoblotted with rabbit anti-MYC. b Cellular colocalization of RFP-ASC and EGFP-NLRC3 in HEK293FT cells. A total of 1 × 106 HEK293FT cells were cotransfected with 2 μg of pEGFP C3-NLRC3 and 1 μg of pTagRFP C3-ASC vectors. c Caspase-1 is an interaction partner of NLRC3. HEK293FT cells were cotransfected with 2.5 μg of FLAG-ECFP-pro-caspase-1 and 10 μg of pcDNA3-MYC-NLRC3. Immunoprecipitation and immunoblotting were performed with rabbit anti-MYC and anti-human caspase-1 antibodies, respectively. d Cellular colocalization of FLAG-ECFP-pro-caspase-1 and EGFP-NLRC3 in HEK293FT cells. A total of 1 × 106 HEK293FT cells were cotransfected with 1 μg of pFLAG-ECFP C3-pro-caspase-1 and 2 μg of pEGFP C3-NLRC3. e NLRC3 but not NLRP13 interacts with caspase-5. A total of 5 × 106 HEK293FT cells were cotransfected with 2.5 μg of pcDNA3-FLAG-caspase-5 and 10 μg of pcDNA3-MYC-NLRC3 or NLRP13 plasmid. Immunoprecipitation and immunoblotting were performed with rabbit anti-FLAG and anti-MYC antibodies, respectively. f EGFP-caspase-5 colocalized with RFP-NLRC3 in HEK293FT cells. pTagRFP C3-NLRC3 (2 μg) and pEGFP C3-caspase-5 (1 μg) plasmids were cotransfected into 1 × 106 HEK293FT cells. g Inflammasome-like complexes composed of NLRC3, ASC and caspase-1. A total of 1 × 106 HEK293FT cells were cotransfected with 2 μg of pEGFP C3-NLRC3, 1 μg of pTagRFP C3-ASC and 1 μg of pFLAG-ECFP C3-pro-caspase-1. IP = Immunoprecipitation; WB = Western blotting; Co-IP = co-immunoprecipitation; neg = negative.
To provide another piece of evidence for the possible interaction between NLRC3 and ASC proteins, colocalization analyses of EGFP-NLRC3 and RFP-ASC were carried out after coexpression in HEK293FT cells. The two proteins EGFP-NLRC3 and RFP-ASC showed distinct points of colocalization, which bolstered the idea of their potential interaction (fig. 2b). Based on the confocal imaging data, it is clear that NLRC3 and ASC colocalization is only partial, since there were also several RFP-ASC specks which did not appear to have EGFP-NLRC3 signal in them (at least 12%). It is interesting to point out that inside the cells there is a lot of free cytosolic NLRC3, possibly allowing the protein to carry out interactions with other partners.
Interaction of NLRC3 with Inflammasome Components: NLRC3 Interacts with Caspase-1 and Caspase-5 in the Presence and/or Absence of ASC
To investigate the possible interaction between NLRC3 and caspase-1 via their CARD domains, MYC-NLRC3 and FLAG-ECFP-pro-caspase-1 proteins were coexpressed in HEK293FT cells, and immunoprecipitation was performed using an anti-caspase-1 antibody and immunoblotting with an anti-MYC antibody. Our co-immunoprecipitation results highlight the physical association of NLRC3 with pro-caspase-1 (fig. 2c). To verify this interaction, overexpression of FLAG-ECFP-pro-caspase-1 and EGFP-NLRC3 proteins was induced in HEK293FT cells, and we found that EGFP-NLRC3 colocalized perfectly with FLAG-ECFP-pro-caspase-1 in the cytoplasm and apparently did not form any speck-like aggregations in the absence of ASC (fig. 2d).
Next, we coexpressed MYC-NLRC3 and FLAG-caspase-5 in HEK293FT cells and carried out co-immunoprecipitation experiments to test whether caspase-5 and NLRC3 can interact. Our results show clearly that NLRC3 interacts with caspase-5 as well as with caspase-1 (fig. 2e). For further validation, EGFP-caspase-5 and RFP-NLRC3 were transfected into HEK293FT cells. We observed that RFP-NLRC3 colocalized completely with EGFP-caspase-5 in the cytoplasm, and similarly, there were no speck-like aggregations in the absence of ASC (fig. 2b, d, f).
In the NLRP3/cryopyrin inflammasome, homotypic interactions occur between the PYRIN domains of NLRP3/cryopyrin and the adaptor protein ASC, and CARD-CARD interactions occur between ASC and caspase-1. Our data indicate that NLRC3 can interact with ASC and caspase-1 individually, without the need for the third component. It is thus suggested that NLRC3 can assemble an inflammasome-like complex via ASC and/or caspase-1 associations. We tested whether NLRC3 mediates inflammasome assembly when coexpressed with ASC and caspase-1. Similar to NLRP3/cryopyrin, EGFP-NLRC3 forms inflammasome complexes with RFP-ASC and FLAG-ECFP-pro-caspase-1 (fig. 2g, online suppl. fig. 2).
NLRC3 Negatively Regulates NLRP3/Cryopyrin-Induced Speck Formation
Under physiological conditions in healthy cells, the adaptor ASC is found as a soluble protein in the cytosol, whereas in apoptotic cells or under inflammasome-activating stimuli it forms ring-like speck structures [11, 12]. It has been previously shown that NLRP3/cryopyrin can enhance the speck formation of ASC when overexpressed. Since we had seen that NLRC3 interacts with ASC, we wondered about its effect on ASC speck formation.
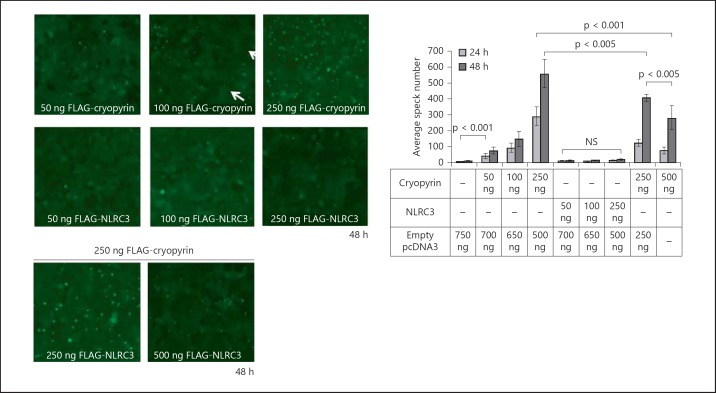
To test the effect of NLRC3 on ASC speck formation, stable EGFP-ASC-expressing HEK293FT cells were transfected with increasing amounts of NLRC3 and/or NLRP3/cryopyrin plasmids. As expected, overexpression of NLRP3/cryopyrin induced speck formation of ASC in a concentration- and time-dependent manner. Whereas empty vector-transfected cells only formed 5.88 ± 1.36 specks at 24 h and 12.625 ± 4 specks at 48 h, transfection with 50 ng of NLRP3/cryopyrin plasmid resulted in 41.5 ± 16.5 ASC speck formations at 24 h and 75.63 ± 20.28 at 48 h. Moreover, transfection with 100 ng of NLRP3/cryopyrin plasmid led to 91.75 ± 28.11 specks at 24 h and 146.86 ± 47.35 at 48 h. When the concentration of the transfected NLRP3/cryopyrin plasmid was raised to 250 ng, stable EGFP-ASC-expressing HEK293FT cells formed 290.375 ± 56.71 specks at 24 h and 556.5 ± 88.76 at 48 h after transfection (fig. 3). In contrast, NLRC3 transfection did not induce speck formation compared to NLRP3/cryopyrin (13 ± 4.34 specks at 24 h and 13.25 ± 4.06 at 48 h for 50 ng of NLRC3; 8.88 ± 1.81 specks at 24 h and 12.13 ± 4.55 at 48 h for 100 ng of NLRC3; 14.25 ± 3.06 specks at 24 h and 17 ± 4.69 at 48 h for 250 ng of NLRC3). However, when cotransfected with 250 ng of NLRP3/cryopyrin, NLRC3 significantly decreased the number of specks in a dose-dependent manner compared to 250 ng of NLRP3/cryopyrin transfected alone (122.88 ± 24.03 specks at 24 h and 405 ± 19.47 at 48 h for 250 ng of NLRP3/cryopyrin cotransfected with 250 ng of NLRC3 and 75.5 ± 19.87 specks at 24 h and 280.71 ± 75.72 at 48 h for 250 ng of NLRP3/cryopyrin cotransfected with 500 ng of NLRC3 compared to 290.375 ± 56.71 specks at 24 h and 556.5 ± 88.76 at 48 h for 250 ng of NLRP3/cryopyrin).
Fig. 3.
NLRC3 reduces NLRP3/cryopyrin-induced ASC speck formation in a dose- and time-dependent manner. A total of 1 × 106 HEK293FT EGFP-ASC stable cells were transfected with 50, 100 and 250 ng of FLAG-NLRP3/cryopyrin or NLRC3 plasmids per well and cotransfected with 250 ng of FLAG-NLRP3/cryopyrin and 250 or 500 ng of FLAG-NLRC3. After 24 and 48 h, cells were analyzed with an inverted fluorescent microscope (Axio Observer Z1 Inverted Microscope, Zeiss, Thornwood, N.Y., USA). Speck number was counted in 4 randomly chosen fields per well (8 fields per condition), and Student's t test for independent samples was performed with SPSS software. Equality of variance was determined with Levene's test, and corresponding two-tailed p values were used. The data represent 1 of 5 independent experiments. Speck formations of ASC are shown with arrows.
To test the correlation of ASC speck formation with inflammasome activation, stable EGFP-ASC-expressing HEK293FT cells were treated with one of the known inflammasome activators, monosodium urate crystals (online suppl. fig. 3). A dose-dependent increase in ASC speck numbers was observed, suggesting that ASC speck formation correlates with inflammasome activation rather than being a random aggregation.
Overall, NLRP3/cryopyrin boosted the aggregations of ASC in a time- and dose-dependent manner, as expected. Although NLRC3 alone had no effect on speck formation, it inhibited speck formation when coexpressed with NLRP3/cryopyrin (fig. 3). These data suggest that NLRC3 may compete with NLRP3/cryopyrin for ASC binding and may maintain ASC in an inactive, diffuse state.
NLRC3 Downregulates NOD1- and NOD2-Induced NF-κB Transcriptional Activity
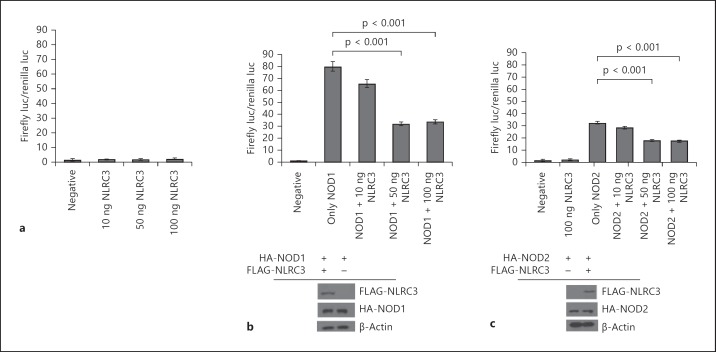
NOD1 and NOD2 are known to upregulate NF-κB signaling [8, 14]. In contrast, NLRC5 and NLRP10 can downregulate this pathway [16, 18, 20, 30]. To determine the effect of NLRC3 protein on NF-κB-dependent transcriptional activity, a luciferase reporter gene assay was performed. The luciferase reporter plasmids pBVIx, pRL-TK and NLRC3 were cotransfected into HEK293FT cells. The firefly luciferase gene has 6 NF-κB binding sites in the promoter sequence. Thus, the induction of the endogenous NF-κB-dependent transcriptional activity in the host cells can be monitored. The data clearly show that NLRC3 does not upregulate NF-κB activity (fig. 4a). As a positive control, we show that 100 ng of NOD1 and NOD2 increase the levels of NF-κB-dependent firefly luciferase activity 35- or 30-fold (fig. 4b, c). Furthermore, we show that NOD1 and NOD2 affected NF-κB-dependent firefly luciferase activity in a concentration-dependent manner (online suppl. fig. 4).
Fig. 4.
NLRC3 suppresses NOD1- and NOD2-induced NF-κB-dependent transcriptional activity. a Luciferase reporter assay for NF-κB activity of NLRC3. b NLRC3 coexpression suppresses NOD1-induced NF-κB-dependent transcription. c NLRC3 coexpression suppresses NOD2-induced NF-κB-dependent transcription. Both NOD1 and NOD2 protein levels did not change when they were cotransfected with NLRC3. Luciferase assays were performed in triplicate, and plates were read at least 3 times. Assays were repeated in 2 independent experiments. Student's t test was used for statistical analysis of the results.
To determine the downregulation potential of NLRC3, 100 ng of NOD1 or NOD2 plasmids were cotransfected with increasing amounts of NLRC3 plasmid. We found that coexpression of NLRC3 with NOD1 or NOD2 proteins results in NF-κB downregulation. While 100 ng of NOD2 plasmid triggers a 32 ± 2.62-fold increase in the NF-κB-dependent firefly luciferase promoter activity, cotransfection with 10 ng of NLRC3 leads to a decrease in activity to 28 ± 4.14-fold, and with 50 ng of NLRC3 plasmid, the promoter activity falls to 18 ± 3.52-fold (fig. 4c). Similarly, transfection with 100 ng of NOD1 results in a 79 ± 7.17-fold increase in NF-κB-dependent promoter activity; cotransfection with 10 ng of NLRC3 decreases the promoter activity to 65 ± 4.52-fold, and it falls further to 31 ± 5.19-fold when 50 ng of NLRC3 plasmid is cotransfected (fig. 4b). It is clear that NLRC3 downregulates NF-κB activity without any effects on NOD1 or NOD2 protein levels.
NLRC3 Suppresses Processing of pro-IL-1β
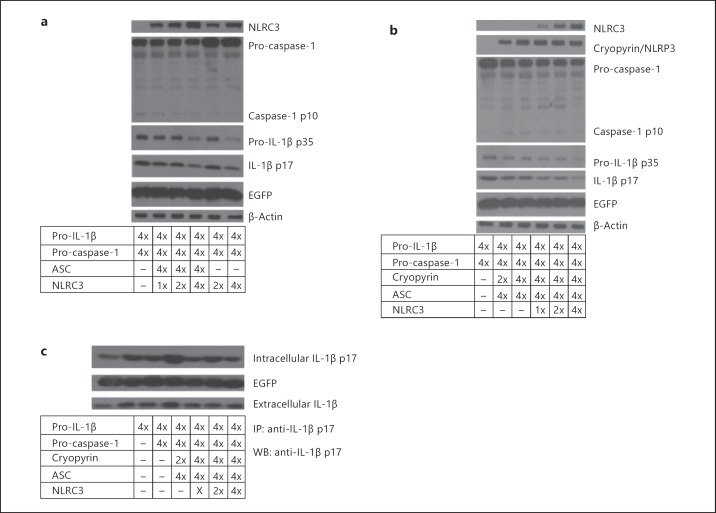
We have shown above that NLRC3 interacts with inflammasome components and assembles inflammasome-like complexes with ASC and caspase-1. To understand how NLRC3 affects pro-caspase-1 cleavage and caspase-1-dependent pro-IL-1β maturation in this platform, we coexpressed NLRC3 with pro-IL-1β, pro-caspase-1 and with or without ASC in HEK293T cells. The Western blotting data suggest that NLRC3 negatively affects pro-IL-1β maturation. When ASC is not coexpressed in HEK293T cells together with pro-caspase-1 and pro-IL-1β, pro-caspase-1 protein accumulates, and cleaved caspase-1 levels are low. When the concentration of transfected NLRC3 is increased, the expression and accumulation of pro-caspase-1 is similar, but cleaved caspase-1 levels decrease (fig. 5a). Furthermore, to assess the suppression effect of NLRC3 on the NLR-triggered inflammatory response, we overexpressed all of the inflammasome components as well as NLRP3/cryopyrin. NLRC3 suppresses pro-caspase-1 cleavage and p17 processing of pro-IL-1β in the cytoplasm. Similar to NLRP3/cryopyrin data, when NOD1 or NOD2 were coexpressed with NLRC3, caspase-1 activation and IL-1β maturation decreased significantly in a dose-dependent manner (online suppl. fig. 5).
Fig. 5.
NLRC3 negatively regulates IL-1β maturation and caspase-1 cleavage. a NLRC3 inhibits pro-IL-1β protein expression, IL-1β maturation and caspase-1 cleavage. b NLRC3 inhibits NLRP3/cryopyrin inflammasome-induced IL-1β maturation and caspase-1 cleavage. c NLRC3 expression leads to a moderate reduction in extracellular IL-1β levels. In brief, 24 h after transfection, supernatant and transfected cells were collected. Cells were lysed to profile the expression of intracellular IL-1β-p17. Supernatants, on the other hand, were precipitated to test the effect of NLRC3 on IL-1β secretion. Immunoprecipitation was performed with rabbit anti-human IL-1β-p17 antibody, and immunoblotting was performed using rabbit anti-human IL-1β-p17. IP = Immunoprecipitation; WB = Western blotting; x = 250 ng of plasmid.
Interestingly, NLRC3 also leads to a reduction in accumulated pro-IL-1β protein (p35) levels. However, NLRC3 expression does not affect the expression of the EGFP gene, which was under the control of CMV promoter in the IRES-EGFP plasmid (fig. 5a, b). This system permits us to translate pro-IL-1β and EGFP genes from a single bicistronic mRNA. Hence, we expect both EGFP and pro-IL-1β genes to be expressed similarly. In spite of the fact that EGFP expression is stable under different conditions, pro-IL-1β protein levels decrease when NLRC3 levels increase. Hence, we propose that NLRC3 may modulate pro-IL-1β protein at the posttranscriptional or posttranslational stages. It seems likely that NLRC3 suppresses processing of both p17 and pro-caspase-1. To address how NLRC3 influences the release of IL-1β, NLRC3 was coexpressed with inflammasome components, and after 24 h, intracellular p17 levels were profiled via Western blotting and extracellular released p17 levels were immunoprecipitated using an anti-IL-1β-p17 antibody. The moderate reduction observed in released IL-1β levels strengthens the idea that NLRC3 may negatively regulate NLR-triggered IL-1β processing (fig. 5c).
Discussion
In this study, we show for the first time that the cytoplasmic NLRC3 protein interacts with the inflammasome components ASC, caspase-1 and caspase-5. This interaction is particularly interesting because it leads to the suppression of p17 processing of pro-IL-1β (fig. 5). Furthermore, NLRC3 overexpression results in the suppression of NF-κB-dependent transcriptional activity and signaling (fig. 4).
In the first publication about NLRC3, Conti et al. [25] showed that NLRC3 is expressed in T cells, where it apparently suppressed activation of the cells after stimulation with CD3 and CD28. Following T cell stimulation, the presence of NLRC3 led to a decrease in the levels of endogenous IL-2 and CD25 transcripts, known as central proteins for the maintenance of T cell activation and prevention of T cell anergy. This reduction was concurrent with a delay of IκBα degradation [25]. This group also showed a negative effect of NLRC3 on NF-κB signaling in human cells. We provide further evidence to support their data under overexpression conditions.
With computational and biochemical analysis, Schneider et al. [24] demonstrated that NLRC3 has an evolutionarily conserved Ser-Leu-Gln TRAF binding motif in the NOD domain which mediates the interaction between NLRC3 and TRAF6. TRAF6 has two lysine (K) residues to be ubiquitinated. K48 is the auto-ubiquitination residue that leads to activation of the inflammatory response, whereas K68 ubiquitination is the target for proteasome-based degradation. They indicated that the interaction between NLRC3 and TRAF6 prevents the K48 ubiquitination of TRAF6, and finally, loss of activated TRAF6 results in the downregulation of the NF-κB signaling in mouse cells [24]. Evidence from the Nlrc3−/- mouse model supports the importance of NLRC3 for TRAF6 inhibition and degradation since there was not only much more active TRAF6 and NF-κB signaling but also more signs of inflammation, such as more nuclear NF-κB and proinflammatory cytokines, in LPS-treatedNlrc3−/- macrophages [24]. Hence, Schneider et al. [24] proposed that NLRC3 and Toll-like receptor constitute a negative feedback loop and described the NLR-TRAF interactions as the formation of a ‘TRAFasome’ complex.
It has also been described that NLRX1 interacts with TRAF6 and attenuates its function in NF-κB signaling [31, 32]. NLRX1 is located in the mitochondria and regulates the antiviral immune response by interacting with RIG-I and MAVS proteins [29]. When Nlrx1 is deleted, MAVS constitutively interacts with RIG-I, and that enhances the expression of antiviral signaling molecules such as IFN-β, STAT2 and OAS1 following the infection of influenza virus [31]. Furthermore, NLRX1 represents another inhibitory function by interacting with TRAF6 and IKK complexes, leading to the inhibition of NF-κB signaling during LPS stimulation of macrophages [31, 32]. Hence, it is possibly suggested that both NLRC3 and NLRX1 share a similar mechanism to negatively regulate the immune response.
Our data favor a model whereby the inhibitory effects of NLRC3 occur in two branches, i.e. the NF-κB arm and the inflammasome arm. NLRC3 overexpression results in the suppression of NF-κB trancriptional activity and hence signaling. Schneider et al. [24] defined well the NF-κB branch of the NLRC3-mediated negative regulation of the immune response. Therefore, it can be suggested that NLRC3 interactions with target proteins on the important pathways of inflammation lead to attenuation of the immune response. This suggestion can be supported by further evidence. Recently, Zhang et al. [33] demonstrated that NLRC3 negatively regulates the activity of STING (the protein stimulator of interferon genes). STING senses intracellular DNA molecules and triggers type I interferon through its interaction with TANK-binding kinase 1 [34]. It has been noted that NLRC3-mediated STING inhibition stems from the interaction between the NOD domain of NLRC3 and STING. This prevents the STING-TANK-binding kinase 1 interaction; hence, type I interferon production is inhibited [33].
NLRC3 can possibly regulate the inflammasome machinery by interacting with its components. Hence, our data are unique in the sense that they provide a link between NLRC3 protein and the inflammasomes. We propose that there is competition between NLRC3 and inflammasome components (NLRP3/cryopyrin, ASC and caspases 1 and 5) to bind to each other, apparently putting a brake on proinflammatory cytokine maturation and probably secretion. The observed reduction in both the intracellular and released forms of IL-1β provides evidence for this mechanistic view. Additionally, reduction in NLRP3/cryopyrin-induced ASC speck formation in a dose- and time-dependent manner after overexpression of NLRC3 hints at a possible molecular mechanism of inhibition of NLRC3 on inflammasomes (fig. 3).
As a last piece of evidence to support our model for the inflammasome branch of NLRC3 inhibition, Shiau et al. [35] investigated the requirement of zebra fish NLRC3 for microglia development. They found that Nlrc3 mutation in zebra fish leads to activation of primitive macrophages that results in systemic inflammation, increased proinflammatory cytokines and macrophage aggregation rather than their migration into the brain to form microglia [35]. Their further biochemical investigation revealed that zebra fish NLRC3 interacts with the adaptor protein ASC. Therefore, they have suggested that zebra fish NLRC3 competes with proinflammatory NLRs for ASC binding to trigger the inflammation [35]. This suggestion clearly supports our data and proposed model.
When NLRC3 is coexpressed with ASC and caspase-1, they assemble inflammasome-like structures. However, NLRC3-assembled molecular platforms do not seem to induce inflammasome activity. Both pro-caspase-1 and pro-IL-1β processes were inhibited. On the other hand, our Western blotting data suggest that NLRC3 has an impact on the regulation of pro-IL-1β (fig. 5). This regulation seems to be posttranscriptional or posttranslational.
In summary, our data support the existence of molecular mechanisms whereby NLRC3 negatively regulates NLR-mediated inflammatory responses, possibly by competing to bind inflammasome components.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgements
We thank all the members of the Apoptosis and Cancer Immunology Lab (AKiL) for helpful suggestions. The authors thank Assist. Prof. Stefan Fuss (Bogazici University-MBG, Istanbul, Turkey) for confocal microscopy training, Prof. Maria S. Soengas (Spanish National Cancer Research Center, Madrid, Spain) for HEK293FT cell lines, Prof. Gabriel Nunez (Department of Pathology, University of Michigan, Ann Arbor, Mich., USA) for pcDNA3-FLAG-caspase-1, Prof. Dr. Ahmet Koman (Bogazici University-MBG) for DsRED MTS vector and Assoc. Prof. Batu Erman (Faculty of Biological Sciences and Bioengineering, Sabancı University, Istanbul, Turkey) for pECFP-ER, pECFP-Golgi, pCFP-Rab5, pCFP-Rab9 and pCFP-Rab11 plasmids. We especially appreciate the contribution of Ali Can Sahillioğlu, M.Sc. (AKiL), in generating pro-IL-1β-IRES2-EGFP, pTagRFP C3-ASC and pFLAG-ECFP-pro-caspase-1 plasmids and stable EGFP-ASC HEK293FT cells. We thank Dr. Omer Hidir Yilmaz, Dr. Kivanc Birsoy and Dr. Paul Koenig (Whitehead Institute for Biomedical Research, MIT, Cambridge, Mass., USA) for their support. This study was supported by EMBO-SDIG 1468 and Bogazici University Research Fund (BAP 6526-11B01D10) grants to N.Ö. and a TEV award to Y.G.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, Carlson A, Merriam S, Lora JM, Briskin M, DiStefano PS, Bertin J. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-κB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 3.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Franchi L, Warner N, Viani K, Nuñez G. Function of NOD-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 8.Bryant C, Fitzgerald AK. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Dinarello CA. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann NY Acad Sci. 1998;856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 10.Dowds AT, Maumoto J, Zhu L, Inohara N, Nunez G. Cryopyrin-induced interleukin 1β in monocytic cells. J Biol Chem. 2004;279:21924–21928. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Alnemri ES. Assembly, purification, and assay of the activity of the ASC pyroptosome. Methods Enzymol. 2008;442:251–270. doi: 10.1016/S0076-6879(08)01413-4. [DOI] [PubMed] [Google Scholar]
- 13.Schroder K, Tschopp J. The inflammasomes. Cell. 2008;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Chaofeng H, Liping S, Yiling H, Daxiang L, Huadong W, Suisheng T. Functional characterization of the NF-κB binding site in the human NOD2 promoter. Cell Mol Immunol. 2010;7:288–295. doi: 10.1038/cmi.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui J, Zhu L, Xia X, Wang YH, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen JZ, Wang RF. NLRC5 negatively regulates the NF-κB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura R, Wang Y, Kinoshita T, Suzuki M, Noda T, Sagara J, Taniguchi S, Okamoto H, Suda T. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J Immunol. 2010;184:5874–5884. doi: 10.4049/jimmunol.0900779. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Hasegawa M, Imamura R, Kinoshita T, Kondo C, Konaka K, Suda T. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int Immunol. 2004;16:777–786. doi: 10.1093/intimm/dxh081. [DOI] [PubMed] [Google Scholar]
- 18.Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF. NLRC5 negatively regulates the NF-κB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1β secretion. J Biol Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- 20.Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, Ting PYJ. Cutting Edge: Monarch-1 suppresses non-canonical NF-κB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 21.Lich JD, Ting PYJ. Monarch-1/PYPAF7 and other CATERPILLER (CLR, NOD, NLR) proteins with negative regulatory functions. Microbes Infect. 2007;9:672–676. doi: 10.1016/j.micinf.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z, Lich DJ, Moore BC, Duncan AJ, Williams LK, Ting PYJ. ATP binding by Monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–1850. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouchier-Hayes L, Martin JS. CARD games in apoptosis and immunity. EMBO Rep. 2002;3:616–621. doi: 10.1093/embo-reports/kvf139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider M, Zimmermann GA, Roberts AR, Zhang L, Swanson VK, Wen H, Davis KB, Allen CI, Holl KE, Ye Z, Rahman AH, Conti BJ, Eitas TK, Koller BH, Ting JP. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti BJ, Davis BK, Zhang J, O'connor W, Jr, Williams KL, Ting JP. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J Biol Chem. 2005;280:18375–18385. doi: 10.1074/jbc.M413169200. [DOI] [PubMed] [Google Scholar]
- 26.Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulosc D, van den Elsen JP, Kobayashi SK. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West AP, Gerald S, Ghosh SS. Mitochondria in innate immune responses. Nature. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, Lich JD, Heise MT, Chen Z, Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 30.Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hösel M, Büning H, Schwarzenbacher R, Kufer TA. A role for the human NLR family member NLRC5 in antiviral response. J Biol Chem. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, Ranjan P, Monroe KM, Pickles RJ, Sambhara S, Ting JP. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, Wang RF. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, Ouyang S, Liu ZJ, Damania B, Shu HB, Duncan JA, Ting JP. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangan MS, Latz E. NLRC3 puts the brakes on STING. Immunity. 2014;40:305–306. doi: 10.1016/j.immuni.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiau CE, Monk KR, Joo W, Talbot WS. An anti-inflammatory NOD-like receptor is required for microglia development. Cell Rep. 2013;5:1342–1352. doi: 10.1016/j.celrep.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data