Abstract
Transmembrane 9 (TM9) proteins, or nonaspanins, are a family of proteins conserved throughout evolution and characterized by 9 transmembrane domains. In Drosophila, TM9 superfamily protein member 4 (TM9SF4) and its closest paralogue, TM9SF2, contribute to phagocytosis of various types of particles, while TM9SF4 displays non-redundant requirement in Gram-negative bacteria engulfment. In addition, the two TM9 proteins control the actin cytoskeleton in larval haemocytes and in Drosophila S2 cells. Here, we show that TM9SF4 and TM9SF2 co-immunoprecipitate with the peptidoglycan recognition protein (PGRP)-LC, which triggers the Drosophila immune response to bacterial infection. Furthermore, both TM9 proteins co-localize with this receptor in intracellular vesicles and at the plasma membrane in Drosophila S2 cells in culture and in the fly fat body. Silencing TM9SF4 prevents plasma membrane localization of PGRP-LC, whereas silencing TM9SF2 does not, which may account for the non-redundant role of TM9SF4 in phagocytosis of Gram-negative bacteria. Finally, we provide a set of data suggesting that TM9 proteins can prevent inappropriate signalling from the unstimulated receptor.
Key Words: Drosophila, Innate immunity, Innate immune receptor, Innate immune signalling, Imd, Nonaspanins, Plasma membrane receptor, Phagocytosis, Transmembrane 9 proteins
Introduction
Innate immune receptors detect microorganisms and altered-self signals to activate a set of stress and immune responses that result in clearance of the recognized entities, thus maintaining organism integrity [1]. They include various classes of receptors that may be secreted in body fluids, expressed at the surface of immune cells or in their cytoplasm. Among them, the peptidoglycan recognition proteins (PGRPs) recognize the bacterial cell wall component peptidoglycan. PGRPs are present in most invertebrates and vertebrates, but whereas in mammals they are mainly secreted and directly bactericidal, in flies they are either secreted or transmembrane proteins acting as receptor molecules that transmit a signal to the interior of the cell. In Drosophila, the transmembrane protein PGRP-LC is one major actor in the immune response to Gram-negative bacteria that activates the immune deficiency (Imd) pathway in the fat body [2, 3, 4]. The signal from PGRP-LC is transmitted via the receptor-bound scaffolding protein Imd [5, 6]. Imd activation induces a signalling cascade resulting in the Relish (NF-κB like transcription factor)-dependent activation of stress and immune response genes including those encoding antimicrobial peptides (AMPs) such as Diptericin(Dipt) and Attacin A(AttA)[7]. In addition, PGRP-LC specifically triggers phagocytosis of Gram-negative bacteria [4], while other phagocytic receptors in Drosophila, such as Eater and NimC1, contribute to the engulfment of various types of particles [8, 9].
The transmembrane 9 (TM9) proteins (also known as nonaspanins) are a group of highly conserved proteins with 9 transmembrane domains [10, 11, 12, 13]. They include 3 members in the amoeba Dictyostelium discoideum (Phg1A, B, C), in the yeast Saccharomyces cerevisiae (TMN1-3) and in Drosophila melanogaster flies (TM9SF2, TM9SF3, TM9SF4) and 4 members in humans (TM9 superfamily proteins TM9SF1 to TM9SF4) [10, 14, 15, 16, 17]. They are found in the endosomal compartments of yeast, Dictyostelium and human cells where they possibly contribute to cell migration, vesicular transport, endocytic trafficking and autophagy [11, 12, 17, 18, 19, 20, 21]. In Dictyostelium, TM9SF4/Phg1A is required for the phagocytosis and killing of bacteria [16, 22, 23]. Moreover, the two Dictyostelium TM9 proteins Phg1A and Phg1B synergistically contribute to the expression and/or localization of transmembrane proteins [14, 24]. The function of TM9SF4 in phagocytosis is conserved in human immune cells, where TM9SF4 overexpression contributes to enhanced phagocytic activity of metastatic tumour cells [13, 25], and in Drosophila [15, 26]. In Drosophila, TM9SF4 mutant macrophages and TM9SF4-silenced S2 cells are notably defective in phagocytosis of Escherichia coli-derived particles, a phenotype reminiscent of that of pgrp-lc-silenced cells [4, 15]. Also, the closely related protein TM9SF2 acts redundantly with TM9SF4 in the phagocytosis of various types of particles and in the control of the actin cytoskeleton [15].
In this study, we show that TM9SF2 and TM9SF4 are important for the subcellular localization and signalling activity of PGRP-LC in Drosophila. These two TM9 proteins interact with PGRP-LC and co-localize with the receptor in both intracellular punctate structures and at the plasma membrane. TM9SF4, but not TM9SF2, is required for PGRP-LC localization at the cell surface, which might account for the specific function of TM9SF4 in internalization of Gram-negative bacteria. Moreover, TM9SF2 and to a lesser extent TM9SF4 mutant flies showed constitutive activation of AMP gene expression, suggesting a negative regulatory function of these two TM9 proteins on the unstimulated receptor. Since expression of both TM9SF2 and TM9SF4 inhibits PGRP-LC but not Imd signalling activity, mediated by their overexpression in S2 cells, these two TM9 proteins likely directly prevent inappropriate PGRP-LC signalling activity by interacting with the receptor.
Materials and Methods
Fly Strains
Flies were raised at 25°C. The TM9SF41 null mutant is described in the report of Bergeret et al. [15]. P[UAS-PGRP-LCx-Flag] (lines 16B and 77A) are described in the report of Schmidt et al. [27]. The P[EP]CG9318EP2088 designed in this study as TM9SF2EP2088 was obtained from the Exelixis Collection at the Harvard Medical School (https://drosophila.med.harvard.edu/). The transgenic lines P[UAS-TM9SF2-GFP] and P[UAS-TM9SF4-GFP] were obtained by germ-line-mediated integration using standard methods. With regard to the FLPout GAL4/UAS method, spontaneous activation of the GAL4 transcription factor without heat shock has been reported by Hennig et al. [28].
Cell Culture
Drosophila S2 cells were maintained in Schneider's medium supplemented with 10% heat-inactivated fetal calf serum (Invitrogen). Gene inactivation was achieved as described by Clemens et al. [29]. The methodology and primers used are described in the legend to online supplementary figure 2 (for all online suppl. material, see www.karger.com/doi/10.1159/000365112). Activation of the Attacin promoter in S2 cells and induction of AMP genes in vivo were monitored as described by Thevenon et al. [30].
DNA Constructs
cDNA clones for TM9SF2 (LD44273) and TM9SF4 (GH02822) were purchased from Drosophila Genomics Resource Center (DGRC). The following primer sets were used for PCR amplification: TM9SF2 forward, 5′-ggggtaccATGATCCTGCTATCCGGACTT-3′, TM9SF2 reverse, 5′-ctagtctagaATCCACCTTGACAACACTGTA-3′; TM9SF4 forward, 5′-ggggaattcCACTCCCACACACCACCAACA-3′, and TM9SF4 reverse, 5′-gcggatccGTCGATCTTCACAGCTCCGTA-3′.
Full-length PCR products were cloned into pAc5.1/V5/HisB vector (Invitrogen) or pAc-GFP vectors, allowing for the expression of corresponding tagged proteins. Full-length and truncated pAc-PGRP-LC-V5 constructs and pAc-Imd-V5 constructs were made from the corresponding pMT vectors described in the report of Choe et al. [5].
Immunoprecipitation
Co-immunoprecipitation of GFP-tagged TM9 protein with V5- or Flag-tagged PGRP-LC was performed following standard procedures.
Immunofluorescence Microscopy and Clonal Analysis
Immunofluorescence microscopy of S2 cells and dissected fat body were performed as described by Bergeret et al. [15] and Taillebourg et al. [31], respectively.
Fluorescence-Activated Cell Sorting Analysis
S2 cells were transfected with pAc-PGRP-LC-V5 and double-stranded RNA (dsTM9SF4, dsGFP or dsPGRP-LC). Then, 48 h after transfection, cells were washed in PBS buffer containing 3% fetal calf serum and 0.05% NaN3 and incubated for 30 min at 4°C with anti-V5 antibody (Invitrogen) then with secondary antibody Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen). Samples of 20,000 cells suspended in PBS buffer with 0.05% NaN3 were analysed using a FACSCalibur flow cytometer (Becton Dickinson).
Results
TM9SF2 and TM9SF4 Display Similar Subcellular Localization to PGRP-LC
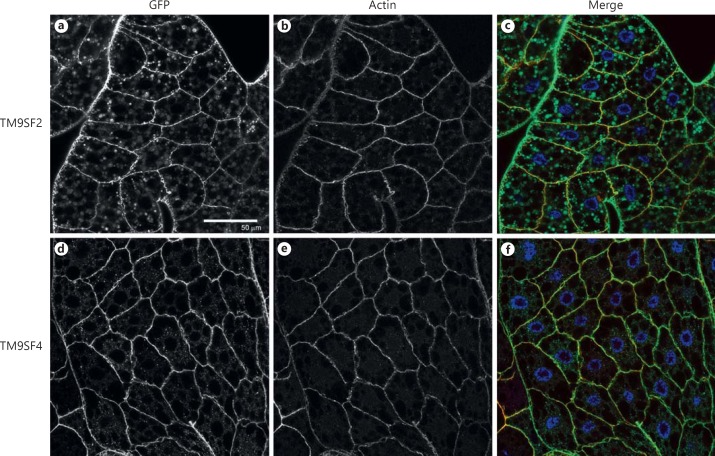
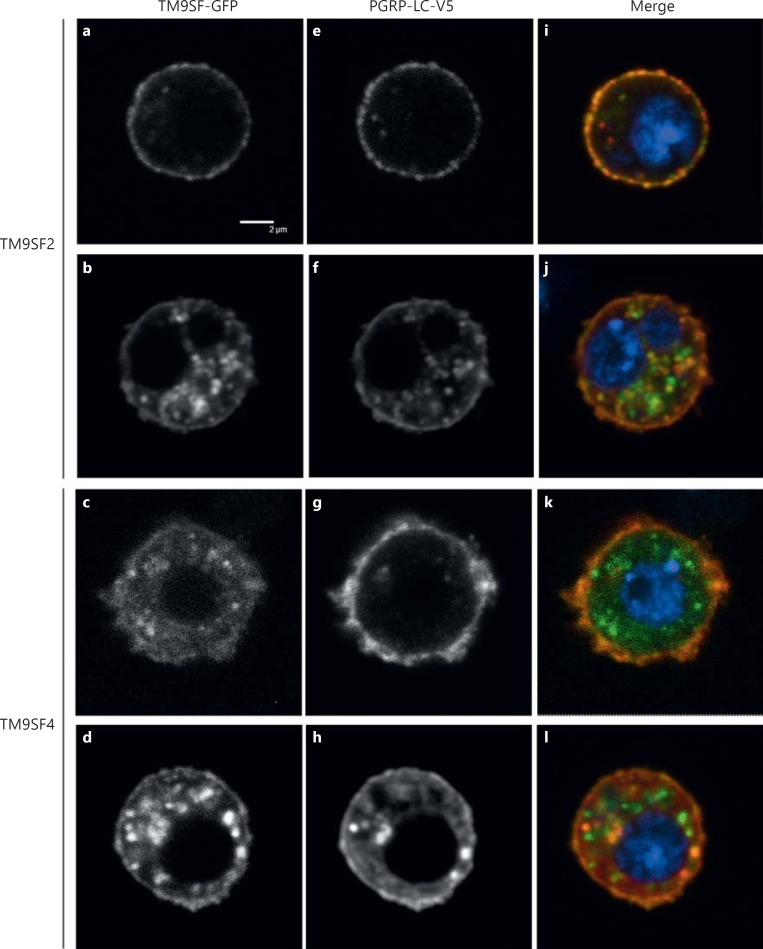
To examine the expression and the subcellular localization of TM9SF proteins in vivo, we constructed transgenic lines expressing TM9SF2-GFP or TM9SF4-GFP fusion proteins. When expressed specifically in the fat body through the specific CgGal4 driver line, both TM9-GFP fusion proteins were present mainly at the plasma membrane and to a lesser extent in cytoplasmic punctate structures (fig. 1). We used the FLPout GAL4/UAS method [32] to induce the expression of the Flag-tagged PGRP-LC (PGRP-LC-Flag) [27] concomitantly with either TM9SF2-GFP or TM9SF4-GFP specifically in clones of fat body cells. PGRP-LC-Flag, TM9SF2-GFP or TM9SF4-GFP fusion proteins were all observed both at the cell surface and intracellularly in these clones (online suppl. fig. 1). Partial co-localization of PGRP-LC with TM9SF2 and TM9SF4 was observed mainly at the plasma membrane (online suppl. fig. 1). We then co-expressed fluorescent TM9SF-GFP fusion proteins (TM9SF2-GFP or TM9SF4-GFP) with a PGRP-LC protein tagged with a V5 epitope (PGRP-LC-V5) [5] in cultured Drosophila S2 cells. TM9SF2-GFP and TM9SF4-GFP proteins localized both in intracellular round structures and at the cell membrane (fig. 2a, b, c, d). Similar staining was observed for PGRP-LC-V5, with about half of the cells showing mainly membrane-bound PGRP-LC (fig. 2e-g) and the second half presenting both membrane-bound and cytosolic staining (fig. 2f, h). These experiments revealed a close co-localization of each TM9SF protein with PGRP-LC (fig. 2i-l). We conclude from these observations that TM9SF2 and TM9SF4 co-localize with PGRP-LC in both intracellular and plasma membrane compartments.
Fig. 1.
TM9SF2 and TM9SF4 subcellular localization in fat body cells. Confocal microscopy images of fat bodies from third instar larvae expressing TM9SF2-GFP (a-c) or TM9SF4-GFP (d-f) in the fat body cells through the CgGal4 driver line. a, d TM9SF-GFP-fusion proteins are visualized through GFP fluorescence. b, e The F-actin network was labelled with Texas red-phalloidin. c, f Merge images in which GFP is green, the actin network is red and the nuclei are stained with Hoechst in blue. Genotypes: w1118;CgGal4/+;UAS-TM9SF2-GFP (a-c), w1118;CgGal4/+;UAS-TM9SF4-GFP (d-f). Scale bar = 50 µm.
Fig. 2.
TM9SF2 and TM9SF4 co-localize with PGRP-LC in S2 cultured cells. Confocal microscopy images of fixed Drosophila S2 cells expressing either TM9SF2-GFP (a, b, e, f, i, j) or TM9SF4-GFP (c, d, g, h, k, l) together with PGRP-LC-V5. a-d TM9SF-GFP fusion proteins are visualized through GFP autofluorescence. e-h PGRP-LC-V5 tagged protein is visualized through immunofluorescence staining with an anti-V5 antibody. i-l Merge images with GFP in green, PGRP-LC-V5 in red and DNA stained with Hoechst in blue. Scale bar = 2 μm.
TM9SF2 and TM9SF4 Interact with PGRP-LC
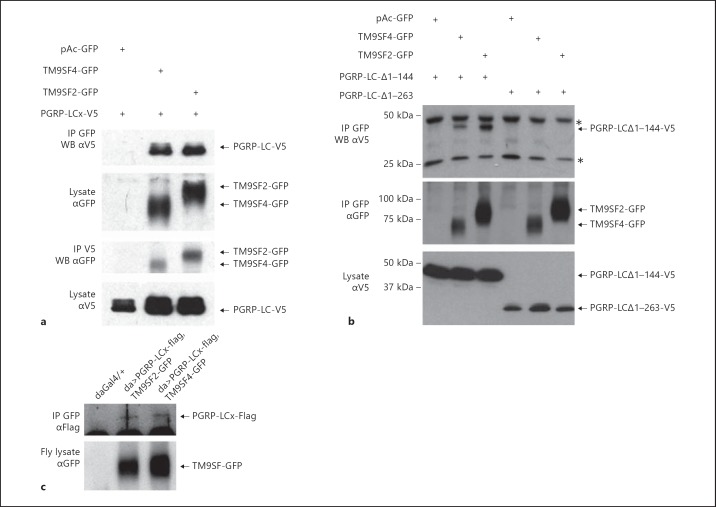
To assess whether TM9SF4 and TM9SF2 interact with PGRP-LC, S2 cells were co-transfected with either TM9SF2-GFP or TM9SF4-GFP and PGRP-LC-V5. Cell lysates were immunoprecipitated with anti-GFP or anti-V5 antibodies, and the immunoprecipitated product was analysed by Western blotting with anti-V5 or anti-GFP antibodies, respectively. In both cases, we observed co-immunoprecipitation of each of the TM9 proteins with PGRP-LC (fig. 3a). Deletion of the entire PGRP-LC cytoplasmic domain (PGRP-LC-Δ1-263) disrupted the interaction between PGRP-LC and the TM9 proteins, whereas deletion of only half of the cytoplasmic domain (PGRP-LC-Δ1-144) still allowed the interaction with both TM9 proteins (fig. 3b). Thus, amino acids 144–263 of the PGRP-LC cytoplasmic domain are required for the interaction of PGRP-LC with TM9SF2 and TM9SF4. To investigate if this interaction occurs in vivo, we expressed each TM9SF-GFP protein in combination with PGRP-LC-Flag in transgenic flies. Indeed, PGRP-LC co-immunoprecipitated from fly extracts with both TM9SF-GFP proteins (fig. 3c). These experiments thus demonstrate that TM9 proteins interact with PGRP-LC.
Fig. 3.
TM9SF2 and TM9SF4 interact with PGRP-LC. a, bDrosophila S2 cells were transfected with pAc-PGRP-LC-V5 expressing full (a) or truncated (b) forms of the isoform PGRP-LCx and either pAc-TM9SF4-GFP or pAc-TM9SF2-GFP, as indicated. Cell extracts were immunoprecipitated (IP) with either anti-GFP (αGFP) or anti-V5 (αV5) antibodies, separated on SDS-PAGE and Western blotted (WB) with αV5 or αGFP, as indicated. b Recombinant protein expression was assessed by Western blot of cell lysates with αGFP or αV5. Asterisks indicate unspecific bands. c Extracts of flies expressing the Flag-tagged PGRP-LCx isoform with either TM9SF2-GFP or TM9SF4-GFP were immunoprecipitated with an anti-GFP antibody. Co-immunoprecipitated product was revealed on a Western blot with anti-Flag (αFlag) antibody (upper panel). Expression of TM9-GFP fusion proteins was assessed by Western blotting of fly lysates with anti-GFP antibody (αGFP; bottom panel). Genotypes are as follows: w1118;dagal4/+ (daughterless-gal4), w1118;daGal4/+;PGRP-LCx-Flag#16B, TM9SF2-GFP and w1118;daGal4/+;PGRP-LCx-Flag#16B, TM9SF4-GFP.
TM9SF4 Is Required for PGRP-LC Localization at the Plasma Membrane
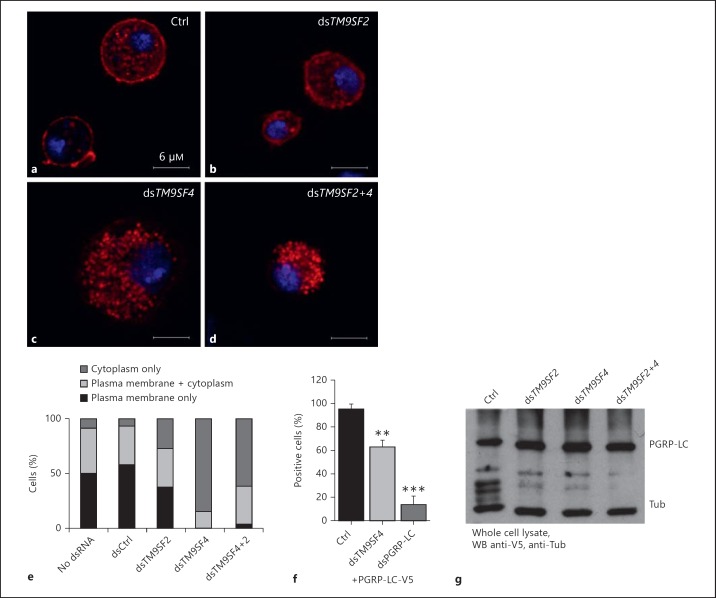
To test whether TM9SF2 and/or TM9SF4 regulate PGRP-LC subcellular localization, we depleted S2 cells of either TM9SF2 or TM9SF4 by dsRNA interference-mediated gene silencing [15] (online suppl. fig. 2). Silencing TM9SF2 had no significant effect on the subcellular localization of PGRP-LC-V5, whereas silencing TM9SF4 provoked a significant loss of PGRP-LC-V5 at the cell surface (fig. 4a-c). Co-silencing both TM9SF2 and TM9SF4 resulted in a similar loss of PGRP-LC-V5 from the cell membrane to silencing TM9SF4 alone, suggesting no significant contribution of TM9SF2 to PGRP-LC membrane localization or stabilization (fig. 4d). Monitoring cells displaying PGRP-LC at the cell membrane or not confirmed that silencing TM9SF4 resulted in a strong reduction in the proportion of cells displaying PGRP-LC-V5 at the plasma membrane or both at the plasma membrane and in the cytoplasm compared to cells displaying only cytoplasmic PGRP-LC, namely from 75% in control cells to 15% in the silenced cells (fig. 4e). By comparison, expression of another phagocytosis receptor, NimC1 [9], was unchanged in cells in which TM9SF2,TM9SF4 or both were silenced (online suppl. fig. 3). To quantify the amount of plasma membrane-associated PGRP-LC, we expressed a PGRP-LC-V5 construct in which the V5 tag was fused to the C terminus of the protein, which is on the extracellular face of the plasma membrane, and we stained the cells without permeabilizing them. Hence, only the plasma membrane PGRP-LC-V5 was accessible to the primary antibody. FACS quantification of stained cells revealed a significant reduction in the proportion of fluorescent cells (63%) in TM9SF4-silenced cells compared to control cells (93%; fig. 4f). Negative control cells treated with a PGRP-LC-silencing dsRNA showed, as expected, a drastic reduction of fluorescent cells (14%; fig. 4f). Loss of PGRP-LC staining at the cell surface was not the result of a decrease in the total amount of cellular PGRP-LC, as observed on a Western blot of cell lysate (fig. 4g). These data indicate that TM9SF4 is required for PGRP-LC localization at the plasma membrane.
Fig. 4.
TM9SF4 is required for PGRP-LC localization at the plasma membrane. a-dDrosophila S2 cells expressing PGRP-LC-V5 together with a control dsRNA (a) or a dsRNA to silence TM9SF2 (b) or TM9SF4 (c) or the two dsRNAs (d). PGRP-LC-V5 was stained with an anti-V5 antibody (red). Nuclei were stained with Hoechst (blue). e The percentage of cells expressing PGRP-LC exclusively at the cell membrane, both at the cell membrane and in the cytoplasm or only in the cytoplasm was determined from the observation of a hundred cells per condition. f FACS analysis of the percentage of positive cells expressing PGRP-LC-V5 on the surface, as determined by staining unpermeabilized cells with αV5 in either control or TM9SF4- or pgrp-lc-silenced cells. FACS analysis was performed on 20,000 cells. Histograms show the percentage of fluorescent cells relative to control cells. Error bars indicate the standard deviation between 3 independent experiments. ** p < 0.02, *** p < 0.01: significant differences (t test). g Lysates of cells as in a-d were analysed by Western blotting with anti-V5 and anti-tubulin (Tub) antibodies. Ctrl = Control.
TM9SF2 and TM9SF4 Prevent Inappropriate PGRP-LC Signalling
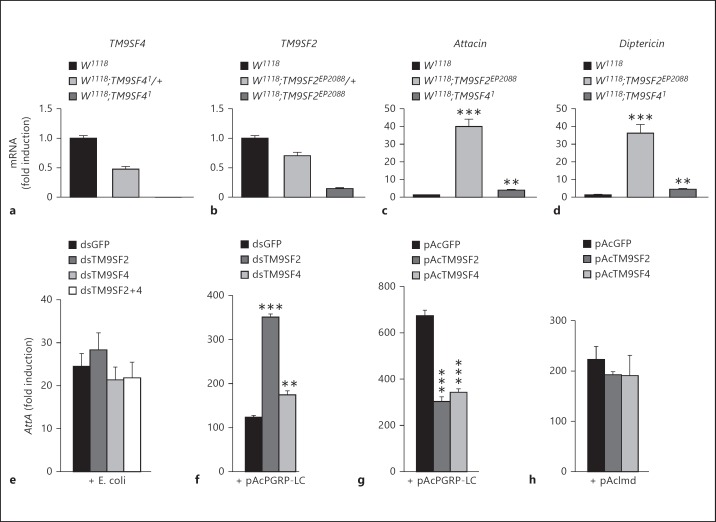
Since PGRP-LC localization is perturbed in TM9SF4 mutant cells, activation of the Imd pathway could be affected when TM9SF4 is absent. However, TM9SF41 null mutant flies (fig. 5a) displayed no major changes in AMP gene induction in response to bacterial infection [15] (online suppl. fig. 4). Similarly, a P element insertion located in the TM9SF2 coding sequence that reduced TM9SF2 expression by more than 80% (fig. 5b) did not show a major change in Dipt or AttA expression in response to E. coli infection (online suppl. fig. 4). However, in non-infected flies we show here that TM9SF2 mutant flies displayed a 30- and 40-fold higher level of Dipt and AttA mRNA gene expression, while TM9SF4 mutant flies displayed a modest but significant increase of 3.6- and 4.2-fold compared to control flies (fig. 5c, d). We then used dsRNA-mediated gene silencing in S2 cells and measured the activation of the Imd pathway by using an AttA-Luc[33] reporter gene in the presence of heat-killed E. coli in the culture media. Actually, silencing TM9SF4 or TM9SF2 or both genes had no significant effect on activation of the AttA-Luc reporter gene in response to E. coli (fig. 5e). By contrast, in S2 cells transfected with a pAcPGRP-LC expression construct, silencing TM9SF2 or TM9SF4 enhanced the PGRP-LC-mediated AttA-Luc gene induction, with a particularly strong increase in the case of TM9SF2-silenced cells (fig. 5f). Moreover, co-expressing TM9SF2 or TM9SF4 with PGRP-LC strongly inhibited the induction of AttA-Luc induced by PGRP-LC (fig. 5g) but not that induced by Imd (fig. 5h). This indicates that TM9SF2 and TM9SF4 exert a negative regulatory function at the level of PGRP-LC.
Fig. 5.
TM9SF2 and TM9SF4 down-regulate unstimulated PGRP-LC. a-d Quantitative analysis of mRNA levels of TM9SF4 (a), TM9SF2 (b) and the AMP genes AttA (c) and Dipt (d) by real-time quantitative PCR of fly mRNA in control (w1118), heterozygous (w1118;TM9SF41/+ or w1118;TM9SF2EP2088/+) or homozygous mutant flies (w1118;TM9SF41 or w1118;TM9SF2EP2088) as indicated. e-h S2 cells were co-transfected with the pAttA-Luc-Firefly reporter gene and pAc-Luc-Renilla normalizer to monitor activation of the Imd pathway. Histograms represent the fold induction compared to control cells transfected with empty pAc vector. e, f Imd pathway signalling was induced by heat-killed E. coli (e) or by expressing pAc-PGRP-LC-V5 (f) in control cells (dsGFP) or TM9SF2 (dsTM9SF2)- and/or TM9SF4 (dsTM9SF4)-silenced cells. g, h S2 cells were transfected with pAc-PGRP-LC-V5 (g) or pAc-Imd-V5 (h) and either pAc-GFP (control), pAc-TM9SF2-GFP or pAc-TM9SF4-GFP constructs as indicated. Error bars indicate the standard deviation between technical triplicates. ** p < 0.005, *** p < 0.0001: significant differences (t test).
Finally, expressing PGRP-LC-Flag [27] specifically in the larval fat body was lethal when flies were raised at 25°C, and strongly perturbed the structure of the tissue by provoking cell rounding and detachment when flies were raised at 18°C to reduce the efficiency of the UAS-Gal4 expression system (online suppl. fig. 5A, B). This phenotype is in accordance with a previous report indicating that PGRP-LC overexpression induces apoptosis [34]. Co-expression of TM9SF2 and TM9SF4 together with PGRP-LC in the fat body partially rescued the fat body defects induced by PGRP-LC overexpression (online suppl. fig. 5C, D), demonstrating that both TM9SF2 and TM9SF4 counteract the deleterious activity of overexpressed PGRP-LC in vivo.
Discussion
Our results demonstrate the differential and common requirement of TM9SF2 and TM9SF4 in the control of PGRP-LC subcellular localization and signalling activity. Both TM9SF2 and TM9SF4 co-localize and interact with PGRP-LC, a receptor which can mediate Gram-negative internalization in S2 cells [4, 15]. As we previously showed that TM9SF2 and TM9SF4 synergistically promote bacterial phagocytosis and actin cytoskeleton network reorganization [15], they possibly contribute to the internalization of Gram-negative bacteria by PGRP-LC through their direct interaction with this receptor, thus coupling bacterial recognition with cytoskeleton reorganization. In addition, TM9SF4 is non-redundantly required for PGRP-LC localization at the cell membrane; specific loss of PGRP-LC at the plasma membrane in TM9SF4 knock-down mutant cells thus likely accounts for the previously observed non-redundant function of TM9SF4 in Gram-negative bacterial internalization [15].
PGRP-LC is essentially known for its signalling activity, which enables Imd pathway activation in various cell types and tissues. The observation that PGRP-LC is weakly present at the cell membrane in TM9SF4-silenced cells while these cells still retain normal signalling activity indicates that in the absence of TM9SF4, PGRP-LC may signal from the cytoplasmic compartment. Alternatively, the amount of remaining PGRP-LC at the plasma cell membrane may be sufficient to ensure signalling.
Similarly to S2 cells, we could not detect major changes in AttA and Dipt antimicrobial gene expression in either TM9SF4 or TM9SF2 mutant flies. This suggests that TM9 proteins are not required for the up- or down-regulation of PGRP-LC activation by peptidoglycan. Alternatively, redundancy between TM9 proteins may mask their function. Indeed, we observed that the TM9SF2/TM9SF4 double mutant is lethal at the embryonic stage, while TM9SF41 null mutant flies are normally viable and TM9SF2EP2088 are poorly viable. Finally, we still have no information on the putative role of the third TM9 protein, TM9SF3 (CG10590). The question of the contribution of TM9 proteins to Imd pathway activation in response to infection should therefore be addressed in the future with conditional double or triple knock-out mutant flies.
Interestingly, TM9SF2 mutant flies and, to a lesser extent, TM9SF4 null mutant flies displayed moderate but significant constitutive activation of AttA and Dipt genes - two main targets of the Imd pathway. Therefore, TM9 proteins may act as negative regulators of the unstimulated PGRP-LC receptor, preventing inappropriate activation of the Imd pathway in the absence of infection. Indeed, complementary experiments in S2 cells showed that both TM9SF2 and TM9SF4 counteract PGRP-LC signalling activity when the receptor was overexpressed in either S2 cells or fly tissues; TM9 protein expression notably rescued the fat body phenotypic defects induced by PGRP-LC overexpression and inhibited PGRP-LC-dependent (but not Imd-dependent) AMP gene activation in S2 cells. Moreover, silencing either TM9SF2 or TM9SF4 greatly enhanced the activation of the Imd pathway mediated by PGRP-LC overexpression in S2 cells. In these conditions, PGRP-LC likely mediates signal activation by auto-activation through self-oligomerization, independently of peptidoglycan binding and stimulation. Thus, our results suggest that TM9SF2 and TM9SF4 prevent inappropriate PGRP-LC signalling in the cytoplasm and/or at the plasma membrane by counteracting auto-activation of the receptor that possibly occurs through self-oligomerization.
In conclusion, our results together with previous studies show that this widely conserved family of TM9 proteins plays an essential function in membrane receptor trafficking and regulation of signalling activity, putatively exerting a chaperone function both in the intracytoplasmic and the cell plasma membrane compartment. They also demonstrate the differential requirement of these two TM9 proteins; TM9SF2 seems to be mostly required to prevent inappropriate signalling from the unstimulated receptor while TM9SF4 has a major role in PGRP-LC subcellular localization at the plasma membrane.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We thank Dr. A.H. Tang for providing PGRP-LC transgenic flies, Dr. K.V. Anderson for sending Imd and PGRP-LC constructs, Dr. E. Kurucz for sending NimC1 antibodies, Dr. J.-L. Imler for sending the AttA-luciferase construct and Dr. P. Cosson for helpful discussions. We thank Dr. D. Grunwald for training in confocal microscopy, E. Bergeret for former help on the project, and C. Bama and I. Jeannin for preparing the fly food. Bloomington Drosophila Stock Center (BDSC) and NIG-Fly contributed to this work by sending flies. Drosophila Genomics Resource Center (DGRC) provided TM9SF cDNAs. The FlyBase and Drosophila RNAi Screening Center (DRSC) provided important information. This work was supported by the Fondation ARC (to J.P.), Région Rhône-Alpes (to J.P.) and the FINOVI foundation (to M.-O.F.).
References
- 1.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 2.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 3.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 4.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 5.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 7.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 8.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart JM, Ferrandon D, Ramet M, Ezekowitz RA. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 9.Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Ando I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Chluba-de Tapia J, de Tapia M, Jaggin V, Eberle AN. Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene. 1997;197:195–204. doi: 10.1016/s0378-1119(97)00263-1. [DOI] [PubMed] [Google Scholar]
- 11.Schimmoller F, Diaz E, Muhlbauer B, Pfeffer SR. Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene. 1998;216:311–318. doi: 10.1016/s0378-1119(98)00349-7. [DOI] [PubMed] [Google Scholar]
- 12.Pruvot B, Laurens V, Salvadori F, Solary E, Pichon L, Chluba J. Comparative analysis of nonaspanin protein sequences and expression studies in zebrafish. Immunogenetics. 2010;62:681–699. doi: 10.1007/s00251-010-0472-x. [DOI] [PubMed] [Google Scholar]
- 13.Lozupone F, Perdicchio M, Brambilla D, Borghi M, Meschini S, Barca S, Marino ML, Logozzi M, Federici C, Iessi E, de Milito A, Fais S. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep. 2009;10:1348–1354. doi: 10.1038/embor.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benghezal M, Cornillon S, Gebbie L, Alibaud L, Bruckert F, Letourneur F, Cosson P. Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol Biol Cell. 2003;14:2890–2899. doi: 10.1091/mbc.E02-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergeret E, Perrin J, Williams M, Grunwald D, Engel E, Thevenon D, Taillebourg E, Bruckert F, Cosson P, Fauvarque MO. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci. 2008;121:3325–3334. doi: 10.1242/jcs.030163. [DOI] [PubMed] [Google Scholar]
- 16.Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275:34287–34292. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- 17.Froquet R, Cherix N, Birke R, Benghezal M, Cameroni E, Letourneur F, Mosch HU, De Virgilio C, Cosson P. Control of cellular physiology by TM9 proteins in yeast and Dictyostelium. J Biol Chem. 2008;283:6764–6772. doi: 10.1074/jbc.M704484200. [DOI] [PubMed] [Google Scholar]
- 18.Bagshaw RD, Mahuran DJ, Callahan JW. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol Cell Proteomics. 2005;4:133–143. doi: 10.1074/mcp.M400128-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Diaz E, Schimmoller F, Pfeffer SR. A novel Rab9 effector required for endosome-to-TGN transport. J Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He P, Peng Z, Luo Y, Wang L, Yu P, Deng W, An Y, Shi T, Ma D. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy. 2009;5:52–60. doi: 10.4161/auto.5.1.7247. [DOI] [PubMed] [Google Scholar]
- 21.Singer-Kruger B, Frank R, Crausaz F, Riezman H. Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J Biol Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- 22.Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, Lardy B, Klein G, Sansonetti P, Charette SJ, Cosson P. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 23.Le Coadic M, Froquet R, Lima WC, Dias M, Marchetti A, Cosson P. Phg1/TM9 proteins control intracellular killing of bacteria by determining cellular levels of the Kil1 sulfotransferase in Dictyostelium. PLoS One. 2013;8:e53259. doi: 10.1371/journal.pone.0053259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Froquet R, le Coadic M, Perrin J, Cherix N, Cornillon S, Cosson P. TM9/Phg1 and SadA proteins control surface expression and stability of SibA adhesion molecules in Dictyostelium. Mol Biol Cell. 2012;23:679–686. doi: 10.1091/mbc.E11-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fais S, Fauvarque MO. TM9 and cannibalism: how to learn more about cancer by studying amoebae and invertebrates. Trends Mol Med. 2012;18:4–5. doi: 10.1016/j.molmed.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt RL, Trejo TR, Plummer TB, Platt JL, Tang AH. Infection-induced proteolysis of PGRP-LC controls the IMD activation and melanization cascades in Drosophila. FASEB J. 2008;22:918–929. doi: 10.1096/fj.06-7907com. [DOI] [PubMed] [Google Scholar]
- 28.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Benaud C, Baudier J, Taillebourg E, Fauvarque MO. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6:309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy. 2012;8:767–779. doi: 10.4161/auto.19381. [DOI] [PubMed] [Google Scholar]
- 32.Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 33.Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- 34.Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data